Abstract
Numerous types of transcripts perform multiple functions in cells, and these functions are mainly facilitated by the interactions of the RNA with various proteins and other RNAs. Insight into the dynamics of RNA biosynthesis, processing and cellular activities is highly desirable because this knowledge will deepen our understanding of cell physiology and help explain the mechanisms of RNA-mediated pathologies. In this review, we discuss the live RNA imaging systems that have been developed to date. We highlight information on the design of these systems, briefly discuss their advantages and limitations and provide examples of their numerous applications in various organisms and cell types. We present a detailed examination of one application of RNA imaging systems: this application aims to explain the role of mutant transcripts in human disease pathogenesis caused by triplet repeat expansions. Thus, this review introduces live RNA imaging systems and provides a glimpse into their various applications.
Keywords: live-cell imaging systems, RNA nuclear foci, RNA localization, RNA fluorescence imaging, triplet repeat diseases
Introduction
RNA molecules are synthesized in cells through highly regulated biogenesis pathways, and when these molecules mature, they participate in and regulate fundamental cellular processes. To gain better insight into the variety of RNA functions, the entire cellular life of transcripts, from their synthesis to their decay, needs to be examined. Molecular methods widely used in RNA biology can identify the length, sequence and structure of an RNA molecule and can be used to determine the mean cellular RNA levels in cell populations. Pull-down assays provide insight into the proteins interacting with RNAs but do not elucidate the spatial and temporal changes that ribonucleoprotein complexes undergo. Fluorescence in situ hybridization (FISH) enables the observation of RNA localization but captures only a single time point in the cellular RNA pathway. The main advantage of live RNA imaging systems is that they allow the study of RNA movement and temporal changes in RNAs and RNPs. These techniques include direct RNA labeling,1,2 the labeling of endogenous transcripts with molecular beacons,3-6 multiply labeled tetravalent RNA imaging probes (MTRIPs)7 or Pumilio,8 and vector-based systems that employ highly specific RNA-protein and RNA-dye interactions. RNA live imaging systems substantially broaden the types of information that can be attained compared with static in situ analyses, by adding a dynamic dimension.
Live imaging systems enable researchers to investigate many cellular processes involving RNAs. Among these processes are the biosynthesis, function and decay of eukaryotic mRNAs, which include multiple steps: i.e., Pol II transcription, primary transcript (pre-mRNA) modifications and splicing, nuclear transport and export of mature mRNA, cytoplasmic mRNA transport to its localization site, mRNA translation and, finally, degradation. With live RNA imaging systems, the entire cellular route of a transcript may be observed in a single experiment, and each step of this pathway may be analyzed separately and in more detail.
In this article, we describe the vector-based systems for RNA imaging in living cells and the adaptations of these systems to various applications in bacterial, fungal, plant and animal cells. We provide details regarding the design of these systems and their important features and critically discuss the advantages and limitations of the individual systems. In the application section, we briefly refer to relevant publications, organizing them according to the cellular processes investigated. We present a detailed discussion of 2 applications of live RNA imaging systems that address the role of RNA nuclear foci in the pathogenesis and treatment of human neurological diseases caused by triplet repeat expansions.
Aptamer-Based Transcript Imaging Systems
Nearly all aptamer-based systems that have been developed for the fluorescence microscopy imaging of single transcripts in living cells require the expression of an exogenous transcript of interest fused to an aptamer sequence (chimeric transcript). The aptamer itself is not fluorescent, but when it binds a specific protein partner fused to an autofluorescent protein (chimeric protein), the entire system becomes fluorescent after excitation at appropriate wavelength. Alternatively, the chimeric transcript may be imaged using a suitable organic dye whose fluorescence can be activated after binding to an aptamer. The rigorous high-affinity and high-specificity criteria are fulfilled by a number of systems that have thus far been devised for use in RNA imaging (Table 1).
Table 1.
Characteristics of vector-based systems for RNA live imaging. Affinity is presented as KD (dissociation constant) – the propensity of a complex to dissociate into parts. Legend: * - Eukaryota/Prokaryota, ** - threshold not estimated, ‘-’ - not applicable, NA - not analyzed, app. – approximately, ? - not specified by authors. Unless stated otherwise, data were obtained from studies cited in the systems description paragraph
| RNA-dye systems |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name Feature | MS2 | λN22 | BglG | PP7 | U1Ap | HTLV-1 Rex | TAT-TAR | REV-RRE | eiF4A | Spinach | Spinach2 | Malachite Green | SRB-2 |
| RNA size | 19 nt | 15 nt | 29 nt | 25 nt | 21 nt | 36 nt | 23 nt?38 | 30 nt?38 | 58 nt | 98 nt | 95 nt | 38 nt | 54 nt |
| number of hairpins | 1–96 | 1–25 | 18 | 1–24 | 4–16 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| number of hairpins required for single molecule imaging | 24/96* | ≤12** | ≤18** | ≤24 ** | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| minimal peptide size used (aa) | 117 | 22 | 58?20 | 128 | 94 | 16 | 9–86?38 | 14–116?38 | 215 +191 | — | — | — | — |
| protein dimerization | YES | NO | NO | YES | NO | NO | NO | NO | — | — | — | — | — |
| affinity of ligands to the aptamer(KD) | 5 nM | 22 nM | NA | 1 nM | < 1 nM?33 | 30 nM | 0.31–2.1 nM?40 | 1–3 nM?39 | 44 nM | 450 nM | 430 nM | 110 nM–1.8 μM | 1.4 ± 0.1 μM |
| BiFC | NO | NO | NO | NO | NO | NO | NO | NO | YES | — | — | — | — |
| background problem | YES | YES | YES | YES | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| comment | — | — | BglG is a bacterial protein | — | U1Ap is a human protein | — | Only tested in TriFC experi-ment | Only tested in TriFC experi-ment | eiF4A is a murine protein | thermal instability | + tRNA sequence | — | + tRNA sequence |
Aptamer-protein systems
The aptamer-protein systems are based on naturally occurring high-affinity interactions between specific RNA structure motifs and their binding proteins. Both the RNA and protein components used in these systems are typically engineered to optimize their sequences for tighter binding. Often, only the RNA binding domain of the protein is retained to minimize the size of the protein-RNA complex. The main drawback of aptamer-protein systems is the strong background signal produced by the constant fluorescence of the chimeric protein not bound to the aptamer, which we address further in the text as a background problem.
MS2 system
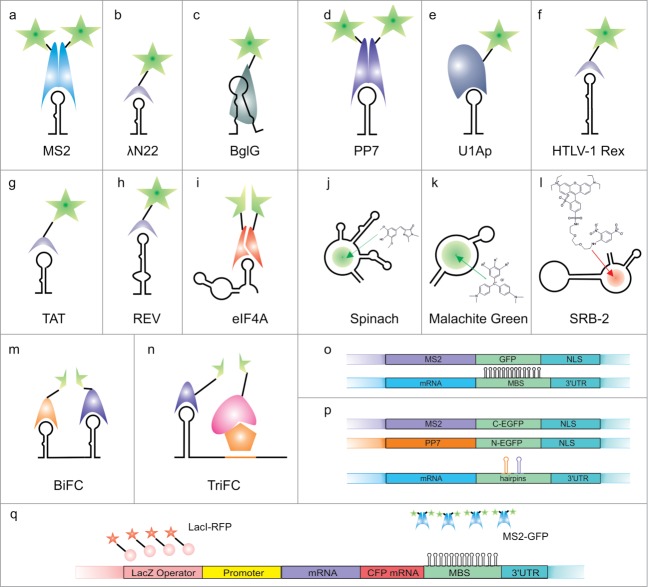
The MS2 system is the prototypical RNA imaging system design introduced by Singer and colleagues in the study of ASH1 mRNA localization in yeast cells9 and first applied by Bloom and colleagues to study the localization of mRNAs in budding yeast.10 In the MS2 bacteriophage, the genomic RNA and its coat protein form a viral capsid through protein multimerization and RNA-protein interactions. The MS2 system (Fig. 1a, Supplementary Table 1) consists of the RNA operator of the MS2 bacteriophage genome and an engineered MS2 protein that binds to this RNA as a dimer and has an enhanced RNA-protein affinity.11,12 The aptamer fused to a transcript of interest forms a 19 nt RNA hairpin with a 4 nt loop and a 7 bp stem that harbors a single adenine bulge. The U>C mutation in the natural loop sequence increased protein binding 50 times.13 Because a single MS2 RNA hairpin is not sufficient to image a single molecule of transcript, multiple hairpins are typically used. The advantages of the MS2 system, such as high specificity and sensitivity, led to its position as the most frequently used system to label and image RNA in living cells (Supplementary Table 2). To date, more than one hundred publications have described the use of MS2 in different biological systems imaging a broad range of RNAs.
Figure 1.
