Abstract
The commensal microbiota of the human gastrointestinal tract live in a largely stable community structure, assisting in host physiological and immunological functions. Changes to this structure can be injurious to the health of the host, a concept termed dysbiosis. Psychological stress is a factor that has been implicated in causing dysbiosis, and studies performed by our lab have shown that restraint stress can indeed shift the cecal microbiota structure as well as increase the severity of a colonic infection caused by Citrobacter rodentium. However, this study, like many others, have focused on fecal contents when examining the effect of dysbiosis-causing stimuli (e.g. psychological stress) upon the microbiota. Since the mucosa-associated microbiota have unique properties and functions that can act upon the host, it is important to understand how stressor exposure might affect this niche of bacteria. To begin to understand whether chronic restraint stress changes the mucosa-associated and/or luminal microbiota mice underwent 7 16-hour cycles of restraint stress, and the microbiota of both colonic tissue and fecal contents were analyzed by sequencing using next-gen bacterial tag-encoded FLX amplicon technology (bTEFAP) pyrosequencing. Both control and stress groups had significantly different mucosa-associated and luminal microbiota communities, highlighting the importance of focusing gastrointestinal community structure analysis by microbial niche. Furthermore, restraint stress was able to disrupt both the mucosa-associated and luminally-associated colonic microbiota by shifting the relative abundances of multiple groups of bacteria. Among these changes, there was a significant reduction in the immunomodulatory commensal genus Lactobacillus associated with colonic mucosa. The relative abundance of Lactobacillus spp. was not affected in the lumen. These results indicate that stressor-exposure can have distinct effects upon the colonic microbiota situated at the mucosal epithelium in comparison to the luminal-associated microbiota.
Keywords: inflammation, microbiota, Psychological stress, pyrosequencing
Introduction
There are extensive bidirectional interactions between the brain and the gut, and it is well recognized that during a stress response, the central and enteric nervous systems have a large influence on gastrointestinal (GI) motility, secretion, blood flow and immune reactivity.1-3 Thus, it is not surprising that stressor exposure has been linked to exacerbations of intestinal diseases, like the inflammatory bowel diseases (IBD), including ulcerative colitis (UC) and Crohn's disease (CD), and irritable bowel syndrome (IBS).4,5 The mechanisms through which the stress response impacts these diseases are not yet completely understood, but it is possible that stressor-induced changes to the intestinal microbiota are involved. Altered profiles of intestinal microbiota have been identified in both IBD and in IBS patients.6,7 Thus, understanding factors that maintain microbiota community stability as well as those that can disrupt the structure could lead to a deeper understanding of how factors such as stress can affect intestinal disease.
The microbiota that colonize the human body collectively outnumber cells of the body by a factor of 10.8 The vast majority of these microbes are bacteria that reside within the intestines as part of the intestinal microbiota, with microbiota levels ranging from <105 bacteria per gram of digesta in the upper parts of the small intestine, to >1012 bacteria per gram of digesta in the large intestine.7 The microbiota reside as a largely stable community that develops as a result of a series of ecological successions involving the selection of species best adapted for the available niche.9 This climax community is relatively resistant and resilient to long-term disruptions in community structure,10 but many factors, such as antibiotic use or colonic inflammation can cause alterations in their community structure.11-14 Whether these types of alterations impact the function of the microbiota is not completely understood, but it is recognized that disrupting the microbiota through the use of antibiotics can enhance pathogen colonization and proliferation.15
Exposure to stressors has been shown to significantly change microbial populations in the gastrointestinal tract of both humans and laboratory animals. This was first demonstrated in Russian cosmonauts preparing for space flight, who were found to have significantly different levels of shed microbiota, with later studies suggesting that the changes could be due to the stress of confinement.16,17 In a more recent study, it was demonstrated that lactobacilli levels shed in the stool of college students were lower during final examinations than during quiescent periods, suggesting that the stress of the examinations affected the microbiota.18 These findings are consistent with findings in laboratory animals, where stressor exposure has been shown to significantly impact the populations of certain bacteria shed in the stool of nonhuman primates, as well as rodents.17,19-22 For the most part, these studies have relied on culture-based techniques and have only examined bacteria in the lumen of the intestines or shed in the feces. Although this approach is informative, there are several disadvantages. Approximately 90% of the intestinal microbiota are strict anaerobes that have not been characterized by traditional culture-based methods due to undefined culture conditions.23 In addition, the diversity of the communities present in the lumen of the intestines is different than those found in the mucous layer.24,25 Thus, it is possible that the stressor-induced alterations to the microbiota might differ between GI compartment.
A recent study using culture-independent 16s rRNA based sequencing has shown that only 2 hours of acute exposure to the social disruption stressor (SDR) altered the mucosa-associated microbiota of C57BL/6 mice (Galley et al, under review). Acute and chronic stressors can have distinct effects upon gastrointestinal physiology, particularly in mucus secretion, wherein acute stressors tend to increase mucus production, while repeated chronic stressors can reduce mucin gene expression.26-28 It is unknown if chronic stress might also have unique impacts upon the colonic microbiota, or if such impacts might be specific to luminal or mucosa-associated microbial niches. Prolonged restraint (RST) is a widely used chronic murine stressor that has been extensively characterized in the literature and is the most commonly used murine stressor in biomedical and biobehavioral research.29 This stressor involves both physical and psychological components. While the physical components are obvious (i.e., physical confinement), the psychological components are more complex and are thought to reflect the animal's perception of burrow collapse and confinement.30 The autonomic nervous system and the hypothalamic-pituitary-adrenal axis are stimulated during the stressor as part of a robust physiological stress response.31-33 SDR and RST, 2 prime representatives of acute and chronic stressors respectively, have contrasting effects on host immunity and cellular stress responses,32-38 so it is important to evaluate how these stressors might differentially affect the gastrointestinal microbiota. In the present study, it was hypothesized that exposure to the prolonged stressor, RST, would have a unique impact on the colonic mucosa-associated microbiota in comparison to the luminal microbiota. To test this hypothesis, bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) was performed in this study to comprehensively characterize chronic stressor-induced changes to community structure in both lumen and tissue of the mouse colon.
Results
The colonic mucosal microbiome and the luminal microbiome are significantly different
Initial analysis of 16s rRNA gene pyrosequencing data showed that regardless of exposure to the stressor, food and water deprivation, or control conditions, colonic mucosa and the lumen each have a distinct microbiota (Fig. 1). This was first manifest as a significant increase in α diversity in mucosa-associated microbial communities. Rarefaction using the observed_species analysis showed that sufficient depth was reached in the sequencing for both luminal and tissue-associated samples (data not shown). Shannon Diversity Index (SDI) was significantly different in the lumen vs. the mucosa (P < .05) demonstrating that the colonic mucosa has significantly higher overall α diversity than in the lumen (Fig. 1A). Microbial richness, assessed using Chao1, was similar in the luminal and mucosa-associated microbiota (Fig. 1B). However, there was significantly higher evenness in the mucosa-associated microbiota compared to the luminal-associated microbiota (P < .05 as assessed using the equitability measurement) (Fig. 1C).
Figure 1.
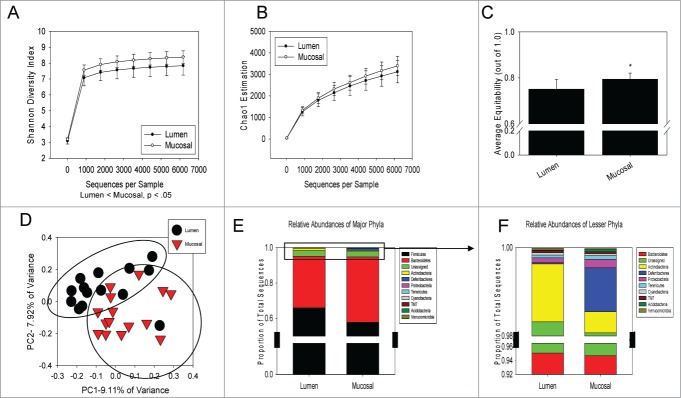
The mucosa-associated and luminal-associated microbiota communities are significantly different from each other. Alpha diversity measurements,including Shannon Diversity, Chao1, and equitability were calculated using QIIME and compared. (A) The Shannon Diversity Index was significantly increased in mucosal samples over luminal samples as measured with QIIME and averaged by group. (B) Richness, estimated with Chao1, was unchanged between both compartments. (C) Evenness, using the equitability measurement, was also increased in the mucosal samples over luminal samples. All data in A-C are mean ± SD. All α diversity measurements were analyzed using a parametric T-test on QIIME at a sequence depth of 6177. (D) Luminal samples and mucosal samples clustered independently of one another based upon unweighted Unifrac distances on a Principle Coordinate Analysis using the ANOSIM statistic (P < 0.0005). (E) Major phyla, including Firmicutes and Bacteroidetes, were unchanged when compared between luminal and mucosal-associated samples. (F) There were significant differences in the lesser phyla when compared between mucosa-associated microbiota and luminally-associated microbiota. The relative abundance of Actinobacteria, Deferribacteres, Proteobacteria, and Acidobacteria were all significantly different when compared between both gastrointestinal compartments. Phyla-level relative abundance was compared using Mann-Whitney U non-parametric tests. Mucosal-associated samples are n = 15; luminal-associated samples are n = 14.
