Abstract
Introduction
The accuracy and utility of electronic health record (EHR)-derived phenotypes in replicating genotype-phenotype relationships has been infrequently examined. Low circulating vitamin D levels are associated with severe outcomes in inflammatory bowel disease (IBD); however, the genetic basis for vitamin D insufficiency in this population has not been examined previously.
Methods
We compared the accuracy of physician-assigned phenotypes in a large prospective IBD registry to that identified by an EHR-algorithm incorporating codified and structured data. Genotyping for IBD risk alleles was performed on the Immunochip and a genetic risk score calculated and compared between EHR-defined patients and those in the registry. Additionally, four vitamin D risk alleles were genotyped and serum 25-hydroxy vitamin D [25(OH)D] levels compared across genotypes.
Results
A total of 1,131 patients captured by our EHR algorithm were also included in our prospective registry (656 Crohn's disease (CD), 475 ulcerative colitis (UC)). The overall genetic risk score for CD (p=0.13) and UC (p=0.32) was similar between EHR-defined patients and a prospective registry. Three of the four vitamin D risk alleles were associated with low vitamin D levels in patients with IBD and contributed an additional 3% of the variance explained. Vitamin D genetic risk score did not predict normalization of vitamin D levels.
Discussion
EHR cohorts form valuable data sources for examining genotype-phenotype relationships. Vitamin D risk alleles explain 3% of the variance in vitamin D levels in patients with IBD.
Keywords: Crohn's disease, ulcerative colitis, vitamin D, electronic health records, genetics
INTRODUCTION
Inflammatory bowel diseases (IBD; Crohn's disease (CD), ulcerative colitis (UC)) are chronic inflammatory diseases affecting an estimated 1.5 million Americans and leading to annual direct costs in excess of $6 billion1, 2. Three decades since the recognition of familial contribution to disease, genome wide association studies (GWAS) led to the discovery of the first risk allele, NOD2, on chromosome 163-5 followed by identification of 163 distinct genetic loci6. However, this progress has not been paralleled by an understanding of the phenotypic implications of these loci in established disease. Key barriers to performing such analyses are the lack of detailed disease-related and co-morbid phenotypic information collected as part of large genetic studies and lack of longitudinal follow-up to capture disease evolution.
Electronic health records (EHR) are increasingly being adopted in healthcare and may soon be utilized in virtually every hospital or large practice7, 8. They function as repositories for a tremendous amount of valuable data generated during routine clinical care. However, challenges in utilizing such data for genetic discovery research include heterogeneity in accuracy and depth of content that is not ‘research-grade’ 9, 10. It has been previously demonstrated that algorithms incorporating structured codified and free text data identified using natural language processing can be used to accurately define diseases, retain portability across institutions, and replicate genetic associations with disease9, 11-28. However, there has been limited examination of the accuracy of such cohorts to identify genotype-phenotype associations that require deeper characterization than merely assign disease status.
In addition, the EHR cohort offers a unique opportunity to examine genetic contribution of laboratory parameters relevant to IBD that are routinely obtained during clinical care but not measured in large cohorts. There is growing recognition of the immunologic role of vitamin D29-31. In patients with CD, vitamin D deficiency may precede diagnosis and increase disease risk32, potentially through co-localization of the vitamin D receptor with the NOD2 locus33. Furthermore, low serum 25-hydroxy vitamin D [25(OH)D] in established CD is associated with more aggressive disease course34. Vitamin D supplementation may reduce risk of relapses consistent with animal models demonstrating amelioration of colitis35, 36. However, the reasons for the high prevalence of vitamin D deficiency in IBD patients is unclear as region of residence, dietary absorption or disease extent have not been consistent determinants37, 38. Recent GWAS identified several genetic variants that influence serum levels of vitamin D39, 40. The extent to which such genetic factors play a role in determining circulating vitamin D levels in IBD and normalization of with supplementation has not been examined previously. Linking routinely obtained vitamin D levels from the EHR to genotype data in a prospective registry will exemplify the utility of EHR cohorts for such translational questions.
We performed this study with the following aims: (1) To determine the accuracy of EHR-derived algorithms in assigning IBD type and disease sub-phenotypes in comparison to physician-defined labels in a prospective registry; (2) To compare the genetic architecture of EHR-algorithm defined IBD patients with those prospectively recruited from a specialty IBD clinic; and (3) define the genetic basis of vitamin D insufficiency in IBD by examining relative contribution of demographic, disease, and genetic risk factors.