Vector-based systems for RNA live imaging. Schematic structures of RNAs with protein partners or fluorescent dyes are presented (a–l). Additionally, examples of genetic constructs used for imaging experiments are depicted (o–r). (a) MS2 systems, (b) λN22 system, (c) BglG systems, (d) PP7 system, (e) U1Ap system, (f) HTLV-1 Rex system, (g) TAT system, (h) REV system, (i) eIF4A system, BiFC with the use of 2 domains, (j) Spinach system, (k) Malachite green system, (l) SRB-2 system, (m) BiFC with the use of 2 systems, (n) TriFC, (o) DNA construct for MS2 system, (p) DNA constructs for BiFC with 2 systems and (r) system for gene locus, mRNA and protein product imaging.
λN22 system
The λN22 system developed by Daigle and Ellenberg, which was first described in mammalian cells,14 takes advantage of the phage lambda transcription antitermination signal (box B RNA) and its binding N protein (Fig. 1b, Supplementary Table 1). In the λN22 system, only the 22 aa arginine-rich fragment of the N protein binds the aptamer, which is a 15 nt hairpin composed of a 5 nt loop and a 5 bp stem (Table 1). The hairpin loop of the box B RNA contains a GNRA fold (R = purine, N = any nucleotide), which is tightly bound by the N protein.15 The small size of the fused N protein fragment is a significant advantage of the system because this feature minimizes the risk of influencing the cellular properties of the investigated transcript.14 The background problem found in the MS2 system, which is partially caused by the proteolysis of the peptide linker between MS2 and GFP, was reduced in the λN22 system by increasing the stability of the chimeric λN-GFP protein.16 The λN22 system is the second most frequently implemented system after MS2, and it has been used to image transcripts in bacterial, fungal, plant and mammalian cells.
BglG system
The BglG system (Fig. 1c, Supplementary Table 1) developed by Hu and colleagues17 was used for the first time together with the MS2 system to demonstrate the heterozygosity of HIV-1 virions in human cells. Two genomes were labeled, one with MS2 and one with a BglG aptamer, and they were imaged simultaneously to differentiate the virus variants and to study virion formation. The system is based on the E. coli transcription antitermination protein, which in principle limits its applications in bacterial cells. The BglG protein binds to an imperfectly palindromic 29 nt hairpin containing a 4 nt terminal loop and 2 bulges in the hairpin stem18,19 (Table 1). In contrast to most other systems, the RNA-protein specific recognition triggers structural changes leading to hairpin destabilization. The minimum peptide size that is able to efficiently bind the RNA structure is 58 aa and spans the N-terminal RNA binding domain of the BglG protein.20 So far, this system has been used only in studies of viruses.21
PP7 system
The PP7 system (Fig. 1d, Supplementary Table 1) was introduced in 2011 by Singer and colleagues to study transcription initiation and elongation in yeast.22 It is based on a similar principle as the MS2 system. The dimer of the PP7 bacteriophage coat protein binds to its cognate RNA structure with very high affinity (Table 1). The aptamer is built from an RNA hairpin containing a 6 nt loop and an 8 bp stem harboring a purine bulge on its 5’ side.23 The PP7 system was used together with the MS2 system to enable bimolecular fluorescence complementation (BiFC) through the reconstruction of the EGFP protein from its fragments (Fig. 1m, p).24,25
U1Ap system
The U1Ap system (Fig. 1e, Supplementary Table 1) was developed simultaneously by Silver and Brodsky26 and by Takizawa and Vale,27 and has been used thus far in only 4 studies carried out in yeast cells.28-31 The system is based on a fragment of the human splicing protein U1Ap and the RNA signal it recognizes. To reduce the size of the protein, a fragment containing the first 94 aa, which comprises the RRM domain (the domain that binds RNA), was fused to a fluorescent protein (Table 1). This polypeptide shows a high affinity for a 21 nt hairpin structure containing a 10 nt loop.32,33 U1Ap was shown to be advantageous in the investigation of the nuclear export of RNA.26
HTLV-1 Rex system
The HTLV-1 Rex system (Fig. 1f, Supplementary Table 1), proposed by Broude and colleagues,34 was used together with λN22 in a BiFC experiment performed in E. coli. The viral peptide used in this system is only 16 aa, which makes it the smallest peptide used in an imaging system (Table 1). The peptide binds specifically to a 36 nt hairpin based on the HTLV-1 Rex responsive element (RxRE), which contains a 4 nt loop and 2 bulges in the stem.35 Physiologically, the RxRE present in the viral RNA is bound by the HTLV-1 Rex protein, which enhances its export from the nucleus.36 It remains to be established whether a chimeric HTLV-1 Rex protein also affects mRNA export.
TAT-TAR and REV-RRE systems
Two novel systems based on sequences present in HIV were introduced recently by Cui and colleagues.37 The natural affinities of the TAT (Fig. 1g, Supplementary Table 1) and REV (Fig. 1h, Supplementary Table 1) proteins to the TAR and RRE RNAs,38-40 respectively, were used to investigate mRNA-protein interactions. Both systems use unmodified RNA sequences, which means that their RNA components are relatively large. The systems were invented as alternatives to earlier designs, taking advantage of fluorescence complementation, and applied to the study of the nuclear export of viral RNAs.37
eIF4A system
The eIF4A system was first described in 2007 by Broude and colleagues and evaluated for the imaging of mRNA and rRNA.41 The system is based on the mouse version of the eukaryotic initiation factor 4A peptide (Table 1), which contains 2 RNA binding domains and binds with a high affinity to a single relatively large 58 nt aptamer.42 This feature can be exploited using fluorescence complementation, in which the 2 domains are dissected and each is fused with half of a fluorescent protein (Fig. 1i, Supplementary Table 1).43
Aptamer-Dye systems
An alternative to the protein-aptamer RNA imaging methods is the replacement of the protein component with a small organic dye. Fluorescence is generated after excitation only when the dye is captured by a specific RNA aptamer structure because the dye can no longer dissipate its energy through intramolecular motions. In this design, the undesired fluorescence background problem was not observed.44 The replacement of proteins with small organic dyes and shorter sequences fused to the investigated transcripts diminishes concerns that proteins or polypeptides bound to aptamer sequences influence transcript properties. The high potential of aptamer-dye systems to image RNAs in various biological systems has been highlighted by several authors.45-47 There are many aptamer-dye pairs described for in vitro RNA labeling, but so far, they have not been implemented in biological studies. Among them are the II-mini3–4 system,48 dimethyl indole red,49 ASR750 and DCF-MPP.51 The systems described below are the first examples of aptamer-fluorophore techniques, which were recently developed and used in living cells.
Spinach and Spinach2 systems
The Spinach system was introduced for RNA imaging by Jaffrey and colleagues in 2011.44 The organic dye, 4-hydroxybenzylidene imidazolinone (HBI), corresponds to the fragment of GFP responsible for its fluorescence. The fluorescent signal is generated when the fluorophore is captured by an 80 nt aptamer fused to the mRNA of interest (Fig. 1j, Supplementary Table 1). Several derivatives of HBI provide a range of different fluorescence wavelengths (different colors). So far, DFHBI (3,5-difluoro-4-hydroxybenzylidene imidazolinone) has been shown to be the most efficient (Table 1); however, the dye can be further improved.52 More fluorophores could be synthesized based on a range of fluorescent proteins (GFP derivatives) because the chemical structure that determines its fluorescence is known.53 These dyes have reduced photobleaching due to fast kinetics of exchange of DFHBI dye in Spinach aptamer, which imposes some limitations on the application of the system. Recently, Spinach2 was introduced due to problems with the thermal stability of the Spinach aptamer in the cellular environment. The aptamer was mutated and stabilized using the tRNA structure to facilitate proper folding. With these improvements, the RNA containing the CGG repeats was imaged.54
Malachite green system
The malachite green system (Fig. 1k, Supplementary Table 1) with 2-photon excitation (TPE) signal-enhancement was introduced by Nicoud and colleagues.55 The malachite green-binding aptamer (38 nt) is considerably shorter than the Spinach aptamer; therefore, it is even less likely to alter transcript properties (Table 1).55 The interactions of malachite green (MG) with the RNA aptamer were previously studied56-58 but without implementation in the field of RNA imaging. The malachite green system was used in the investigation of RNA degradation in mammalian cells. In this experiment, the malachite green aptamer was stabilized by the pRNA 3WJ sequence.59
SRB-2 system
The most recently introduced system is the SRB-2 system (Fig. 1l, Supplementary Table 1), which was developed by Sunbul and Jaschke.60 It uses a well-described 54 nt SRB-2 aptamer and Sulforhodamine B (SR) as the dye (Table 1). Because of the constant fluorescence of SR, a dinitroaniline (DN) quencher was added to create the small molecule marker SR-DN for the SRB-2 aptamer. The optimal length of the linker placed between the fluorophore and the quencher was shown to be 2–3 ethylene glycol units. After the aptamer binds the small dye, the labeled RNA is observed as a red signal. The performance of the system has been tested only in E. coli cells so far.