In addition to the changes in the α diversity, the β diversity of luminal and mucosa-associated microbiota communities were significantly different independently of whether the mice were exposed to the stressor, food and water deprivation or control conditions. Luminal samples clustered separately from the mucosa-associated samples on a principal coordinate analysis (PCoA) plot of the unweighted UniFrac distance matrix for all samples combined (Fig. 1D). The clustering was statistically significant (P < 0.0005) based on ANOSIM indicating the mucosal and luminal microbial populations are significantly different. These overall differences extended to significant differences in Actinobacteria, Acidobacteria, Deferribacteres, and Proteobacteria at the phyla level (Figs. 1E and F). Actinobacteria was significantly increased in the luminal group over the mucosal group (1.31% vs. 0.48%) (P < 0.05), while Acidobacteria (0.01% vs. 0.05%) (P < 0.05), Deferribacteres (0.03% vs. 1.00%) (P < 0.001) and Proteobacteria (0.11% vs. 0.20%) (P < 0.05) were all significantly enriched in the mucosal group (Fig. 1F). The 2 colonic compartment's contrasting profiles provided the rationale for investigating the effect of stressor exposure on the 2 microbiomes independently of one another.
Restraint Stress alters the β-diversity, but not the α-diversity of the luminal-associated microbiota
Upon separating the luminal-associated microbiota from the tissue-associated microbiota, the effect of restraint-stress exposure on luminal populations alone was examined. SDI was used to discern changes in overall α diversity as a product of stressor exposure, but was unchanged in the luminal microbiome due to restraint stress (Fig. 2A). This was further confirmed by measuring evenness (using the equitability measurement) and richness (using Chao1) in which there no changes due to restraint stress (Figs. 2B and C).
Figure 2.
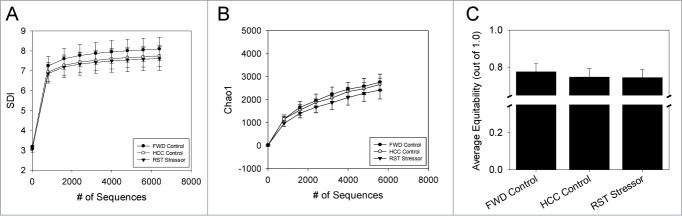
Stressor exposure was not associated with shifts in α diversity in the luminally-associated microbiota. (A) The Shannon Diversity index (SDI), (B) richness (using Chao1) and (C). Evenness (using equitability measurement) was measured for each sample using QIIME and averaged by group. None of the α diversity measurements were affected by stressor exposure. Data are mean ± SD. N = 5 for all groups, with the exception of HCC-Control, which had n = 4. Groups were compared using parametric T-tests on QIIME with a modified Bonferroni correction for multiple comparisons.
The major phyla, Firmicutes and Bacteroidetes, were unaffected by either restraint stress or food and water deprivation in the luminal-associated microbiota (Fig. 3A). However, restraint-stressor exposed groups (RST Stressor) had significantly lower levels of the phylum Actinobacteria compared to both the food and water deprivation control group (FWD Control) and the undisturbed control group (HCC Control) (P < 0.05). Deferribacteres, another phylum was significantly increased in the RST Stressor group compared to both control groups in the luminally-associated populations (P < 0.05) (Fig. 3B). HCC Control and FWD Control were not significantly different among the phyla abundances in the luminally-associated microbiota. In β diversity analysis, the PCoA of the unweighted UniFrac distance matrix for the luminal samples showed significant clustering of the RST Stressor group compared to the FWD Control group (P < 0.01), but RST Stressor did not cluster separately from the HCC Control group. In addition, FWD Control and HCC Control groups did not cluster significantly differently from each other (Fig. 3C). Though RST-5 was separated from the other samples, it was not an outlier using the 2x standard deviation method when the absolute abundance of Firmicutes and Bacteroidetes in RST-5 were compared to other RST samples.
Figure 3.
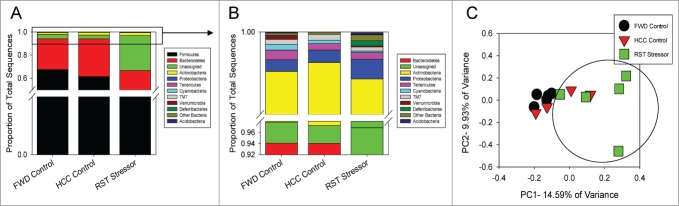
Stressor exposure significantly affects the community structure of the luminally-associated microbiota. (A) Major phyla were unchanged between any group. (B) Among the lesser phyla, Actinobacteria was significantly reduced in RST Stressor mice compared to both HCC Control and FWD Control, while Deferribacteres was significantly reduced in RST Stressor mice compared to HCC Control and FWD Control mice. Abundances were compared using non-parametric Kruskal-Wallis tests, and post-hoc testing was performed with non-parametric Mann-Whitney U tests. (C) Principle coordinate analysis was used to visualize stressor exposure-induced community profile clustering based upon unweighted Unifrac distances. RST Stressor shifted the community structure of the luminally-associated microbiota compared to FWD Control mice (P < 0.01) significantly using the ANOSIM statistic, but not significantly compared to HCC Control mice. FWD and HCC Controls were unchanged compared to each other. Data are from n = 5 for RST Stressor and FWD Control groups, and n = 4 for HCC Control.
Restraint Stress shifts both α and β diversity of the mucosa-associated microbiota
In contrast to the lack of α diversity changes in the luminal associated microbiota, restraint stress altered both the α and β diversity in the mucosa-associated microbiota. Separate analysis of the mucosa-associated microbiota made evident significant shifts in the RST Stressor group compared to FWD Control and HCC Control groups. RST Stressor had a significant reduction compared to FWD Control in α diversity using the SDI (P < 0.01). SDI was not different between RST Stressor and HCC Controls or between FWD and HCC Control groups (Fig. 4A). Further α diversity analysis revealed that there were no differences between any groups in richness estimated using Chao1 (Fig. 4B), but the RST Stressor group was associated with a reduction in equitability in comparison with FWD Control (P< 0.05) but not HCC Control (Fig. 4C). HCC Control and FWD Control groups did not differ in either Chao1 or equitability.
Figure 4.
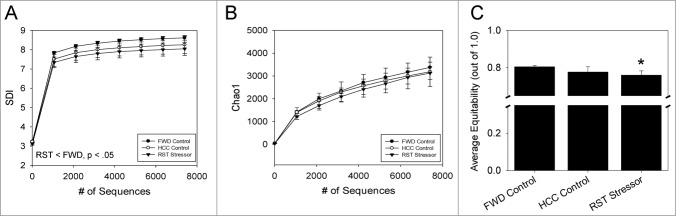
Stressor exposure significantly reduces α diversity in the mucosa-associated microbiota. (A) The Shannon Diversity index (SDI) was measured for each sample and averaged by group. RST Stressor was significantly lower than FWD Control mice (P < 0.05), but was not significantly different than HCC Control SDI values. (B) Richness (by Chao1 measurement) and (C) evenness (by equitability) were then calculated using QIIME and averaged by group. RST Stressor did not affect overall richness, but evenness was reduced in RST Stressor-exposed mice when compared to FWD Control mice (P< 0.05). Data are mean ± SD for n = 5 for all groups. For α diversity, groups were compared using parametric T-tests on QIIME with a modified Bonferroni correction for multiple comparisons.