METHODS
Study Populations
The main population for this retrospective cohort study was patients recruited into the Prospective Registry for the Study of IBD at MGH (PRISM). This ongoing cohort initiated in 2005 recruits eligible adult (18 years and older) patients with a diagnosis of CD, UC, or IBD-unspecified (IBDU) seeking care at the MGH Crohn's and Colitis center. This study included patients recruited prior to August 2013. All patients seeking care at the center for their IBD were approached and offered enrollment in the registry. After informed consent, trained research coordinators obtained details regarding demographics, disease characteristics, medical and surgical treatments, and co-morbidity. Disease location and behavior in CD and disease extent in UC were classified according to the Montreal classification and confirmed by the treating physician. All information was ascertained by patient interview and review of medical records by trained research coordinators and confirmed by the treating gastroenterologist. Consented patients provide blood, urine, stool, and/or biopsies. The prospective registry consisted of a mix of incident and prevalent disease with varying durations of time since diagnosis prior to enrollment.
The creation of our EHR-IBD cohort has been described previously11, 34. Briefly, this consisted of all patients with at least one International Classification of Diseases, 9th edition, (ICD-9-CM) diagnosis code for CD or UC who initiated care prior to July 2010. Using structured codified data and free text concepts identified using natural language processing, we developed a classification algorithm that assigned a disease label of CD or UC with a 97% positive predictive value, resulting in a final cohort of 11,001 IBD patients. The sensitivity and specificity for the CD algorithm was 69% and 98% and for the UC algorithm was 79% and 97% respectively11. Using an institutional review board approved honest broker, we mapped the intersection of patients in the prospective registry (PRISM) who were also captured by our EHR-algorithm. This yielded two distinct groups of patients for each disease – overlap patients from the prospective registry who were also captured by our EHR algorithm (EHR-CD and EHR-UC) and those who were part of the prospective registry alone and not in our EHR cohort. The latter consisted primarily of those establishing care at our center after the implementation of the EHR algorithm (REG-CD and REG-UC) but also in part of patients with lack of sufficient detail in the EHR to allow ascertainment of CD or UC with a high PPV at the time of the algorithm run. Among the patient included in this study, the date of first ICD-9 code for CD or UC ranged from 9/1990 to 6/2010; only EHR data collected prior to 6/30/2010 was eligible for inclusion in our datamart.
Genetic burden of IBD
Patients enrolled in PRISM provided 10mL of blood from which genomic DNA was extracted using standard procedures using the QIAGEN DNeasy 96 kit. All patients were genotyped on the Illumina Immunochip at the Broad Institute (Cambridge, MA). The Immunochip is a custom-designed chip that includes nearly 200,000 fine-mapping loci relevant to immune-mediated diseases. Allele frequencies were extracted for the 163 IBD risk loci6 and as previously described, a weighted genetic risk score was calculated multiplying the natural log of the odds ratio for strength of association with CD or UC by the frequency of variant alleles (∑ [log(OR) × allele dose])41. Genotyping was performed in three batches, one each in 2010, 2012, and 2013. There was no batch-to-batch variation in minor allele frequency of the risk alleles. To examine the association with disease phenotype, we extracted information on the three common NOD2 variants (R702W, G908R, and 1007fs).
Genetic contribution to serum vitamin D
Vitamin D status was assessed using serum 25(OH)D obtained during routine clinical care from the EHR. For patients with ≥ 2 time points of measurement, we examined change in vitamin D between initial and subsequent assessment and defined normalization as initially insufficient levels (< 30ng/mL) subsequent achieving sufficiency (≥30ng/mL). Patients were genotyped for four vitamin D risk alleles previously described to influence serum levels in healthy adults39, 40. These vitamin D risk alleles were described in Caucasian populations. These included a locus each at the cytochrome P450 2R1 (CYP2R1) (rs10741657; chromosome 11, position 14893332), cytochrome P450 24A1 (rs6013897; chromosome 20, position 54125940), nicotinamide adenine dinucleotide synthetase 1 (NADSYN1) (rs2060793; chromosome 11, position 14893764) and 7-dehydrocholesterol reductase (DHCR7) (rs12785878; chromosome 11, 71456403). Genotyping was performed on a Sequenom platform (Sequenom Inc, San Diego, CA) at the Broad Institute in a single batch. In addition to analyzing each locus separately, a vitamin D genetic risk score defined as the sum of risk alleles for all four loci was calculated (range 0-8). Weighting was not performed for this score given similar effect sizes for all four loci.