Critical issues in aptamer-based systems design
A crucial issue in the experimental design of RNA imaging systems is the site of the aptamer insertion within the transcript. The most common practice in mRNA trafficking studies is to place the aptamer sequence between the ORF and the 3’ UTR (Supplementary Table 2). This location is considered to be the safest and the most likely to preserve the properties of the endogenous transcript because mRNA localization signals are not altered and mRNA translation is expected to occur normally. For some research purposes, the experimental system may be simplified: e.g., for localization analysis only, the 3’ UTR of the mRNA may be used.
Some applications may require aptamers to be placed at positions other than between the ORF and the 3’ UTR (Table 2). However, it has been shown that the site of aptamer insertion may affect the level of protein translated from the chimeric mRNA,61 influence transcript localization and interactions with endogenous proteins62 and disturb pre-mRNA splicing63 (Table 2). It is therefore hard to provide general guidelines regarding the optimal placing of the aptamer sequence within the investigated transcript. To ensure that the cellular pathway and the final localization of the mRNA are not affected by the aptamers, FISH experiments are typically performed.
Table 2.
Localization of hairpins within transcripts: opportunities and concerns
| localization | research purposes | disadvantages | |
|---|---|---|---|
| in the 5’ UTR | protein binding | nonsense peptide production, decreased protein level | |
| in the ORF | intron | splicing localization | splicing alteration |
| exon | alternative splicing | protein alteration | |
| between the ORF and the 3’ UTR | mRNA localization | decreased protein level | |
| in the 3’ UTR | protein binding | protein binding prevention, transport alteration | |
A second important issue is the background problem, which occurs when autofluorescent proteins are used. To solve this problem and increase the signal-to-noise ratio, several options have been considered. One possible solution is to increase the number of GFP units attached to a single aptamer-binding protein64; another is to multiply the aptamer sequences in the transcript. For a system based on the GFP-MS2-NLS protein, 24 MS2 hairpins enabled the imaging of a single mRNA molecule in mammalian cells.65 Concerns that the attachment of large RNA-protein complexes can limit the use of these systems to only large RNAs were dispelled by the successful application of live imaging techniques to small RNAs.66,67 In studies of cytoplasmic events, a nuclear localization signal (NLS) may be added to a chimeric protein to direct the unbound protein to the nucleus9,14 However, it must be verified that the addition of an NLS does not alter the localization of the imaged RNA. Yet another way to resolve the background problem is the use of fluorescence complementation systems43,68,69 as reviewed by Tyagi70 (Fig. 1i, m, p).
A transcript with an aptamer sequence and a chimeric protein is typically delivered to cells using genetic vectors. Two standard plasmids harboring relevant expression cassettes are used as depicted in Figure 1o. Using differently designed cassettes, the mRNA, protein and gene locus can be imaged in one experiment (Fig. 1r).71 In the case of aptamer-dye systems, only one vector is needed to image a single mRNA. Small fluorophores easily enter the cell; however, some of them: e.g., MG, cause considerable cytotoxicity.44 The use of 2 different systems allows researchers to image transcripts from both alleles of the same gene72 or to demonstrate the co-localization of distinct mRNAs62 in a single cell.
Examples of Applications of Live RNA Imaging Systems
Numerous types of RNAs have been imaged in both prokaryotic and eukaryotic cells (Supplementary Table 2), including mRNAs, non-coding RNAs and RNA viruses. However, the most frequently investigated topic is the life cycle of mRNA in mammalian cells. In the following section, we describe which aspects of the cellular mRNA journey can be and already have been explored using live RNA imaging systems. We discuss in more detail the results of RNA imaging experiments focused on the roles of mutant RNAs in neurodegenerative diseases caused by triplet repeat expansions.
Nuclear journey of mRNA
Monitoring transcript synthesis begins with the measurement of gene activity. This analysis includes studying transcription initiation and single transcription events.73 The number of newly synthesized transcripts can be counted in a time-lapse manner, and temporal changes in transcriptional activity can be measured under changing conditions. Variations in the transcription dynamics of the investigated promoter have been analyzed during the cell cycle74,75 and in response to various transcription inducers or inhibitors.76-78 Additionally, the time period elapsed from the recruitment of initiation factors to the start of transcription has been be measured.79
Live imaging systems allow the determination of the transcript elongation rate and a detailed description of RNA polymerase II activity.22 To distinguish between newly synthesized transcripts and transcripts already released from the transcription site, the Fluorescence Recovery After Photobleaching (FRAP) technique was used.78,80 The transcript elongation rate was measured with dually labeled RNA, in which PP7 hairpins were inserted into the 5’ UTR and MS2 hairpins were inserted into the 3’ UTR. The time between capturing the signals from these tags corresponds to the time required for the elongation of the sequence between the PP7 and MS2 hairpins.72
The nuclear steps accompanying and following transcription include splicing and 3’ UTR formation. Both steps can influence the retention of pre-mRNA at a transcription site. In contrast to transcripts devoid of introns, transcripts containing introns stay at the transcription site longer than the polymerase, which implies that splicing stops the pre-mRNA at the transcription site.81 In studies of splicing dynamics, the aptamer sequence was placed inside the intron, and the duration of its signal was measured.63 Using RNA imaging systems, it was also shown that mRNA is released from the transcription site after 3’end formation.82
After the nuclear processing of the transcript is completed, the mature mRNA is transported to the nuclear borders to be exported from the nucleus. By imaging the mobility of RNA molecules, it was shown that transcript movement is not directional but is influenced by energy.83 Transcripts leave the nucleus in different ways,84 typically through nuclear pores. Export was shown to be faster than the nucleoplasmic diffusion rates of mRNA.85 The retention of mRNAs was observed in cells with decreased ATP levels,83 disturbed splicing, 3’end processing26 and triplet repeat expansion mutations.54,86
Cytoplasmic journey of mRNA
Transcripts accompanied by various proteins are transported in the cytoplasm in many ways, including along actin filaments, along microtubules or through simple diffusion. The transport of numerous transcripts has been investigated with live imaging systems to answer questions regarding the velocity,83 efficiency,87 direction28,88 and continuity88 of transcript movement. These features have been shown in many cases to differ substantially between the analyzed transcripts. Therefore, instead of attempting to provide the reader with specific answers and numbers, which is beyond the scope of this review, we only provide references to the relevant papers.
Numerous analyses were performed to study the proteins involved in transcript transport.9,89 Interactions between RNAs and proteins are typically analyzed using double-labeling co-localization experiments. The influence of specific proteins on RNA transport can also be investigated in cells with silenced expression of the implicated protein.90 Additionally, live imaging systems allow precise analyses of the mRNA sequences responsible for transport and localization. Two approaches for such analyses were proposed: the use of a construct containing only the transport signal, and mutating this signal to disturb transcript transport.17,91,92
The use of the Trimolecular Fluorescence Complementation (TriFC) technique has shown that some protein-protein interactions are observed only in the presence of specific RNA molecules.93 A fluorescent protein is split into halves; one half is fused with the protein required for RNA imaging, and the other is fused with a protein indirectly bound to the RNA. The fluorescent signal is restored only when all components co-localize (Fig. 1n).
The retention of mRNAs in specific cytoplasmic bodies occurs when the transcript is stored for delayed translation or degradation.94,95 The exchange of mRNAs between the motile fraction and stable granules can be analyzed using the FRAP technique.96 Disturbances in transcript localization caused by various environmental factors, e.g., amino acid starvation or temperature changes, were also monitored with RNA imaging systems.97,98
The labeling of both mRNAs and their protein products has allowed researchers to answer several questions about translation. It has been shown that transcripts are not translationally active during transport in the cytoplasm.99 Addressing the question of whether transcript localization depends on protein translation resulted in the finding that the localization of some mRNAs defines proper protein localization99 and in the converse direction, translation activity can define transcript localization in an SRP-dependent manner.100 Further analyses demonstrated that several proteins and microRNAs regulate RNA stability101 and influence the translation90 102,103 of the investigated transcripts.
The cellular life of RNA is terminated in several ways. The main mechanism of mRNA decay usually starts with deadenylation, continues through 5’ decapping and ends with 5’→3’ exonucleolytic degradation.104 The analysis of mRNA degradation using live imaging systems was conducted by analyzing transcript co-localization with the cellular compartments responsible for degradation: e.g., P bodies.31,105,106 The site where transcript degradation occurs was observed after inhibiting its nucleolytic decay,107 and the dynamics of RNA degradation were monitored in cells after the global inhibition of transcription.108
RNA imaging in diseases caused by simple repeat expansions
The pathology of a group of genetic neurodegenerative diseases associated with a simple repeat expansion may be caused by toxic effects induced by the mutant transcript, mutant protein or both. Another mechanism of pathogenesis is the decrease in the amount of functional protein product translated from the mutant gene. The earliest symptom is often neuronal cell dysfunction; therefore, studies examining the pathogenesis of these diseases should be performed in relevant experimental models. Many experiments taking advantage of RNA imaging in living cells have been conducted thus far in neuronal cell lines (see Table 3). In this section, we summarize the results of imaging experiments that explore the aberrant life of transcripts from mutant alleles of the genes involved in triplet repeat expansion diseases.