At the phyla level in the mucosa-associated populations, the Firmicutes and Bacteroidetes were unaffected by exposure to the restraint stressor (Fig. 5A). Actinobacteria was significantly reduced in the RST Stressor group compared to both FWD Control and HCC Control mice, while Deferribacteres was significantly increased in RST Stressor mice over HCC Control only (Fig. 5B) (P < 0.05). There were no significant differences between FWD Control and HCC Control in phyla abundances in the mucosa-associated microbiota. PCoA of the unweighted UniFrac distance matrix for the mucosa-associated samples illustrated the clustering of the RST Stressor group from the FWD Control and HCC Control groups (Fig. 5C). The clustering was significant, using ANOSIM (HCC Control vs. RST Stressor, P < 0.01) (FWD Control vs. RST Stressor, P < 0.01). HCC Control and FWD Control also clustered separately (HCC Control vs. FWD Control, P < 0.05). Overall, these data highlight that restraint stress as well as food and water deprivation cause changes in the microbiota community membership and composition at the mucosal tissue level.
Figure 5.
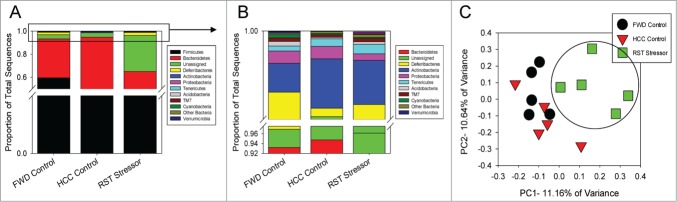
Stressor exposure significantly alters the community structure of the mucosa-associated microbiota. (A) Firmicutes and Bacteroidetes were unchanged between any of the groups in the experiment. (B) Upon examining the lesser phyla, Actinobacteria was significantly reduced in the RST Stressor group compared to FWD Control and HCC Control mice. Deferribacteres was significantly increased in RST Stressor mice compared to HCC Control mice, but not FWD Control mice. Phyla abundance data were compared first using non-parametric Kruskal-Wallis test, and Mann-Whitney U tests were used post-hoc. (C) Principle coordinate analysis was used to visualize clustering of similar community profiles. Restraint stressor-exposed mice significantly clustered using the ANOSIM statistic (HCC Control vs. RST Stressor, P < 0.01) (FWD Control vs. RST Stressor, P < 0.01). FWD Control and HCC Control also clustered significantly apart from each other (P < 0.05). Data are for N = 5 for all groups.
Restraint Stress alters the relative abundance of colonic microbiota bacterial groups
Larger taxonomic comparisons were performed to distinguish stressor- induced shifts in the microbial groups associated with the mucosa and lumen. In the lumen, the family S24-7 of the order Bacteroidales was significantly reduced in RST Stressor mice (P < 0.05) compared to HCC Control mice, but not FWD Control mice (Table 1). At genus level, Adlercreutzia was decreased in RST Stressor mice (P < 0.05) in comparison to HCC Control and FWD Control mice (Table 2). Additionally, an unclassified genus in the family S24-7 was significantly decreased in RST Stressor mice compared to HCC Control mice only (P < 0.05)
Table 1.
Top 10 Most Abundant Colonic Luminal-Associated Bacterial Families
| HCC | FWD | RST | ||
|---|---|---|---|---|
| Lactobacillaceae | 30.06 ± 8.32 | 37.56 ± 8.19 | 20.50 ± 5.27 | |
| S24-7 | 22.54 ± 2.82 | 18.81 ± 2.30 | 13.16 ± 2.25** | RST vs. HCC |
| Clostridiales;f__ | 12.41 ± 1.50 | 14.25 ± 2.83 | 20.75 ± 6.60 | |
| Lachnospiraceae | 12.80 ± 4.79 | 9.21 ± 1.70 | 17.41 ± 3.83** | RST vs. FWD |
| Rikenellaceae | 4.52 ± 0.43 | 4.71 ± 0.80 | 7.18 ± 0.70** | HCC vs. FWD |
| Bacteroidaceae | 3.56 ± 0.92 | 2.20 ± 0.29 | 7.74 ± 2.13 | |
| Ruminococcaceae | 2.42 ± 0.49 | 3.19 ± 0.53 | 5.04 ± 1.32 | |
| Unassigned | 3.32 ± 0.38 | 3.74 ± 0.27 | 2.57 ± 0.20 | |
| Clostridiaceae | 4.36 ± 0.94 | 1.23 ± 0.43 | 1.64 ± 0.51 | |
| Prevotellaceae | 0.38 ± 0.19 | 0.90 ± 0.30 | 2.04 ± 1.28 |
Data are the mean relative abundance ± standard error.
* P < .05, * p = .05
Table 2.
Top 15 Most Abundant Luminal-Associated Bacterial Genera
| HCC | FWD | RST | ||
|---|---|---|---|---|
| Lactobacillus spp. | 30.06 ± 9.61 | 37.56 ± 8.19 | 20.50 ± 5.27 | |
| S24-7, g__ | 22.54 ± 3.26 | 18.81 ± 2.30 | 13.16 ± 2.25** | RST vs. HCC |
| Clostridiales; f__; g__ | 12.42 ± 1.73 | 14.25 ± 2.83 | 20.75 ± 6.60 | |
| Lachnospiraceae; g__ | 9.90 ± 4.55 | 7.11 ± 1.34 | 15.57 ± 3.61 | |
| Rikenellaceae; g__ | 4.52 ± 0.50 | 4.71 ± 0.80 | 7.17 ± 0.70 | |
| Bacteroides spp | 3.56 ± 1.06 | 2.20 ± 0.29 | 7.74 ± 2.13 | |
| Unassigned | 3.32 ± 0.43 | 3.74 ± 0.27 | 2.57 ± 0.21** | RST vs. FWD |
| Candidatus Athromitus | 4.19 ± 1.08 | 1.02 ± 0.40 | 1.50 ± 0.51** | HCC vs. FWD |
| Oscillospira spp. | 1.06 ± 0.22 | 0.88 ± 0.15 | 2.10 ± 0.61 | |
| Prevotella spp | 0.38 ± 0.22 | 0.90 ± 0.30 | 2.04 ± 1.28 | |
| Adlercreutzia spp. | 2.11 ± 0.38 | 1.61 ± 0.30 | 0.20 ± 0.02** | RST vs. HCC/FWD |
| Ruminococcaceae; g__ | 0.89 ± 0.20 | 1.23 ± 0.28 | 1.90 ± 0.56 | |
| Lachnospiraceae; Ruminococcus | 1.62 ± 0.81 | 1.08 ± 0.30 | 0.63 ± 0.25 | |
| Lachnospiraceae; Other | 0.75 ± 0.29 | 0.48 ± 0.06 | 0.64 ± 0.25 | |
| Ruminococcaceae; Ruminococcus | 0.21 ± 0.09 | 0.91 ± 0.21 | 0.55 ± 0.11** | HCC vs. FWD |
Data are the mean relative abundance ± standard error.
* P < .05, * p = .05
In the tissue, the family Ruminococcaceae were also increased in RST Stressor over HCC Control (P<0.05), and the family S24-7 of the order Bacteroidales was reduced in RST Stressor compared to both HCC Control and FWD Control (p = 0.05). The family Lactobacillaceae was also significantly reduced in RST Stressor-exposed mice (P < 0.05) (Table 3) compared to both FWD Control (P < 0.05) and HCC Control groups (p = 0.05). When genera were examined in the Lactobacillacea family, Lactobacillus spp was the only genus significantly reduced in stressor exposed mice compared to both HCC Control (p = 0.05) and FWD Control (P < 0.05) (Table 4 ); HCC Control and FWD Control were not significantly different from one another. The effects of the stressor on the lactobacilli were limited to mucosa-associated populations, and RST Stressor exposure did not significantly reduce the relative abundance of luminal-associated Lactobacillus. The relative abundance of the genus Oscillospira was also significantly increased in the mucosa of RST Stressor-exposed mice and FWD Control mice compared to HCC control (P < 0.05). There were not significant differences in Oscillospira relative abundance in the mucosa of RST Stressor and FWD Control mice (Table 4). Food and water deprivation also had effects on bacterial abundances that were independent of stressor exposure. In the lumen, Candidatus Arthromitus, a group of segmented filamentous bacteria, and the genus Ruminococcus of the Ruminococcaceae family were decreased in FWD Control compared to HCC Control (P < 0.05) (Table 2). The relative abundances of bacterial families and genera were not significantly different between the FWD Control and HCC Control mice when assessed on the mucosal tissue.
Table 3.