Statistical Analysis
All SNPs passed our threshold of Hardy–Weinberg p-value <0.001 and a call rate >99%. Individuals with <80% genotyping success rate were excluded. Genotype extraction and calculation of the CD and UC genetic risk score was performed using Plink V1.07. Continuous variables were summarized using means and standard deviations when normal and compared with a t-test. Non-normally distributed variables were summarized using medians and interquartile ranges and compared using the Mann-Whitney test. Categorical variables were expressed as proportions and compared using chi-square. The difference between the weighted CD and UC risk scores between the EHR- and prospective registry cohorts was tested using the t-test while the frequency of distribution of the disease risk alleles was compared using chi-square tests. A two-sided p-value < 0.05 indicated statistical significance. The association of heterozygosity and homozygosity at the NOD2 locus with disease subphenotype was examined using logistic regression models using physician-defined outcomes of fistulizing disease and surgery in the registry cohort (REG-CD), and an EHR-derived definition (≥ 1 ICD-9 code or free text mention for abdominal abscesses or fistulae and CD-related surgery). The estimates from this analysis were compared to published literature42.
Serum 25(OH)D levels were compared across the various allele distributions at each of the four vitamin D risk loci. Multivariate linear regression with serum 25(OH)D as the outcome determined the independent contribution of genetic, disease related, and demographic variables to serum 25(OH)D status. The fraction of variance in serum 25(OH)D levels explained by each of these factors was assessed using the model pseudo-R2 and the independent contribution of each parameter established using likelihood ratio tests. The study was approved by the Institutional Review Board of Partners Healthcare.
RESULTS
Overlap between EHR-cohort and prospective registry
A total of 1,131 patients captured by our EHR algorithm were also included in our prospective registry (656 CD, 475 UC). An additional 364 CD and 272 UC patients were part of the prospective registry alone (REG-CD, REG-UC). There was no difference in disease location, but CD patients captured by our EHR algorithm more frequently had stricturing, penetrating, or perianal disease (Table 1). UC patients captured by the EHR algorithm were younger at diagnosis and more likely to be women but similar in disease extent. While statistically significant differences were observed for some medications, this did not consistently favor a more severe phenotype in REG or EHR patients.
Table 1.
A comparison of the characteristics of patients in an electronic health record (EHR) inflammatory bowel disease cohort and a prospective patient registry
| Characteristic | EHR cohort (n = 656) | Prospective registry (n = 364) | p-value |
|---|---|---|---|
| Crohn's disease | |||
| Age at diagnosis | 23 (18 – 33) | 25 (19-35) | 0.06 |
| Female | 52 | 57 | 0.20 |
| Smoking | 0.83 | ||
| Never | 63 | 63 | |
| Past | 28 | 29 | |
| Current | 9 | 8 | |
| Disease location | 0.16 | ||
| Ileum | 24 | 24 | |
| Colon | 22 | 29 | |
| Ileocolon | 53 | 47 | |
| Upper GI | 0.3 | 0.3 | |
| Disease behavior | < 0.001 | ||
| Inflammatory | 44 | 59 | |
| Stricturing | 24 | 17 | |
| Penetrating | 32 | 25 | |
| Perianal disease | 28 | 19 | 0.001 |
| Medications | |||
| Aminosalicylates | 86 | 79 | 0.005 |
| Steroids | 84 | 82 | 0.36 |
| Immunomodulator | 72 | 62 | 0.001 |
| Anti-TNF | 54 | 61 | 0.03 |
| Surgery | 56 | 45 | 0.001 |
| Ulcerative colitis | (n = 475) | (n = 272) | |
| Age at diagnosis | 26 (20-37) | 28 (21-41) | 0.02 |
| Female | 53 | 45 | 0.03 |
| Smoking | 0.03 | ||
| Never | 65 | 74 | |
| Past | 31 | 23 | |
| Current | 4 | 3 | |
| Extent | 0.74 | ||
| Proctitis | 12 | 14 | |
| Left-sided colitis | 33 | 31 | |
| Pancolitis | 55 | 54 | |
| Medications | |||
| Aminosalicylates | 96 | 90 | 0.005 |
| Steroids | 81 | 74 | 0.03 |
| Immunomodulator | 59 | 48 | 0.004 |
| Anti-TNF | 30 | 37 | 0.06 |
| Surgery | 11 | 11 | 0.98 |
Covariates listed in the table were obtained from the data forms from the prospective patient registry.