Table 3.
Examples of RNA live imaging systems used in studies on neuronal cell line. The other studies are presented in Supplementary Table 2
| mRNA function examined | specific research purpose | system used | imaged mRNA | Ref. |
|---|---|---|---|---|
| transcriptional activity | β-actin mRNA transcription and transport | MS2 | β-actin mRNA | 135 |
| RNA localization | dendritic targeting signals | MS2 | Kv4.2 mRNA | 136 |
| localization signals of nos mRNA | MS2 | nos mRNA | 137 | |
| CaMKIIα mRNA localization | MS2 | CaMKIIα mRNA | 94 | |
| region responsible for dendritic transport | MS2 | ApoE mRNAs | 92 | |
| localization of Arc mRNA | MS2 | Arc mRNA | 88 | |
| 5’ UTR and 3’ UTR transport signals | MS2 | kor, SV40 mRNAs | 138 | |
| altered localization in memory | MS2 | CaMKIIα mRNA | 139 | |
| RNA movement | MMP-9 mRNA movement | MS2 | MMP-9 mRNA | 140 |
| mobility of kor mRNAs | MS2 | kor mRNAs | 141 | |
| RNA transport mechanisms | dynein-dependent transport | MS2 | nos, osk mRNA | 142 |
| Stau2 role in mRNA distribution | MS2 | Map1b, Map2 mRNA | 143 | |
| Kv4.2 transport in dendrites | MS2 | Kv4.2 mRNA | 144 | |
| Htt role in BDNF mRNA transport | MS2 | BDNF mRNA | 145 | |
| Htt role in mRNAs transport | MS2, λN22 | β-actin mRNA | 146 | |
| role of FMRP in mRNA transport | MS2 | CaMKIIα, Fmr1 mRNAs | 147 | |
| Htt role in dendritic transport | MS2, λN22 | IP3R1, β-actin mRNA | 148 | |
| FMRP role in mRNAs transport | MS2 | CG9293, chic mRNAs | 96 | |
| FMRP role in mRNAs transport | MS2 | CaMKIIα mRNA | 149 | |
| RNA-Protein interactions | RNG105 colocalization with NKA mRNAs | MS2 | NKA mRNAs | 150 |
| FMRP interaction with MMP-9 mRNA | MS2 | MMP-9 mRNA | 151 | |
| Copb1 function | MS2 | kor, SV40 mRNAs | 152 | |
| SYNCRIP role in mRNA granules | MS2 | IP3R1 mRNA | 153 | |
| RNA translation | synapse-specific mRNA translation | MS2 | Arc mRNA | 154 |
| FMRP and hnRNP C competitive translation control | MS2 | APP mRNA | 90 | |
| RNA stability | DLK-1 function | MS2 | CEBP-1, UNC-54 mRNA | 101 |
RNA toxicity manifests mainly through the formation of nuclear foci. Ribonuclear inclusions are observed in the cells of patients with neurological diseases caused by trinucleotide (CAG in SCA3 in HD,109,110 CTG in DM1111-113 and HDL2,114 CGG in FXTAS115,116), tetranucleotide (CCTG in DM2117-119), pentanucleotide (TGGAA in SCA31,120 ATTCT in SCA10121) and hexanucleotide (GGGGCC in ALS/FTD122,123) repeat expansions as reviewed by Wojciechowska and Krzyzosiak.124 Transcripts containing expanded repeat tracts form aggregates in which they sequester various proteins. The application of FISH and immunofluorescence techniques allowed for the determination of the sizes and shapes of the inclusions as well as the identification of several sequestered proteins.125,126 FISH imaging was used to estimate the number of foci in a single nucleus127 and to establish whether the foci harbored full-length mutant transcript or only fragments containing expanded repeats.119 The influence of the mRNA expression level on foci formation113,128,129 and the toxicity of cytoplasmic RNA foci were also described.130 However, not all features of RNA aggregates can be described using static methods such as FISH, immunodetection and fluorescent protein labeling.
To gain better insight into the nature of RNA toxicity and its role in pathogenesis, it is crucial to learn more about the differences in the cellular behavior of normal and mutant transcripts. The cellular fates of both transcripts can be followed with the use of one imaging system in different experiments54,86 or using 2 different imaging systems in one experiment.72 The toxicity of the mutant transcripts, understood as abnormal processing or functioning, can be followed at every step of its life.131,,132 In polyglutamine diseases, mutant transcripts temporarily retained in the nucleus can reach nuclear pores and be exported to the cytoplasm. The effectiveness of their translation may differ from that of normal transcripts.
It is not well understood whether aggregation is fully controlled or stochastic. The important issues concerning nuclear foci include foci dynamics and stability, changes in composition, factors influencing stability and therapeutic molecules targeting RNA foci. All aspects concerning the dynamics of foci formation, mobility, stability and interactions with proteins can be investigated using live RNA imaging systems. Using these methods, 2 types of nuclear inclusions containing transcripts with expanded triplet repeats were analyzed. The studies were performed using the MS2 and Spinach2 systems, which were used to label fragments of the DMPK and FMR1 transcripts, containing mutant CUG and CGG repeats, respectively54,86 (Table 4).
Table 4.
Details of RNA imaging experiment design used in studies of CUG and CGG repeat toxicity in DM1 and FXTAS
| CUG-repeat transcript imaging | CGG-repeat transcript imaging | |
|---|---|---|
| Goals | • movement of mutant transcripts • dynamics of foci formation • RNA co-localization with Mbnl1 • Mbnl1 role in foci formation |
• RNA foci formation and stability • foci dynamics during the cell cycle • effect of drugs on foci formation • RNA co-localization with Sam68 |
| System | MS2 | Spinach2 |
| Number of aptamers | 24 | 1 |
| Localization of aptamers | Upstream of 3’UTR | Upstream of the polyA signal |
| Fluorescent protein/dye | GFP/mCherry | DFHBI |
| Delivery method | Retroviral vectors | Plasmid vectors |
| Promoter | TRE (inducible) | CMV (strong, non-inducible) |
| Additional techniques | FRAP, FLIP, imaged simultaneously with protein | imaged simultaneously with protein |
| Cell type | C2C12 myoblasts | COS-7 |
| Temporal resolution | Every 333 msec for 20 sec or every 30 sec for up to 23 min |
Every 20 min for 6 h. |
| Microscopy | Spinning disk confocal microscopy | Epifluorescence microscopy |
| Software | MetaMorph software | NIS-Elements software |
| Comment | • Higher temporal resolution is needed for the analysis of transcript movement. • Standard microscopic filters can be used. • Protein-RNA systems enable photobleaching. • Single RNA granule tracking is possible with 24 MS2 hairpins |
• Longer observation is needed for changes during the cell cycle. • Fast dye exchange reduces photobleaching. • DFHBI exhibits better fluorescence signal stability than GFP. • DFHBI can be replaced with DFHBI-1T for standard microscopic filters |
| Reference | Querido et al. 2011 | Strack et al. 2013 |
The movements of the unsequestered mutant transcripts into foci were followed in the nucleoplasm. The mobility of the transcript with expanded CUG repeats was decreased to half that of the normal transcript.86 The RNA retardation may contribute to the compromised export of the mutant transcript observed in DM1 cells.111 The co-localization of the transcript containing expanded CUG repeats with the MBNL1 protein was detected soon after transcription. This result suggests that the MBNL1 protein actively participates in foci formation rather than being passively sequestered by already-formed foci. In support of this conclusion, the number of foci in the cells decreased after silencing the MBNL1 protein. The interaction of the protein with transcripts present outside of aggregates is consistent with alterations in alternative splicing, which are characteristic of DM1 and also observed in cells devoid of nuclear foci.133
In imaging experiments of transcripts containing CGG repeats, the foci are formed within a few hours from the start of transcription.54 The fast transcript aggregation can be explained by the use of a strong promoter in the RNA imaging construct, which is in accord with the results of the FISH experiments.114 The combination of RNA imaging with the FRAP and Fluorescence Loss in Photobleaching (FLIP) techniques enabled the authors to examine the dynamics of CUG foci formation.86 Inclusions are formed randomly and have a stochastic nature. With the use of Spinach2, the influence of the cell cycle on foci formation was demonstrated for the first time.54 During cell division, all CGG-containing foci combined into one large inclusion, which was then divided between daughter cells. After cell division, the aggregates temporarily dissociated, and the signal from the labeled transcripts was observed across the cytoplasm before rapid foci reconstitution.