Top 10 Most Abundant Colonic Mucosal-Associated Bacterial Families
| HCC | FWD | RST | ||
|---|---|---|---|---|
| Clostridales; f__ | 25.67 ± 6.27 | 31.03 ± 3.58 | 34.92 ± 5.49 | |
| S24-7 | 30.74 ± 7.85 | 20.94 ± 4.22 | 9.85 ± 2.02* | RST vs. FWD/HCC |
| Lachnospiraceae | 8.68 ± 1.62 | 13.15 ± 2.82 | 16.49 ± 4.35 | |
| Rikenellaceae | 8.54 ± 1.60 | 8.74 ± 2.20 | 11.23 ± 1.75 | |
| Ruminococcaceae | 5.23 ± 1.26 | 7.88 ± 0.84 | 10.95 ± 1.39** | RST vs. HCC |
| Bacteroidaceae | 5.51 ± 1.59 | 3.24 ± 0.63 | 7.98 ± 2.01 | |
| Lactobacillaceae | 6.75 ± 3.20 | 5.46 ± 1.39 | 1.25 ± 0.43* | RST vs. FWD/HCC |
| Unassigned | 3.64 ± 0.63 | 3.44 ± 0.15 | 0.23 ± 0.12** | RST vs. FWD |
| Prevotellaceae | 0.98 ± 0.56 | 0.72 ± 0.23 | 1.77 ± 1.26 | |
| Deferribacteraceae | 0.14 ± 0.05 | 2.08 ± 1.71 | 0.74 ± 0.12** | RST vs. HCC |
Data are the mean relative abundance ± standard error.
** P < .05, * p = .05
Table 4.
Top 15 Most Abundant Mucosal-Associated Bacterial Genera
| HCC | FWD | RST | ||
|---|---|---|---|---|
| Clostridiales; f__; g__ | 25.67 ± 6.27 | 31.03 ± 3.58 | 34.92 ± 5.49 | |
| S24-7; g__ | 30.74 ± 7.85 | 20.94 ± 4.22 | 9.85 ± 2.02* | RST vs. FWD/HCC |
| Lachnospiraceae; g__ | 6.64 ± 1.15 | 10.60 ± 2.60 | 14.36 ± 4.02 | |
| Rikenellaceae; g__ | 8.54 ± 1.60 | 8.73 ± 2.20 | 11.22 ± 1.75 | |
| Bacteroides spp | 5.51 ± 1.59 | 3.24 ± 0.63 | 7.98 ± 2.01 | |
| Lactobacillus spp. | 6.75 ± 3.20 | 5.46 ± 1.39 | 1.25 ± 0.43* | RST vs. FWD/HCC |
| Oscillospira spp | 1.75 ± 0.32 | 3.44 ± 0.36 | 4.69 ± 0.69** | HCC vs. FWD/RST |
| Unassigned | 3.64 ± 0.63 | 3.44 ± 0.15 | 2.33 ± 0.12** | RST vs. FWD |
| Ruminococcaceae; g__ | 1.98 ± 0.54 | 2.78 ± 0.18 | 3.81 ± 0.79 | |
| Prevotella spp. | 0.98 ± 0.56 | 0.72 ± 0.23 | 1.77 ± 1.26 | |
| Mucispirillum spp | 0.14 ± 0.05 | 2.08 ± 1.71 | 0.74 ± 0.12** | RST vs. HCC |
| Ruminococcaceae; Other | 0.51 ± 0.11 | 0.73 ± 0.25 | 1.58 ± 0.52 | |
| Ruminococcaceae; Ruminococcus | 0.99 ± 0.45 | 0.91 ± 0.18 | 0.85 ± 0.17 | |
| Lachnospiraceae; Ruminococcus | 0.87 ± 0.32 | 0.93 ± 0.16 | 0.82 ± 0.19 | |
| Clostridiales; Other; Other | 0.69 ± 0.04 | 0.89 ± 0.16 | 0.56 ± 0.09 |
Data are the mean relative abundance ± standard error.
** P < .05, * p = .05
Discussion
Exposure to psychological stress has been associated with exacerbation of IBD and heightened immune responses to enteric pathogens.22,39,40 Some studies suggest that changes to the microbiota could be involved in stressor-induced GI immune dysfunction, but stressor-induced changes in gut microbiota have not been well-characterized.41,42 In this study, mice were exposed to a widely used and well-validated murine model of chronic stress to elucidate the effects of a long-term stressor upon the colonic microbiota. The data show that microbial communities associating with colonic tissue and found in the lumen of the colon have unique community structures that are differentially impacted by psychological stressor exposure. In particular, the restraint stressor significantly reduced the α diversity in the mucosa-associated compartment of stressed mice compared to controls, but did not affect the α diversity in the luminal populations. Furthermore, stressor-induced alterations in β-diversity were demonstrated when compared with both food and water-deprived controls and undisturbed HCC Control mice in the mucosa, but stressor-associated changes were only identified between RST Stressor and FWD Control groups in the lumen. The mucosal community-wide effects were partially due to a significant reduction in the relative abundance of bacteria in the immunomodulatory genus Lactobacillus. Stressor exposure also affected luminal microbiota groups, such as the genus Adlercreutzia that was significantly reduced, but the relative abundance of Lactobacillus was not significantly reduced in the colonic lumen of stressor-exposed mice.
The lack of a stressor effect on the relative abundance of luminal lactobacilli was surprising, because previous studies have shown that stressor exposure can reduce fecal lactobacilli. These studies, however, have primarily involved higher mammals. For example, rhesus monkeys that experienced a chronic prenatal stress or maternal separation stress had reductions in Lactobacillus, with similar effects of stress upon lactobacilli being reported in college students during exam periods.18,20,21 However, our current study is in agreement with several other studies involving laboratory rodents that have failed to find a significant effect of stressor exposure on fecal or luminal lactobacilli levels.19,42,43 However, the finding that colonic mucosa-associated lactobacilli are reduced in mice exposed to the chronic restraint stressor is consistent with a previous study in as little as one 2-hour exposure to a social stressor significantly reduced the relative abundance of colonic mucosa-associated Lactobacillus in C57BL/6 and CD-1 mice.44 Thus, it is apparent that stressor exposure reduces mucosa-associated, but not luminal, lactobacilli in laboratory mice. Additional studies are needed to confirm and extend these findings in higher mammals.
It is important to note that true stratification of luminal and mucosal populations does not exist, because there is substantial crossover between microbes associated with the lumen and those that can adhere to the mucus layer of the gastrointestinal tract. In this study, we analyzed the majority of the luminal population separately from the mucosa-associated populations. The finding that stressor exposure has strong effects on mucosa-associated populations is important, because gut microbes that adhere to the colonic mucosa can have different effects on the host than luminal populations.45 Mucosa-associated lactobacilli may be particularly important, because this bacterial group is well known for its ability to impact mucosal immune responses. Probiotic Lactobacillus-mediated interventions have been shown to downregulate TNF-α in the colon, enhance gut barrier activity, and reduce overall reductions in colitis-related pathology.46-49 Additionally, treatment with probiotic Lactobacillus reuteri in mice abrogates the stressor-induced increases in the severity of the inflammatory response to a colonic pathogen.40 Interestingly, though previous studies have found that Lactobacillus is reduced in the stool,18,20,21,50 this study showed a reduction in mucosal epithelium-associated Lactobacillus but not in luminal lactobacilli, implying that psychological stressor exposure can have a distinct effects upon groups of mucosal microbiota compared to their luminal counterparts. Since Lactobacillus and other commensals can mediate host gut health, this specific reduction might be associated with stressor-induced increases in colitic inflammation.51-53
Despite the constant crossover between luminal and mucosa-associated populations, unique properties and diversity levels can be associated with each niche.24,25,45,54 Descriptive analyses often concentrate on fecal or luminal contents as a read-out for the entire community structure of the gastrointestinal tract, but the current data emphasize the importance of a targeted approach. In this study, stressor-induced changes to bacterial groups and the extent to which stressor-exposure affected diversity were different based upon the colonic compartment that was analyzed. Bacteria in the colonic lumen and associated with the colonic mucosa have evolved to their present relationship symbiotically and as a result have different biological function. For example, it is thought that the luminal-associated microbiota are more heavily involved in metabolism and digestion, while the microbes of the mucosal epithelium assist the host in immunomodulation.45 Thus, characterizing how factors that cause dysbiosis, like psychological stress, affect microbes in these niches can have different implications.
The current study indicates that chronic stress affects the composition of the colonic mucosa-associated microbiota and thus may influence colonic immunity. Many colonic pathogens, like enteropathogenic or enterohemorrhagic Escherichia coli (EPEC or EHEC, respectively), which are often modeled by murine challenge with Citrobacter rodentium, colonize the intestinal epithelium and can cause severe colitis along the epithelial barrier.55,56 The colonic microbiota are known to influence susceptibility and resistance to these attaching and effacing pathogens,57,58 since disrupting the mucosa-associated populations with antibiotics increased disease severity upon pathogen challenge.59,60 Stressor induced shifts to luminal populations might also have downstream effects upon host metabolism. The changes to these niches could be inter-related, wherein possible mechanisms such as stressor-induced reductions or exhaustion of mucous secretion, as well as increased motility could affect the composition of both compartments. Increased shedding of mucosal populations into the lumen would also alter overall community structure. As these populations shift during stressor exposure, it is likely that overall microbiome function is also being impacted in regards to how the microbiota interact with the host and with each other. Proper analysis of these compartment-specific changes could be performed in future studies using whole genome sequencing and metagenomic approaches, as well as metabolomic analyses in order to investigate if the unique stressor-induced changes to lumenal and mucosal population structure extends to distinct changes in functional output.
We have previously shown that exposure to prolonged restraint stressor increases the severity of C. rodentium infection upon oral challenge.22,40 It is not known whether stressor-induced changes to the microbiota were solely responsible for the increased disease severity, but orally treating the mice with probiotic L. reuteri reduced the severity of C. rodentium infection in stressor-exposed mice,40 suggesting that preventing stressor-induced reductions in lactobacilli might attenuate colonic inflammation. However, the effects may not be specific to lactobacilli. This study, along with our previous study, demonstrated that prolonged restraint decreases α diversity in the colonic mucosa, as well as in the cecal lumen. Reduced diversity has previously been associated with increased cecal colonization by Campylobacter jejuni,61 marked by marginally increased pro-inflammatory marker (e.g., IL-1b, IL-6, TNF-α, IFN-g, IL-10) levels, while mouse strains susceptible to C. rodentium infection have lower fecal bacterial counts (particularly in Bacteroidetes) than those that are resistant.61 Thus, it is possible that the shifts in certain bacterial populations evident in the microbiota of stressor-exposed mice in this study contribute to enhanced colonic inflammation. Indeed, a preliminary, unpublished work by Galley et al. indicate that RST-induced changes to the microbiota are directly associated with an colonic inflammatory response to a pathogen challenge, likely due to shifts in microbiome function. Further studies are already underway to identify how compositional changes in the microbiota are related to functional shifts that affect host health.
It is currently not known whether stressor-induced changes to the microbiota contribute to exacerbation of human colonic inflammation, but it has been well-documented that patients with IBD and IBS often have an altered microbiome compared to healthy subjects. Such differences include a reduction in overall diversity, and shifts in major phyla like the Firmicutes and Bacteroidetes, as well as commensals like Bifidobacterium, Faecalibacterium prausnitzii and Roseburia.63-67 It is not yet clear whether the changed microbiome is the cause or the effect of the increased inflammatory flare-ups and sensitivity to pain, but studies are beginning to demonstrate a role of the microbiota in the exacerbation of these diseases.68,69 Of particular importance, exposure to psychological stress often precedes the exacerbation of IBD.4,5 In our experience, the effects of stressor exposure on microbial community structure are more consistently manifest on the mucosal surface versus the luminal or fecal compartments. While it is a goal to understand how changes to microbial community structure in the feces predicts changes to community structure at the mucosal surface, until these relationships are well understood, mucosa-associated populations should be assessed in studies of the brain-gut microbiota axis during periods of health and disease.
In the current study, mice were exposed to repeated daily exposures to the prolonged restraint. Interestingly, this results in partial habituation; corticosterone levels after 1 day of the stressor are higher than corticosterone levels after 6 days of the stressor (even though corticosterone levels are higher than baseline levels on all days of stressor exposure).70 Despite this habituation, past studies show that the effects of stressor exposure upon the microbiota are additive, with significant effects of the stressor on microbiota composition, including absolute levels of Lactobacillus reuteri, only being evident after repeated cycles of the stressor.44 It is not yet known whether alterations in the microbiota contribute to this stressor-induced corticosterone profile, but it is known that microbiota do significantly impact hypothalamic-pituitary-adrenal activity and corticosterone levels.71,72 Thus, it is possible that differences in corticosterone levels and in microbial communities over repeated cycles of stressor exposure are inter-related. However, repeated cycles of stress are not always needed in order for changes in the microbiome to manifest. For example, a single 2-hr exposure to a social stressor was sufficient to significantly shift the mucosa-associated microbiome.44 Further research is needed to determine why a single exposure to some stressors is sufficient to change the microbiome, whereas others require repeated exposures.
Prolonged restraint is a widely used murine stressor that has been extensively characterized in the literature and is one of the most commonly used murine stressors in biomedical and biobehavioral research29, in part because it reliably induces a physiological stress response that results in the elevation of endogenous glucocorticoids.31-33 Because the strong glucocorticoid response can suppress multiple components of the immune response32,73, it seemed plausible that stressor-induced suppression of the mucosal immune response led to the observed changes in microbiota community structure. However, RST did not induce changes in TNF-α, iNOS, or IL-6 message levels in the colonic tissue (data not shown). Moreover, in the absence of pathogen challenge, stressor exposure had no effect on other measures of mucosal immunity (e.g., secretory IgA, β-defensin-1, and leukocyte infiltration into the colon) in previously published work.22 Thus, it is unlikely that RST-induced suppression of immune activity strongly affects the composition of the gut microbiota. However, immunomodulation is not the only way in which stressor exposure may impact the composition of the gut microbiota. In an olfactory bulbectomy model of stressor sensitization, Park et al. (2013) demonstrated that stressor-induced increases in colonic motility impact microbiota structure through a corticotrophin-releasing hormone (CRH)-dependent mechanism.85 Because CRH-induced alterations in colonic motility can be disrupted by cutting the vagal nerve86, and because the vagus nerve is considered a primary route through which the brain and gut microbiota interact87-89, it is possible that the observed stressor-induced changes in the colonic microbiota are vagally-mediated. This hypothesis warrants further attention.
Results from studies involving behavioral stressors in laboratory animals must be cautiously extrapolated to humans due to differences in host perceptions to stressful stimuli, the physiological stress response, and available coping mechanisms. However, microbiota community structure alterations that consistently occur in response to stressors of different type (e.g., physical vs. social), different duration (e.g., a few hours vs. a few days or weeks), and in different host species (e.g.,, rodents vs. non-human primates) can be predicted to occur in human populations. Although the prolonged restraint stressor used in the current study is considered a severe, chronic stressor, results obtained with prolonged restraint are consistent with an increasing number of studies indicating that stressor exposure changes gut microbiota community composition.19,22 This was previously demonstrated with exposure to as little as 2 hrs of a social stressor,44 and now has been demonstrated with a prolonged exposure to a physical stressor. In both cases, exposure to the stressor reduced lactobacilli. Stressor-induced reductions in lactobacilli have also been observed in non-human primates,20,21 suggesting that reductions in lactobacilli are a conserved response to stressor exposure that is likely to occur in human populations. In support of this contention, stressor-induced reductions in lactobacilli have been observed in college students during examination periods.18
Diet can greatly affect the GI microbiota.74,75 The data here demonstrate that food and water deprivation is able to affect the community structure of the mucosal associated microbiota in comparison with undisturbed controls, while also affecting the abundances of select bacterial groups in the lumen. This confirms a previous study that showed separate clustering of control mice from FWD mice after a single 16-hr period of food and water deprivation.22 Thus, it is difficult to fully control for the effects of food and water deprivation in stress studies. However, samples from RST Stressor mice consistently cluster separately from samples from FWD Control (and HCC Control) mice indicating that the psychological and physical aspects of the prolonged stressor have unique effects on the microbiome, including a reduction in Lactobacillus and overall α diversity. These changes cannot simply be accounted for by the fact that mice in restraining tubes do not consume food or water, since the effects are not evident in animals deprived of food and water. We have not found that the overall quantity of food consumption is affected by RST, as the stressor-exposed mice consume an equivalent amount of food as HCC mice, despite having reduced access (unpublished observations). However, since mice are restrained during the dark (i.e., active) cycle, which is when mice typically consume their food, exposure to the RST stressor undoubtedly skews feeding patterns while leaving overall food quantity unchanged. This likely affects overall metabolism, since mice exposed to the RST stressor lose weight with repeated exposures to the stressor. Thus, it is clear that further studies are needed to understand the individual contributions of the stress response and food and water deprivation on stressor-induced alterations of the GI microbiota.
Dysbiotic microbial profiles are associated with inflammation and increases in disease severity. Stressful periods, which can alter microbial community structures, can worsen the pathology of colonic pathogens (e.g., EHEC) and inflammatory disorders (e.g.,, IBD).4 Thus, investigations into the disruptions in the community structure of beneficial GI microbes as a result of external effectors like psychological stress are especially important. These results further develop the relationship between psychological stress and the microbiota by demonstrating that restraint stressor-induced dysbiosis in the colonic microbiota changes the community composition of both the luminal and mucosa-associated populations, but that the stressor-induced disruptions upon each niche are distinct.