The instability of the structure and the composition of the nuclear foci raise hope that drugs targeting nuclear foci could efficiently trigger the disintegration of pathogenic inclusions. With RNA imaging systems, it has been possible to observe how small molecular weight drugs affect foci formation and stability.54 The tested drugs either decreased de novo foci formation or induced their disaggregation.
RNA imaging systems have allowed to observe mutant triplet repeat transcripts in one very important phase of their cellular life that leads to pathology. A detailed comparison of normal and mutant transcripts in their entire cellular pathways (Fig. 2) will likely reveal new RNA-mediated pathogenic mechanisms in this group of neurodegenerative diseases.
Figure 2.
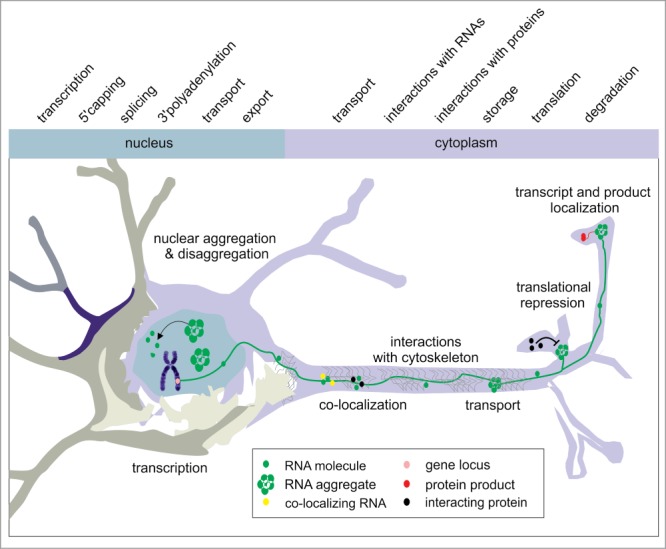
RNA cellular journey in neurons. The figure presents RNA-related processes, which were investigated in neuronal cell lines using live imaging systems. Followed RNA and its aggregates are depicted in green, co-transported RNAs are in yellow and interacting proteins are in black. Protein products are illustrated in red, and gene loci are pink.
Final Remarks and Future Perspectives
Insight into dynamics of transcript birth, maturation, adult life and death is one of the major objectives in cell biology. This very ambitious goal cannot be achieved in the short term considering the multitude of coding and noncoding transcripts and their involvement in countless cellular functions. With the advent of RNA live imaging systems described in the first part of this article, many important questions regarding transcripts synthesis, processing, trafficking and interactions were answered, which is evident from the second part of this review. The field of RNA live imaging is no longer in its infancy, mainly because of the widespread applications of the MS2 system, which was invented by Robert Singer and colleagues in the nineties.
There are numerous RNA-related cellular processes current understanding of which would benefit from the application of RNA live imaging systems. These processes include the complex and strictly regulated process of transcription and the multistep process of transcript maturation, which both engage a variety of cellular proteins. An example of a research area that has not yet strongly benefited from RNA imaging systems is the regulation of protein-coding gene expression by numerous non-coding RNAs. Among the extensively investigated but still unresolved issues are the following: How are the mRNAs regulated by microRNAs? By what mechanism do the small RNAs find their cytoplasmic targets? What is the role of the proteins involved in the RNA interference machinery in the cell nucleus134? Even more vague is our knowledge of the role of long non-coding RNAs in cells and the functions of multiple antisense transcripts. Biomedical research would benefit from an improved understanding of alterations in RNA dynamics in human diseases so that this knowledge could be used in planning therapeutic interventions.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Ministry of Science and Higher Education (grant numbers N N301 569340, N N302 633240), National Science Center (2011/03/B/NZ1/03259, 2012/06/A/NZ1/00094), and European Regional Development Fund within Innovative Economy Program (POIG.01.03.01–30–098/08).
References
- 1. Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol 1993; 123:431-41; PMID:7691830; http://dx.doi.org/ 10.1083/jcb.123.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gagnon JA, Mowry KL. Visualizing RNA localization in Xenopus oocytes. J Vis Exp 2010; PMID:20075839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA.RNA hybridization in living cells. Proc Natl Acad Sci U S A 1998; 95:11538-43; PMID:9751701; http://dx.doi.org/ 10.1073/pnas.95.20.11538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsuo T. In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim Biophys Acta 1998; 1379:178-84; PMID:9528652; http://dx.doi.org/ 10.1016/S0304-4165(97)00090-1 [DOI] [PubMed] [Google Scholar]
- 5. Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 1996; 14:303-8; PMID:9630890; http://dx.doi.org/ 10.1038/nbt0396-303 [DOI] [PubMed] [Google Scholar]
- 6. Santangelo PJ. Molecular beacons and related probes for intracellular RNA imaging. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2010; 2:11-9; PMID:20049827; http://dx.doi.org/ 10.1002/wnan.52 [DOI] [PubMed] [Google Scholar]
- 7. Santangelo PJ, Lifland AW, Curt P, Sasaki Y, Bassell GJ, Lindquist ME, Crowe JE Jr. Single molecule-sensitive probes for imaging RNA in live cells. Nat Methods 2009; 6:347-9; PMID:19349979; http://dx.doi.org/ 10.1038/nmeth.1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods 2007; 4:413-9; PMID:17401370 [DOI] [PubMed] [Google Scholar]
- 9. Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/ 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 10. Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol 1999; 9:569-78; PMID:10359695; http://dx.doi.org/ 10.1016/S0960-9822(99)80260-7 [DOI] [PubMed] [Google Scholar]
- 11. Lim F, Peabody DS. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res 1994; 22:3748-52; PMID:7937087; http://dx.doi.org/ 10.1093/nar/22.18.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Querido E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods Cell Biol 2008; 85:273-92; PMID:18155467; http://dx.doi.org/ 10.1016/S0091-679X(08)85012-1 [DOI] [PubMed] [Google Scholar]
- 13. Lowary PT, Uhlenbeck OC. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res 1987; 15:10483-93; PMID:3697094; http://dx.doi.org/ 10.1093/nar/15.24.10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods 2007; 4:633-6; PMID:17603490; http://dx.doi.org/ 10.1038/nmeth1065 [DOI] [PubMed] [Google Scholar]
- 15. Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptideboxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell 1998; 93:289-99; PMID:9568720; http://dx.doi.org/ 10.1016/S0092-8674(00)81579-2 [DOI] [PubMed] [Google Scholar]
- 16. Schonberger J, Hammes UZ, Dresselhaus T. In vivo visualization of RNA in plants cells using the lambdaN(2)(2) system and a GATEWAY-compatible vector series for candidate RNAs. Plant J 2012; 71:173-81; PMID:22268772; http://dx.doi.org/ 10.1111/j.1365-313X.2012.04923.x [DOI] [PubMed] [Google Scholar]
- 17. Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A 2009; 106:13535-40; PMID:19628694; http://dx.doi.org/ 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglGSacY family. Proc Natl Acad Sci U S A 1992; 89:10410-4; PMID:1279678; http://dx.doi.org/ 10.1073/pnas.89.21.10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 1990; 62:1153-63; PMID:1698125; http://dx.doi.org/ 10.1016/0092-8674(90)90392-R [DOI] [PubMed] [Google Scholar]
- 20. Manival X, Yang Y, Strub MP, Kochoyan M, Steinmetz M, Aymerich S. From genetic to structural characterization of a new class of RNA-binding domain within the SacYBglG family of antiterminator proteins. Embo J 1997; 16:5019-29; PMID:9305643; http://dx.doi.org/ 10.1093/emboj/16.16.5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ni N, Nikolaitchik OA, Dilley KA, Chen J, Galli A, Fu W, Prasad VV, Ptak RG, Pathak VK, Hu WS. Mechanisms of human immunodeficiency virus type 2 RNA packaging: efficient trans packaging and selection of RNA copackaging partners. J Virol 2011; 85:7603-12; PMID:21613401; http://dx.doi.org/ 10.1128/JVI.00562-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 2011; 332:475-8; PMID:21512033; http://dx.doi.org/ 10.1126/science.1202142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lim F, Peabody DS. RNA recognition site of PP7 coat protein. Nucleic Acids Res 2002; 30:4138-44; PMID:12364592; http://dx.doi.org/ 10.1093/nar/gkf552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011; 333:470-4; PMID:21700839; http://dx.doi.org/ 10.1126/science.1206938 [DOI] [PubMed] [Google Scholar]