Materials and Methods
Mice
All experiments were performed using male CD-1 mice, 6-8 weeks of age, from Charles River Laboratories. Upon arrival, mice habituated to surroundings for 1 week. Vivarium was AALAC-approved. Food and water provided ad libitum, unless experimentation was being performed (food and water deprivation or restraint). Mice were kept on a 12:12 hr light:dark schedule (lights on at 0600). All procedures were approved by The Ohio State University's Animal Care and Use Committee.
Stressor Paradigm
Mice were placed in restraint stress (RST Stressor), non-stressed home cage control (HCC Control) or non-stressed food and water deprivation control (FWD Control) groups. For restraint stress, mice were placed inside ventilated 50-ml conical tubes beginning at 1800 (start of mouse active cycle) and removed at 0900 the following morning. This was repeated for a total of 7 consecutive overnight restraint sessions. During the stressor period, the HCC Control had full access to food and water and were not handled, and the FWD Control had access to food and water removed to control for the RST Stressor diet conditions. During non-stress periods, all mice were allowed full access to food and water and were not handled.
Tissue Removal
Immediately following the seventh and final cycle of RST Stressor, all mice were humanely euthanized by CO2 asphyxiation. Colons were sterilely removed with forceps, and all fecal contents were so that mucous layer was not disturbed. Both the colonic contents and tissue were snap-frozen and stored at −80C.
DNA extraction
Samples were centrifuged at 14000 rpm for 30 seconds, then resuspended in 500 μL of RLT buffer with b-mercaptoethanol (Qiagen, Valenica, CA). Samples were further lysed in a QIAgen TissueLyser (30 Hz for 5min) with the use of a single sterile steel bead (5-mm) and 500 μL of glass beads (0.1-mm) (Scientific Industries, Inc.., NY, USA). After the samples were quickly centrifuged, a 100 μL aliquot of sample was combined with 100 μL of 100% ethanol. This combination was added to a Qiagen DNA spin column, and the QIAamp DNA Mini Kit Tissue Protocol was followed, beginning with Step 7. Elution was performed using 30 μL of water. Samples were quantified with a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France). Samples were then diluted to 20 ng/μL.
Pyrosequencing
bTEFAP was performed as described previously at the Research and Testing Laboratory (Shallowater, TX).22
Sequence analysis
The Quantitative Insights Into Microbial Ecology (QIIME) software package was used to analyze the sequences obtained from bTEFAP pyrosequencing.76 In summary, sequences were grouped by unique barcode labeling. The mean of all high-quality sequences among all samples was 9119. Lower bound on sequence length was 200 bases and upper-bound was 600 bases. Barcode lengths were 8. Maximum homopolymer run and ambiguous bases allowed were set at 6. No mismatches were allowed in primer sequences. 89.47% of obtained sequences were selected for analysis based upon these thresholds. The analysis was performed on tissue and lumen samples combined as well as both groups separated.
Operational Taxonomic Units (OTUs) were made using the pick_otus.py command pipeline. UCLUST was used in OTU-picking at a threshold level of 0.97 in order to cluster like-sequences.77 A representative sequence was obtained from each OTU and aligned to the GreenGenes reference database of sequences using PyNAST.78,79 Taxonomy was then assigned to each representative sequence using the Greengenes taxonomy database at a minimum confidence threshold of 0.8.80 A phylogenetic tree was constructed using these aligned sequences for downstream analysis using UniFrac.81
Statistical analyses
Non-parametric Kruskal Wallis Tests were used for comparison of HCC Control, FWD Control, and RST Stressor group abundances. Non-parametric Mann-Whitney U Tests with modified Bonferroni corrections were used as post-hoc tests and were performed using SPSS for Windows (v.21, Chicago, IL). The α diversity measurement, Shannon Diversity Index (SDI) was computed by QIIME. Shannon, equitability and Chao1 significance was calculated using a parametric T-test at a depth of 7402 sequences for mucosal samples and 5589 sequences for luminal samples with a modified Bonferroni correction, and at a depth of 6177 sequences for combined α-diversity comparisons of mucosal and luminal microbiota.82 Unweighted UniFrac principal component analyses and dendograms were created using a rarefaction depth of 8500 for tissue samples alone and 6303 for luminal samples alone and both groups combined. The β-diversity analysis, analysis of similarity (ANOSIM), was used to detect significant differences in the distance matrices between groups. This was performed using the vegan package of R, implemented in QIIME.83,84
Sequence availability
Sequences were deposited on the Metagenomic Analysis Server MG-RAST and are identified as MG-RAST 4573854.3 through 4573884.3.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was funded by NIH grant RO1AT006552-01A1 and NIH/NIDCR grant T32 DE014320.
Reference
- 1. Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 2012; 9:286-94; PMID:22392290; http://dx.doi.org/ 10.1038/nrgastro.2012.32 [DOI] [PubMed] [Google Scholar]
- 2. Holzer P. Efferent-like roles of afferent neurons in the gut: Blood flow regulation and tissue protection. Autonom Neurosci: Basic Clin 2006; 125:70-5; http://dx.doi.org/ 10.1016/j.autneu.2006.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Jonge WJ. The Gut's Little Brain in Control of Intestinal Immunity. ISRN Gastroenterol 2013; 2013:630159; PMID:23691339; http://dx.doi.org/ 10.1155/2013/630159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernstein CN, Singh S, Graff LA, Walker JR, Miller N, Cheang M. A prospective population-based study of triggers of symptomatic flares in IBD. Am J Gastroenterol 2010; 105:1994-2002; PMID:20372115; http://dx.doi.org/ 10.1038/ajg.2010.140 [DOI] [PubMed] [Google Scholar]
- 5. Whitehead WE, Crowell MD, Robinson JC, Heller BR, Schuster MM. Effects of stressful life events on bowel symptoms: subjects with irritable bowel syndrome compared with subjects without bowel dysfunction. Gut 1992; 33:825-30; PMID:1624167; http://dx.doi.org/ 10.1136/gut.33.6.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee KJ, Tack J. Altered intestinal microbiota in irritable bowel syndrome. Neurogastroenterol Motil: Off J Euro Gastrointest Mot Soc 2010; 22:493-8; http://dx.doi.org/ 10.1111/j.1365-2982.2010.01496.x [DOI] [PubMed] [Google Scholar]
- 7. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010; 90:859-904; PMID:20664075; http://dx.doi.org/ 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 8. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449:804-10; PMID:17943116; http://dx.doi.org/ 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huffnagle GB. The microbiota and allergies/asthma. PLoS Pathogens 2010; 6:e1000549; PMID:20523892; http://dx.doi.org/ 10.1371/journal.ppat.1000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 2008; 105(Suppl 1):11512-9; PMID:18695234; http://dx.doi.org/ 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 2009; 77:2367-75; PMID:19307217; http://dx.doi.org/ 10.1128/IAI.01520-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280; PMID:19018661; http://dx.doi.org/ 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hakansson A, Tormo-Badia N, Baridi A, Xu J, Molin G, Hagslatt ML, Karlsson C, Jeppsson B, Cilio CM, Ahrne S. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med 2014; PMID:24414342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwab C, Berry D, Rauch I, Rennisch I, Ramesmayer J, Hainzl E, Heider S, Decker T, Kenner L, Muller M, et al. . Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J 2014; PMID:24401855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun 2011; 79:1536-45; PMID:21321077; http://dx.doi.org/ 10.1128/IAI.01104-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shilov VM, Lizko NN, Borisova OK, Prokhorov VY. Changes in the microflora of man during long-term confinement. Life Sci Space Res 1971; 9:43-9; PMID:11942343 [PubMed] [Google Scholar]
- 17. Lizko NN, Silov VM, Syrych GD. [Events in he development of dysbacteriosis of the intestines in man under extreme conditions]. Die Nahrung 1984; 28:599-605; PMID:6387499; http://dx.doi.org/ 10.1002/food.19840280604 [DOI] [PubMed] [Google Scholar]
- 18. Knowles SR, Nelson EA, Palombo EA. Investigating the role of perceived stress on bacterial flora activity and salivary cortisol secretion: a possible mechanism underlying susceptibility to illness. Biol Psychol 2008; 77:132-7; PMID:18023961; http://dx.doi.org/ 10.1016/j.biopsycho.2007.09.010 [DOI] [PubMed] [Google Scholar]
- 19. Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun 2011; 25:397-407; PMID:21040780; http://dx.doi.org/ 10.1016/j.bbi.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol 1999; 35:146-55; PMID:10461128; http://dx.doi.org/ 10.1002/(SICI)1098-2302(199909)35:2%3c146::AID-DEV7%3e3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 21. Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutri 2004; 38:414-21; http://dx.doi.org/ 10.1097/00005176-200404000-00009 [DOI] [PubMed] [Google Scholar]
- 22. Bailey MT, Dowd SE, Parry NM, Galley JD, Schauer DB, Lyte M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun 2010; 78:1509-19; PMID:20145094; http://dx.doi.org/ 10.1128/IAI.00862-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nocker A, Burr M, Camper AK. Genotypic microbial community profiling: a critical technical review. Microb Ecol 2007; 54:276-89; PMID:17345133; http://dx.doi.org/ 10.1007/s00248-006-9199-5 [DOI] [PubMed] [Google Scholar]
- 24. Hong PY, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PloS one 2011; 6:e25042; PMID:21966408; http://dx.doi.org/ 10.1371/journal.pone.0025042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68:3401-7; PMID:12089021; http://dx.doi.org/ 10.1128/AEM.68.7.3401-3407.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shigeshiro M, Tanabe S, Suzuki T. Repeated exposure to water immersion stress reduces the Muc2 gene level in the rat colon via two distinct mechanisms. Brain Behav Immun 2012; 26:1061-5; PMID:22683765; http://dx.doi.org/ 10.1016/j.bbi.2012.05.016 [DOI] [PubMed] [Google Scholar]
- 27. Castagliuolo I, Lamont JT, Qiu B, Fleming SM, Bhaskar KR, Nikulasson ST, Kornetsky C, Pothoulakis C. Acute stress causes mucin release from rat colon: role of corticotropin releasing factor and mast cells. Am J Physiol 1996; 271:G884-92; PMID:8944704 [DOI] [PubMed] [Google Scholar]
- 28. O'Malley D, Julio-Pieper M, Gibney SM, Dinan TG, Cryan JF. Distinct alterations in colonic morphology and physiology in two rat models of enhanced stress-induced anxiety and depression-like behaviour. Stress 2010; 13:114-22; PMID:20214436; http://dx.doi.org/ 10.3109/10253890903067418 [DOI] [PubMed] [Google Scholar]
- 29. Buynitsky T, Mostofsky DI. Restraint stress in biobehavioral research: Recent developments. Neurosci Biobehav Rev 2009; 33:1089-98; PMID:19463853; http://dx.doi.org/ 10.1016/j.neubiorev.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 30. Cavigelli SA, McClintock MK. Fear of novelty in infant rats predicts adult corticosterone dynamics and an early death. Proc Natl Acad Sci U S A 2003; 100:16131-6; PMID:14673078; http://dx.doi.org/ 10.1073/pnas.2535721100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobbs CM, Vasquez M, Glaser R, Sheridan JF. Mechanisms of stress-induced modulation of viral pathogenesis and immunity. J Neuroimmunol 1993; 48:151-60; PMID:8227313; http://dx.doi.org/ 10.1016/0165-5728(93)90187-4 [DOI] [PubMed] [Google Scholar]
- 32. Dobbs CM, Feng N, Beck FM, Sheridan JF. Neuroendocrine regulation of cytokine production during experimental influenza viral infection: effects of restraint stress-induced elevation in endogenous corticosterone. J Immunol 1996; 157:1870-7; PMID:8757304 [PubMed] [Google Scholar]
- 33. Padgett DA, Marucha PT, Sheridan JF. Restraint stress slows cutaneous wound healing in mice. Brain Behav Immun 1998; 12:64-73; PMID:9570862; http://dx.doi.org/ 10.1006/brbi.1997.0512 [DOI] [PubMed] [Google Scholar]
- 34. Bonneau RH, Sheridan JF, Feng N, Glaser R. Stress-induced modulation of the primary cellular immune response to herpes simplex virus infection is mediated by both adrenal-dependent and independent mechanisms. J Neuroimmunol 1993; 42:167-76; PMID:8429102; http://dx.doi.org/ 10.1016/0165-5728(93)90007-L [DOI] [PubMed] [Google Scholar]
- 35. Dong-Newsom P, Powell ND, Bailey MT, Padgett DA, Sheridan JF. Repeated social stress enhances the innate immune response to a primary HSV-1 infection in the cornea and trigeminal ganglia of Balb/c mice. Brain Behav Immun 2010; 24:273-80; PMID:19822203; http://dx.doi.org/ 10.1016/j.bbi.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stark JL, Avitsur R, Hunzeker J, Padgett DA, Sheridan JF. Interleukin-6 and the development of social disruption-induced glucocorticoid resistance. J Neuroimmunol 2002; 124:9-15; PMID:11958817; http://dx.doi.org/ 10.1016/S0165-5728(02)00004-8 [DOI] [PubMed] [Google Scholar]
- 37. Avitsur R, Stark JL, Dhabhar FS, Padgett DA, Sheridan JF. Social disruption-induced glucocorticoid resistance: kinetics and site specificity. J Neuroimmunol 2002; 124:54-61; PMID:11958822; http://dx.doi.org/ 10.1016/S0165-5728(02)00010-3 [DOI] [PubMed] [Google Scholar]
- 38. Bailey MT, Kierstein S, Sharma S, Spaits M, Kinsey SG, Tliba O, Amrani Y, Sheridan JF, Panettieri RA, Haczku A. Social stress enhances allergen-induced airway inflammation in mice and inhibits corticosteroid responsiveness of cytokine production. J Immunol 2009; 182:7888-96; PMID:19494313; http://dx.doi.org/ 10.4049/jimmunol.0800891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Greene BR, Blanchard EB, Wan CK. Long-term monitoring of psychosocial stress and symptomatology in inflammatory bowel disease. Behav Res Therapy 1994; 32:217-26; http://dx.doi.org/ 10.1016/0005-7967(94)90114-7 [DOI] [PubMed] [Google Scholar]
- 40. Mackos AR, Eubank TD, Parry NM, Bailey MT. Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infect Immun 2013; 81:3253-63; PMID:23798531; http://dx.doi.org/ 10.1128/IAI.00278-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y, Zhang M, Chen CC, Gillilland M, Sun X, El-Zaatari M, Huffnagle GB, Young VB, Zhang J, Hong SC, et al. . Stress-induced corticotropin-releasing hormone-mediated NLRP6 inflammasome inhibition and transmissible enteritis in mice. Gastroenterol 2013; 144:1478-87, 87 e1-8; http://dx.doi.org/ 10.1053/j.gastro.2013.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu D, Gao J, Gillilland M, Wu X, Song I, Kao JY, Owyang C. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterol 2014; 146:484-96 e4; http://dx.doi.org/ 10.1053/j.gastro.2013.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia-Rodenas CL, Bergonzelli GE, Nutten S, Schumann A, Cherbut C, Turini M, Ornstein K, Rochat F, Corthesy-Theulaz I. Nutritional approach to restore impaired intestinal barrier function and growth after neonatal stress in rats. J Pediatric Gastroenterol Nutrit 2006; 43:16-24; http://dx.doi.org/ 10.1097/01.mpg.0000226376.95623.9f [DOI] [PubMed] [Google Scholar]
- 44. Galley JD, Nelson MC, Yu Z, Dowd SE, Walter J, Kumar PS, Lyte M, Bailey MT. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol 2014; 14:189; PMID:25028050; http://dx.doi.org/ 10.1186/1471-2180-14-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol Rev 2011; 35:681-704; PMID:21361997; http://dx.doi.org/ 10.1111/j.1574-6976.2011.00270.x [DOI] [PubMed] [Google Scholar]
- 46. Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PloS One 2012; 7:e31951; PMID:22384111; http://dx.doi.org/ 10.1371/journal.pone.0031951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dicksved J, Schreiber O, Willing B, Petersson J, Rang S, Phillipson M, Holm L, Roos S. Lactobacillus reuteri maintains a functional mucosal barrier during DSS treatment despite mucus layer dysfunction. PloS one 2012; 7:e46399; PMID:23029509; http://dx.doi.org/ 10.1371/journal.pone.0046399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zakostelska Z, Kverka M, Klimesova K, Rossmann P, Mrazek J, Kopecny J, Hornova M, Srutkova D, Hudcovic T, Ridl J, et al. . Lysate of probiotic Lactobacillus casei DN-114 001 ameliorates colitis by strengthening the gut barrier function and changing the gut microenvironment. PloS one 2011; 6:e27961; PMID:22132181; http://dx.doi.org/ 10.1371/journal.pone.0027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jang SE, Hyam SR, Han MJ, Kim SY, Lee BG, Kim DH. Lactobacillus brevis G-101 ameliorates colitis in mice by inhibiting NF-kappaB, MAPK and AKT pathways and by polarizing M1 macrophages to M2-like macrophages. J Appl Microbiol 2013; 115:888-96; PMID:23742179; http://dx.doi.org/ 10.1111/jam.12273 [DOI] [PubMed] [Google Scholar]