- 25. Wu B, Chen J, Singer RH. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci Rep 2014; 4:3615; PMID:24402470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. Rna 2000; 6:1737-49; PMID:11142374; http://dx.doi.org/ 10.1017/S1355838200001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci U S A 2000; 97:5273-8; PMID:10792032; http://dx.doi.org/ 10.1073/pnas.080585897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kilchert C, Spang A. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. Embo J 2011; 30:3567-80; PMID:21792172; http://dx.doi.org/ 10.1038/emboj.2011.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J Cell Biol 2010; 189:755-67; PMID:20457760; http://dx.doi.org/ 10.1083/jcb.200912011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005; 310:486-9; PMID:16141371; http://dx.doi.org/ 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lavut A, Raveh D. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet 2012; 8:e1002527; PMID:22383896; http://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 1994; 372:432-8; PMID:7984237; http://dx.doi.org/ 10.1038/372432a0 [DOI] [PubMed] [Google Scholar]
- 33. Allain FH, Gubser CC, Howe PW, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature 1996; 380:646-50; PMID:8602269; http://dx.doi.org/ 10.1038/380646a0 [DOI] [PubMed] [Google Scholar]
- 34. Yiu H-W, Demidov VV, Toran P, Cantor CR, Broude NE. RNA detection in live bacterial cells using fluorescent protein complementation triggered by interaction of two RNA aptamers with two RNA-binding peptides. Pharmaceuticals 2011; 4:494-508; http://dx.doi.org/ 10.3390/ph4030494 [DOI] [Google Scholar]
- 35. Baskerville S, Zapp M, Ellington AD. Anti-Rex aptamers as mimics of the Rex-binding element. J Virol 1999; 73:4962-71; PMID:10233958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakano K, Watanabe T. HTLV-1 Rex: the courier of viral messages making use of the host vehicle. Front Microbiol 2012; 3:330; PMID:22973269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yin J, Zhu D, Zhang Z, Wang W, Fan J, Men D, Deng J, Wei H, Zhang XE, Cui Z. Imaging of mRNA-protein interactions in live cells using novel mCherry trimolecular fluorescence complementation systems. PLoS One 2013; 8:e80851; PMID:24260494; http://dx.doi.org/ 10.1371/journal.pone.0080851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harada K, Martin SS, Frankel AD. Selection of RNA-binding peptides in vivo. Nature 1996; 380:175-9; PMID:8600395; http://dx.doi.org/ 10.1038/380175a0 [DOI] [PubMed] [Google Scholar]
- 39. Heaphy S, Dingwall C, Ernberg I, Gait MJ, Green SM, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 1990; 60:685-93; PMID:1689218; http://dx.doi.org/ 10.1016/0092-8674(90)90671-Z [DOI] [PubMed] [Google Scholar]
- 40. Cao H, Tamilarasu N, Rana TM. Orientation and affinity of HIV-1 Tat fragments in Tat-TAR complex determined by fluorescence resonance energy transfer. Bioconjug Chem 2006; 17:352-8; PMID:16536465; http://dx.doi.org/ 10.1021/bc050277u [DOI] [PubMed] [Google Scholar]
- 41. Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat Methods 2007; 4:421-7; PMID:17401371 [DOI] [PubMed] [Google Scholar]
- 42. Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. Rna 2003; 9:394-407; PMID:12649492; http://dx.doi.org/ 10.1261/rna.2161303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valencia-Burton M, Broude NE. Visualization of RNA using fluorescence complementation triggered by aptamer-protein interactions (RFAP) in live bacterial cells. Curr Protoc Cell Biol 2007; Chapter 17:Unit 17 1; PMID:18228500 [DOI] [PubMed] [Google Scholar]
- 44. Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science 2011; 333:642-6; PMID:21798953; http://dx.doi.org/ 10.1126/science.1207339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods 2009; 6:331-8; PMID:19404252; http://dx.doi.org/ 10.1038/nmeth.1321 [DOI] [PubMed] [Google Scholar]
- 46. Nevo-Dinur K, Govindarajan S, Amster-Choder O. Subcellular localization of RNA and proteins in prokaryotes. Trends Genet 2012; 28:314-22; PMID:22521614; http://dx.doi.org/ 10.1016/j.tig.2012.03.008 [DOI] [PubMed] [Google Scholar]
- 47. Ehrhardt DW, Frommer WB. New technologies for 21st century plant science. Plant Cell 2012; 24:374-94; PMID:22366161; http://dx.doi.org/ 10.1105/tpc.111.093302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sando S, Narita A, Aoyama Y. Light-up Hoechst-DNA aptamer pair: generation of an aptamer-selective fluorophore from a conventional DNA-staining dye. Chembiochem 2007; 8:1795-803; PMID:17806095; http://dx.doi.org/ 10.1002/cbic.200700325 [DOI] [PubMed] [Google Scholar]
- 49. Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS, Armitage BA. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org Lett 2008; 10:1561-4; PMID:18338898; http://dx.doi.org/ 10.1021/ol702920e [DOI] [PubMed] [Google Scholar]
- 50. Lee J, Lee KH, Jeon J, Dragulescu-Andrasi A, Xiao F, Rao J. Combining SELEX screening and rational design to develop light-up fluorophore-RNA aptamer pairs for RNA tagging. ACS Chem Biol 2010; 5:1065-74; PMID:20809562; http://dx.doi.org/ 10.1021/cb1001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sparano BA, Koide K. Fluorescent sensors for specific RNA: a general paradigm using chemistry and combinatorial biology. J Am Chem Soc 2007; 129:4785-94; PMID:17385867; http://dx.doi.org/ 10.1021/ja070111z [DOI] [PubMed] [Google Scholar]
- 52. Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-play fluorophores extend the spectral properties of spinach. J Am Chem Soc 2014; 136:1198-201; PMID:24393009; http://dx.doi.org/ 10.1021/ja410819x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pakhomov AA, Martynov VI. GFP family: structural insights into spectral tuning. Chem Biol 2008; 15:755-64; PMID:18721746; http://dx.doi.org/ 10.1016/j.chembiol.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 54. Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods 2013; 10:1219-24; PMID:24162923; http://dx.doi.org/ 10.1038/nmeth.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lux J, Pena EJ, Bolze F, Heinlein M, Nicoud JF. Malachite green derivatives for two-photon RNA detection. Chembiochem 2012; 13:1206-13; PMID:22549874; http://dx.doi.org/ 10.1002/cbic.201100747 [DOI] [PubMed] [Google Scholar]
- 56. Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J Mol Biol 2000; 301:117-28; PMID:10926496; http://dx.doi.org/ 10.1006/jmbi.2000.3951 [DOI] [PubMed] [Google Scholar]
- 57. Kolpashchikov DM. Binary malachite green aptamer for fluorescent detection of nucleic acids. J Am Chem Soc 2005; 127:12442-3; PMID:16144363; http://dx.doi.org/ 10.1021/ja0529788 [DOI] [PubMed] [Google Scholar]
- 58. Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc 2003; 125:14716-7; PMID:14640641; http://dx.doi.org/ 10.1021/ja037994o [DOI] [PubMed] [Google Scholar]
- 59. Reif R, Haque F, Guo P. Fluorogenic RNA nanoparticles for monitoring RNA folding and degradation in real time in living cells. Nucleic Acid Ther 2012; 22:428-37; PMID:23113765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sunbul M, Jaschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed Engl 2013; 52:13401-4; PMID:24133044; http://dx.doi.org/ 10.1002/anie.201306622 [DOI] [PubMed] [Google Scholar]
- 61. Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell 2005; 123:1025-36; PMID:16360033; http://dx.doi.org/ 10.1016/j.cell.2005.09.031 [DOI] [PubMed] [Google Scholar]
- 62. Lange S, Katayama Y, Schmid M, Burkacky O, Brauchle C, Lamb DC, Jansen RP. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 2008; 9:1256-67; PMID:18485054; http://dx.doi.org/ 10.1111/j.1600-0854.2008.00763.x [DOI] [PubMed] [Google Scholar]
- 63. Martin RM, Rino J, Carvalho C, Kirchhausen T, Carmo-Fonseca M. Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep 2013; 4:1144-55; PMID:24035393; http://dx.doi.org/ 10.1016/j.celrep.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2009; 106:19848-53; PMID:19903887; http://dx.doi.org/ 10.1073/pnas.0910754106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 2003; 13:161-7; PMID:12546792; http://dx.doi.org/ 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 2004; 16:1047-59; PMID:15037734; http://dx.doi.org/ 10.1105/tpc.019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 2007; 48:1243-53; PMID:17675322; http://dx.doi.org/ 10.1093/pcp/pcm099 [DOI] [PubMed] [Google Scholar]
- 68. Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. Embo J 2004; 23:3346-55; PMID:15282548; http://dx.doi.org/ 10.1038/sj.emboj.7600341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Michnick SW. Protein fragment complementation strategies for biochemical network mapping. Curr Opin Biotechnol 2003; 14:610-7; PMID:14662390; http://dx.doi.org/ 10.1016/j.copbio.2003.10.014 [DOI] [PubMed] [Google Scholar]
- 70. Tyagi S. Splitting or stacking fluorescent proteins to visualize mRNA in living cells. Nat Methods 2007; 4:391-2; PMID:17464293; http://dx.doi.org/ 10.1038/nmeth0507-391 [DOI] [PubMed] [Google Scholar]
- 71. Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y. The life of an mRNA in space and time. J Cell Sci 2010; 123:1761-74; PMID:20427315; http://dx.doi.org/ 10.1242/jcs.062638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods 2013; 10:119-21; PMID:23263691; http://dx.doi.org/ 10.1038/nmeth.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Quantifying the transcriptional output of single alleles in single living mammalian cells. Nat Protoc 2013; 8:393-408; PMID:23424748; http://dx.doi.org/ 10.1038/nprot.2013.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Barberis M, Beck C, Amoussouvi A, Schreiber G, Diener C, Herrmann A, Klipp E. A low number of SIC1 mRNA molecules ensures a low noise level in cell cycle progression of budding yeast. Mol Biosyst 2011; 7:2804-12; PMID:21717009; http://dx.doi.org/ 10.1039/c1mb05073g [DOI] [PubMed] [Google Scholar]
- 75. Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods 2010; 7:631-3; PMID:20639867; http://dx.doi.org/ 10.1038/nmeth.1482 [DOI] [PubMed] [Google Scholar]
- 76. Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One 2010; 5:e10272; PMID:20422051; http://dx.doi.org/ 10.1371/journal.pone.0010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell 2004; 116:683-98; PMID:15006351; http://dx.doi.org/ 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 2007; 14:796-806; PMID:17676063; http://dx.doi.org/ 10.1038/nsmb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dowen JM, Bilodeau S, Orlando DA, Hubner MR, Abraham BJ, Spector DL, Young RA. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Reports 2013; 1:371-8; PMID:24286025; http://dx.doi.org/ 10.1016/j.stemcr.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep 2011; 12:1280-5; PMID:22015688; http://dx.doi.org/ 10.1038/embor.2011.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 2011; 9:e1000573; PMID:21264352; http://dx.doi.org/ 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 2007; 179:291-304; PMID:17954611; http://dx.doi.org/ 10.1083/jcb.200706018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science 2004; 304:1797-800; PMID:15205532; http://dx.doi.org/ 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pitchiaya S, Heinicke LA, Custer TC, Walter NG. Single molecule fluorescence approaches shed light on intracellular RNAs. Chem Rev 2014; 114:3224-65; PMID:24417544; http://dx.doi.org/ 10.1021/cr400496q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 2010; 12:543-52; PMID:20453848; http://dx.doi.org/ 10.1038/ncb2056 [DOI] [PubMed] [Google Scholar]
- 86. Querido E, Gallardo F, Beaudoin M, Menard C, Chartrand P. Stochastic and reversible aggregation of mRNA with expanded CUG-triplet repeats. J Cell Sci 2011; 124:1703-14; PMID:21511730; http://dx.doi.org/ 10.1242/jcs.073270 [DOI] [PubMed] [Google Scholar]
- 87. Yamagishi M, Shirasaki Y, Funatsu T. Size-dependent accumulation of mRNA at the leading edge of chicken embryo fibroblasts. Biochem Biophys Res Commun 2009; 390:750-4; PMID:19835844; http://dx.doi.org/ 10.1016/j.bbrc.2009.10.043 [DOI] [PubMed] [Google Scholar]
- 88. Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol 2007; 500:433-47; PMID:17120280; http://dx.doi.org/ 10.1002/cne.21189 [DOI] [PubMed] [Google Scholar]
- 89. Bookwalter CS, Lord M, Trybus KM. Essential features of the class V myosin from budding yeast for ASH1 mRNA transport. Mol Biol Cell 2009; 20:3414-21; PMID:19477930; http://dx.doi.org/ 10.1091/mbc.E08-08-0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, et al. hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies. Nat Struct Mol Biol 2010; 17:732-9; PMID:20473314; http://dx.doi.org/ 10.1038/nsmb.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science 2011; 331:1081-4; PMID:21350180; http://dx.doi.org/ 10.1126/science.1195691 [DOI] [PubMed] [Google Scholar]
- 92. Oh JY, Nam YJ, Jo A, Cheon HS, Rhee SM, Park JK, Lee JA, Kim HK. Apolipoprotein E mRNA is transported to dendrites and may have a role in synaptic structural plasticity. J Neurochem 2010; 114:685-96; PMID:20456011; http://dx.doi.org/ 10.1111/j.1471-4159.2010.06773.x [DOI] [PubMed] [Google Scholar]
- 93. Ghoujal B, Milev MP, Ajamian L, Abel K, Mouland AJ. ESCRT-II's involvement in HIV-1 genomic RNA trafficking and assembly. Biol Cell 2012; 104:706-21; PMID:22978549; http://dx.doi.org/ 10.1111/boc.201200021 [DOI] [PubMed] [Google Scholar]
- 94. Rook MS, Lu M, Kosik KS. CaMKIIalpha 3’ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 2000; 20:6385-93; PMID:10964944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Karimian Pour N, Adeli K. Insulin silences apolipoprotein B mRNA translation by inducing intracellular traffic into cytoplasmic RNA granules. Biochemistry 2011; 50:6942-50; PMID:21721546; http://dx.doi.org/ 10.1021/bi200711v [DOI] [PubMed] [Google Scholar]
- 96. Estes PS, O’Shea M, Clasen S, Zarnescu DC. Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol Cell Neurosci 2008; 39:170-9; PMID:18655836; http://dx.doi.org/ 10.1016/j.mcn.2008.06.012 [DOI] [PubMed] [Google Scholar]
- 97. Aronov S, Gerst JE. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem 2004; 279:36962-71; PMID:15192110; http://dx.doi.org/ 10.1074/jbc.M402068200 [DOI] [PubMed] [Google Scholar]
- 98. Shimada Y, Burn KM, Niwa R, Cooley L. Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol 2011; 355:250-62; PMID:21570389; http://dx.doi.org/ 10.1016/j.ydbio.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD. Assembling an intermediate filament network by dynamic cotranslation. J Cell Biol 2006; 172:747-58; PMID:16505169; http://dx.doi.org/ 10.1083/jcb.200511033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kraut-Cohen J, Afanasieva E, Haim-Vilmovsky L, Slobodin B, Yosef I, Bibi E, Gerst JE. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2013; 24:3069-84; PMID:23904265; http://dx.doi.org/ 10.1091/mbc.E13-01-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell 2009; 138:1005-18; PMID:19737525; http://dx.doi.org/ 10.1016/j.cell.2009.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Reich J, Snee MJ, Macdonald PM. miRNA-dependent translational repression in the Drosophila ovary. PLoS One 2009; 4:e4669; PMID:19252745; http://dx.doi.org/ 10.1371/journal.pone.0004669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol 2011; 31:4219-31; PMID:21859890; http://dx.doi.org/ 10.1128/MCB.05955-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009; 136:763-76; PMID:19239894; http://dx.doi.org/ 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- 105. Jang LT, Buu LM, Lee FJ. Determinants of Rbp1p localization in specific cytoplasmic mRNA-processing foci, P-bodies. J Biol Chem 2006; 281:29379-90; PMID:16885161; http://dx.doi.org/ 10.1074/jbc.M601573200 [DOI] [PubMed] [Google Scholar]
- 106. Blanco FF, Sanduja S, Deane NG, Blackshear PJ, Dixon DA. Transforming growth factor beta regulates P-body formation through induction of the mRNA decay factor tristetraprolin. Mol Cell Biol 2014; 34:180-95; PMID:24190969; http://dx.doi.org/ 10.1128/MCB.01020-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003; 300:805-8; PMID:12730603; http://dx.doi.org/ 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Muramoto T, Cannon D, Gierlinski M, Corrigan A, Barton GJ, Chubb JR. Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc Natl Acad Sci U S A 2012; 109:7350-5; PMID:22529358; http://dx.doi.org/ 10.1073/pnas.1117603109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res 2011; 39:8938-51; PMID:21795378; http://dx.doi.org/ 10.1093/nar/gkr608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res 2011; 39:3852-63; PMID:21247881; http://dx.doi.org/ 10.1093/nar/gkq1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol 1995; 128:995-1002; PMID:7896884; http://dx.doi.org/ 10.1083/jcb.128.6.995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3’ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A 1997; 94:7388-93; PMID:9207101; http://dx.doi.org/ 10.1073/pnas.94.14.7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol 2003; 54:760-8; PMID:14681885; http://dx.doi.org/ 10.1002/ana.10763 [DOI] [PubMed] [Google Scholar]
- 114. Wilburn B, Rudnicki DD, Zhao J, Weitz TM, Cheng Y, Gu X, Greiner E, Park CS, Wang N, Sopher BL, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron 2011; 70:427-40; PMID:21555070; http://dx.doi.org/ 10.1016/j.neuron.2011.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremorataxia syndrome (FXTAS). RNA Biol 2004; 1:103-5; PMID:17179750; http://dx.doi.org/ 10.4161/rna.1.2.1035 [DOI] [PubMed] [Google Scholar]