- 50. Tannock GW, Savage DC. Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect Immun 1974; 9:591-8; PMID:4593471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011; 34:794-806; PMID:21596591; http://dx.doi.org/ 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- 52. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15:374-81; PMID:24629343; http://dx.doi.org/ 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. Selected commensal-related bacteria and Toll-like receptor 3 agonist combinatorial codes synergistically induce interleukin-12 production by dendritic cells to trigger a T helper type 1 polarizing programme. Immunol 2009; 128:e523-31; http://dx.doi.org/ 10.1111/j.1365-2567.2008.03022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ouwehand AC, Salminen S, Arvola T, Ruuska T, Isolauri E. Microbiota composition of the intestinal mucosa: association with fecal microbiota? Microbiol Immunol 2004; 48:497-500; PMID:15272194; http://dx.doi.org/ 10.1111/j.1348-0421.2004.tb03544.x [DOI] [PubMed] [Google Scholar]
- 55. Bekassy ZD, Calderon Toledo C, Leoj G, Kristoffersson A, Leopold SR, Perez MT, Karpman D. Intestinal damage in enterohemorrhagic Escherichia coli infection. Pediatr Nephrol 2011; 26:2059-71; PMID:20809220; http://dx.doi.org/ 10.1007/s00467-010-1616-9 [DOI] [PubMed] [Google Scholar]
- 56. Moxley RA. Escherichia coli 0157:H7: an update on intestinal colonization and virulence mechanisms. Anim Health Res Rev/Conf Res Workers Anim Dis 2004; 5:15-33; http://dx.doi.org/ 10.1079/AHR200463 [DOI] [PubMed] [Google Scholar]
- 57. Theriot CM, Koenigsknecht MJ, Carlson PE, Jr., Hatton GE, Nelson AM, Li B, Huffnagle GB, J ZL, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 2014; 5:3114; PMID:24445449; http://dx.doi.org/ 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sham HP, Yu EY, Gulen MF, Bhinder G, Stahl M, Chan JM, Brewster L, Morampudi V, Gibson DL, Hughes MR, et al. . SIGIRR, a negative regulator of TLR/IL-1R signalling promotes Microbiota dependent resistance to colonization by enteric bacterial pathogens. PLoS Pathogens 2013; 9:e1003539; PMID:23950714; http://dx.doi.org/ 10.1371/journal.ppat.1003539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bassis CM, Theriot CM, Young VB. Alteration of the Murine Gastrointestinal Microbiota by Tigecycline Leads to Increased Susceptibility to Clostridium difficile Infection. Antimicrob Agents Chemoth 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 2012; 80:62-73; PMID:22006564; http://dx.doi.org/ 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lone AG, Selinger LB, Uwiera RR, Xu Y, Inglis GD. Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PloS one 2013; 8:e75325; PMID:24066174; http://dx.doi.org/ 10.1371/journal.pone.0075325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ghosh S, Dai C, Brown K, Rajendiran E, Makarenko S, Baker J, Ma C, Halder S, Montero M, Ionescu VA, et al. . Colonic microbiota alters host susceptibility to infectious colitis by modulating inflammation, redox status, and ion transporter gene expression. Am J Physiol Gastrointest Liver Physiol 2011; 301:G39-49; PMID:21454446; http://dx.doi.org/ 10.1152/ajpgi.00509.2010 [DOI] [PubMed] [Google Scholar]
- 63. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. . Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55:205-11; PMID:16188921; http://dx.doi.org/ 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. . A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2013; . [DOI] [PubMed] [Google Scholar]
- 65. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007; 104:13780-5; PMID:17699621; http://dx.doi.org/ 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009; 15:1183-9; PMID:19235886; http://dx.doi.org/ 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- 67. Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil: Off J Eur Gastrointest Motil Soc 2012; 24:521-30, e248; http://dx.doi.org/ 10.1111/j.1365-2982.2012.01891.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Allegretti JR, Hamilton MJ. Restoring the gut microbiome for the treatment of inflammatory bowel diseases. World J Gastroenterol : WJG 2014; 20:3468-74; http://dx.doi.org/ 10.3748/wjg.v20.i13.3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kao D, Hotte N, Gillevet P, Madsen K. Fecal Microbiota Transplantation Inducing Remission in Crohn's Colitis and the Associated Changes in Fecal Microbial Profile. J Clin Gastroenterol 2014; PMID:24667590 [DOI] [PubMed] [Google Scholar]
- 70. Hermann G, Tovar CA, Beck FM, Sheridan JF. Kinetics of glucocorticoid response to restraint stress and/or experimental influenza viral infection in two inbred strains of mice. J Neuroimmunol 1994; 49:25-33; PMID:8294561; http://dx.doi.org/ 10.1016/0165-5728(94)90177-5 [DOI] [PubMed] [Google Scholar]
- 71. Crumeyrolle-Arias M, Jaglin M, Bruneau A, Vancassel S, Cardona A, Dauge V, Naudon L, Rabot S. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinol 2014; 42:207-17; http://dx.doi.org/ 10.1016/j.psyneuen.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 72. Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 2004; 558:263-75; PMID:15133062; http://dx.doi.org/ 10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang D, Kishihara K, Wang B, Mizobe K, Kubo C, Nomoto K. Restraint stress-induced immunosuppression by inhibiting leukocyte migration and Th1 cytokine expression during the intraperitoneal infection of Listeria monocytogenes. J Neuroimmunol 1998; 92:139-51; PMID:9916889; http://dx.doi.org/ 10.1016/S0165-5728(98)00197-0 [DOI] [PubMed] [Google Scholar]
- 74. Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PloS one 2012; 7:e47713; PMID:23091640; http://dx.doi.org/ 10.1371/journal.pone.0047713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Daniel H, Moghaddas Gholami A, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A, et al. . High-fat diet alters gut microbiota physiology in mice. ISME J 2014; 8:295-308; PMID:24030595; http://dx.doi.org/ 10.1038/ismej.2013.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. . QIIME allows analysis of high-throughput community sequencing data. Nature methods 2010; 7:335-6; PMID:20383131; http://dx.doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460-1; PMID:20709691; http://dx.doi.org/ 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 78. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-72; PMID:16820507; http://dx.doi.org/ 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26:266-7; PMID:19914921; http://dx.doi.org/ 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610-8; PMID:22134646; http://dx.doi.org/ 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 2010; 5:e9490; PMID:20224823; http://dx.doi.org/ 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shannon CE, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1949.; . [Google Scholar]
- 83. Oksanen J BF, Kindt R, Legendre R, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. .R package version 2.0-3. [Google Scholar]
- 84. Development RCT. R: a language and environment for statistical computing. Coventry, United Kingdom: R Foundation for Statistical Computing.; . [Google Scholar]
- 85. Park AJ, Collins J, Blennerhassett PA, Ghia JE, Verdu EF, Bercik P, Collins SM. Altered colonic function and microbiota profile in a mouse model of chronic depression. Neurogastroenterol Motilil 2013;25:733-e575; http://dx.doi.org/ 10.1111/nmo.12153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tsukamoto K, Nakade Y, Mantyh C, Ludwig K, Pappas TN, Takahashi T. Peripherally administered CRF stimulates colonic motility via central CRF receptors and vagal pathways in conscious rats. Am J Physiol; Regulat Integr Comp Physiol 2006; 290:R1537-R1541; http://dx.doi.org/ 10.1152/ajpregu.00713.2005 [DOI] [PubMed] [Google Scholar]
- 87. Goehler LE, Gaykema RP, Opitz N, Reddaway R, Badr N, Lyte M. Activation in vagal afferents and central autonomic pathways: early responses to intestinal infection with Campylobacter jejuni. Brain Behav Immun 2005;19:334-344; PMID:15944073; http://dx.doi.org/ 10.1016/j.bbi.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 88. Foster JA, Vey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 2013;36:305-312; PMID:23384445; http://dx.doi.org/ 10.1016/j.tins.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 89. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 2011;108:16050-16055; PMID:21876150; http://dx.doi.org/ 10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]