- 116. Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. Embo J 2010; 29:1248-61; PMID:20186122; http://dx.doi.org/ 10.1038/emboj.2010.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet 2006; 15:1808-15; PMID:16624843; http://dx.doi.org/ 10.1093/hmg/ddl103 [DOI] [PubMed] [Google Scholar]
- 118. Schoser BG, Ricker K, Schneider-Gold C, Hengstenberg C, Durre J, Bultmann B, Kress W, Day JW, Ranum LP. Sudden cardiac death in myotonic dystrophy type 2. Neurology 2004; 63:2402-4; PMID:15623712; http://dx.doi.org/ 10.1212/01.WNL.0000147335.10783.E4 [DOI] [PubMed] [Google Scholar]
- 119. Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 2001; 293:864-7; PMID:11486088; http://dx.doi.org/ 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- 120. Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S, et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet 2009; 85:544-57; PMID:19878914; http://dx.doi.org/ 10.1016/j.ajhg.2009.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. White MC, Gao R, Xu W, Mandal SM, Lim JG, Hazra TK, Wakamiya M, Edwards SF, Raskin S, Teive HA, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet 2010; 6:e1000984; PMID:20548952; http://dx.doi.org/ 10.1371/journal.pgen.1000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 2013; 126:881-93; PMID:24132570; http://dx.doi.org/ 10.1007/s00401-013-1189-3 [DOI] [PubMed] [Google Scholar]
- 123. Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 2013; 5:208ra149; PMID:24154603; http://dx.doi.org/ 10.1126/scitranslmed.3007529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet 2011; 20:3811-21; PMID:21729883; http://dx.doi.org/ 10.1093/hmg/ddr299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 2002; 11:805-14; PMID:11929853; http://dx.doi.org/ 10.1093/hmg/11.7.805 [DOI] [PubMed] [Google Scholar]
- 126. Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J 2000; 19:4439-48; PMID:10970838; http://dx.doi.org/ 10.1093/emboj/19.17.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cardani R, Mancinelli E, Giagnacovo M, Sansone V, Meola G. Ribonuclear inclusions as biomarker of myotonic dystrophy type 2, even in improperly frozen or defrozen skeletal muscle biopsies. Eur J Histochem 2009; 53:107-11; PMID:19683984; http://dx.doi.org/ 10.4081/ejh.2009.e13 [DOI] [PubMed] [Google Scholar]
- 128. Furling D, Coiffier L, Mouly V, Barbet JP, St Guily JL, Taneja K, Gourdon G, Junien C, Butler-Browne GS. Defective satellite cells in congenital myotonic dystrophy. Hum Mol Genet 2001; 10:2079-87; PMID:11590125; http://dx.doi.org/ 10.1093/hmg/10.19.2079 [DOI] [PubMed] [Google Scholar]
- 129. Holt I, Jacquemin V, Fardaei M, Sewry CA, Butler-Browne GS, Furling D, Brook JD, Morris GE. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am J Pathol 2009; 174:216-27; PMID:19095965; http://dx.doi.org/ 10.2353/ajpath.2009.080520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dansithong W, Wolf CM, Sarkar P, Paul S, Chiang A, Holt I, Morris GE, Branco D, Sherwood MC, Comai L, et al. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One 2008; 3:e3968; PMID:19092997; http://dx.doi.org/ 10.1371/journal.pone.0003968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 2012; 40:11-26; PMID:21908410; http://dx.doi.org/ 10.1093/nar/gkr729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Galka-Marciniak P, Urbanek MO, Krzyzosiak WJ. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol Chem 2013; 393:1299-315 [DOI] [PubMed] [Google Scholar]
- 133. Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet 2006; 38:1066-70; PMID:16878132; http://dx.doi.org/ 10.1038/ng1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Stroynowska-Czerwinska A, Fiszer A, Krzyzosiak WJ. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell Mol Life Sci 2014; 71:2253-70; PMID:24468964; http://dx.doi.org/ 10.1007/s00018-013-1551-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 2011; 8:165-70; PMID:21240280; http://dx.doi.org/ 10.1038/nmeth.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lee HY, Ge WP, Huang W, He Y, Wang GX, Rowson-Baldwin A, Smith SJ, Jan YN, Jan LY. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron 2011; 72:630-42; PMID:22099464; http://dx.doi.org/ 10.1016/j.neuron.2011.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol 2008; 18:745-50; PMID:18472422; http://dx.doi.org/ 10.1016/j.cub.2008.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci U S A 2006; 103:19919-24; PMID:17167054; http://dx.doi.org/ 10.1073/pnas.0607394104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 2006; 124:191-205; PMID:16413491; http://dx.doi.org/ 10.1016/j.cell.2005.12.017 [DOI] [PubMed] [Google Scholar]
- 140. Dziembowska M, Milek J, Janusz A, Rejmak E, Romanowska E, Gorkiewicz T, Tiron A, Bramham CR, Kaczmarek L. Activity-dependent local translation of matrix metalloproteinase-9. J Neurosci 2012; 32:14538-47; PMID:23077039; http://dx.doi.org/ 10.1523/JNEUROSCI.6028-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bi J, Hu X, Loh HH, Wei LN. Mouse kappa-opioid receptor mRNA differential transport in neurons. Mol Pharmacol 2003; 64:594-9; PMID:12920195; http://dx.doi.org/ 10.1124/mol.64.3.594 [DOI] [PubMed] [Google Scholar]
- 142. Xu X, Brechbiel JL, Gavis ER. Dynein-dependent transport of nanos RNA in Drosophila sensory neurons requires Rumpelstiltskin and the germ plasm organizer Oskar. J Neurosci 2013; 33:14791-800; PMID:24027279; http://dx.doi.org/ 10.1523/JNEUROSCI.5864-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lebeau G, Miller LC, Tartas M, McAdam R, Laplante I, Badeaux F, DesGroseillers L, Sossin WS, Lacaille JC. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn Mem 2011; 18:314-26; PMID:21508097; http://dx.doi.org/ 10.1101/lm.2100611 [DOI] [PubMed] [Google Scholar]
- 144. Jo A, Kim HK. Up-regulation of dendritic Kv4.2 mRNA by activation of the NMDA receptor. Neurosci Lett 2011; 496:129-34; PMID:21511008; http://dx.doi.org/ 10.1016/j.neulet.2011.03.099 [DOI] [PubMed] [Google Scholar]
- 145. Ma B, Culver BP, Baj G, Tongiorgi E, Chao MV, Tanese N. Localization of BDNF mRNA with the Huntington's disease protein in rat brain. Mol Neurodegener 2010; 5:22; PMID:20507609; http://dx.doi.org/ 10.1186/1750-1326-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ma B, Savas JN, Yu MS, Culver BP, Chao MV, Tanese N. Huntingtin mediates dendritic transport of beta-actin mRNA in rat neurons. Sci Rep 2011; 1:140; PMID:22355657; http://dx.doi.org/ 10.1038/srep00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kao DI, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci U S A 2010; 107:15601-6; PMID:20713728; http://dx.doi.org/ 10.1073/pnas.1010564107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. A role for huntington disease protein in dendritic RNA granules. J Biol Chem 2010; 285:13142-53; PMID:20185826; http://dx.doi.org/ 10.1074/jbc.M110.114561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell 2008; 14:926-39; PMID:18539120; http://dx.doi.org/ 10.1016/j.devcel.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Shiina N, Yamaguchi K, Tokunaga M. RNG105 deficiency impairs the dendritic localization of mRNAs for Na+K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J Neurosci 2010; 30:12816-30; PMID:20861386; http://dx.doi.org/ 10.1523/JNEUROSCI.6386-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Janusz A, Milek J, Perycz M, Pacini L, Bagni C, Kaczmarek L, Dziembowska M. The Fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci 2013; 33:18234-41; PMID:24227732; http://dx.doi.org/ 10.1523/JNEUROSCI.2207-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Bi J, Tsai NP, Lu HY, Loh HH, Wei LN. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc Natl Acad Sci U S A 2007; 104:13810-5; PMID:17698811; http://dx.doi.org/ 10.1073/pnas.0703805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, Inoue T, Mikoshiba K. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J Biol Chem 2004; 279:53427-34; PMID:15475564; http://dx.doi.org/ 10.1074/jbc.M409732200 [DOI] [PubMed] [Google Scholar]
- 154. Dynes JL, Steward O. Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol 2012; 520:3105-19; PMID:22350812; http://dx.doi.org/ 10.1002/cne.23073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.