EHR – electronic health record; immunomodulator includes azathioprine, 6-mercaptopurine, and methotrexate; Anti-TNF - monoclonal antibodies against tumor necrosis factor α (infliximab, adalimumab, certolizumab pegol)
Among the EHR cohort, there were 12 CD (1.8%) and 45 UC (9.4%) patients whose algorithm-assigned diagnosis was discordant with our registry assigned diagnosis (Table 2). Among the 12 CD patients who were ‘misclassified’, the largest group comprised those with IBD-U who had interchangeable use of terms for CD and UC in the medical record. Similarly, among the 45 UC patients who had discordant classification, the vast majority had IBD-U (n=18) with interchangeable use of terms for CD and UC, while the second largest subgroup consisted of those who developed CD of the J-pouch after initial surgery for UC. Also of note, in our registry, disease type was assigned at the time of enrollment while the EHR-algorithm included all EHR data till June 2010. Consequently, some of the misclassification observed reflects this difference in time points making our observed discordance frequency an over-estimate of the true value. Thus true misclassification by our algorithm was infrequent, and discordance reflected true uncertainty in diagnosis or evolution of disease.
Table 2.
Discordant results between disease classification algorithms in the EHR cohort and prospective patient registry
| Algorithm-assigned diagnosis | Prospective registry diagnosis | Clinical diagnosis on chart review | Explanation |
|---|---|---|---|
| Crohn's disease (n=12) | IBD-U (n = 7) | IBD-U | Labeled variably as both CD and UC in the medical record |
| Ulcerative colitis (n = 1) | Ulcerative colitis | Initially self-reported as CD in the medical record; diagnosis subsequently changed | |
| Ulcerative colitis (n = 2) | Crohn's disease | Wrong diagnosis in prospective registry | |
| Ulcerative colitis (n = 2) | Crohn's disease of the J pouch | Crohn's disease of the J-pouch following proctocolectomy for ulcerative colitis | |
| Ulcerative colitis (n = 45) | IBD-U (n = 18) | IBD-U | |
| Crohn's disease (n = 11) | Crohn's disease | Crohn's disease of the J-pouch following proctocolectomy for ulcerative colitis | |
| Crohn's disease (n = 1) | Crohn's disease | Developed Crohn's disease in the terminal ileum after ileostomy for ulcerative colitis | |
| Crohn's disease (n = 6) | Crohn's disease | Initial self-reported diagnosis as ulcerative colitis, subsequently modified to Crohn's disease | |
| Crohn's disease (n = 1) | Ulcerative colitis | Wrong diagnosis label in the prospective registry | |
| Crohn's disease (n = 11) | IBD-U | Wrong diagnosis label in the prospective registry |
IBD-U – inflammatory bowel disease unspecified
Genetic architecture of EHR-cohort and prospective registry
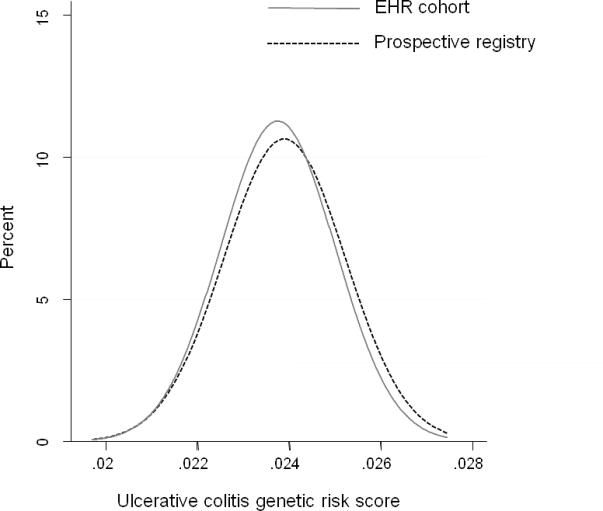
We compared distribution of overall disease-related genetic burden as well as frequency of occurrence of specific disease risk alleles in our EHR-cohort and prospective registry. As noted in Figure 1a, there was no difference in the distribution of the CD risk score derived from the 140 Immunochip risk alleles between 592 EHR-CD and 190 REG-CD patients (p=0.13). Similarly, the distribution of the UC genetic risk score derived from 133 risk alleles was similar between 412 EHR-UC patients and 111 REG-UC patients (p=0.32).
Figure 1. Comparison of genetic risk score between an electronic health record (EHR) inflammatory bowel disease cohort and patients recruited from a prospective registry.
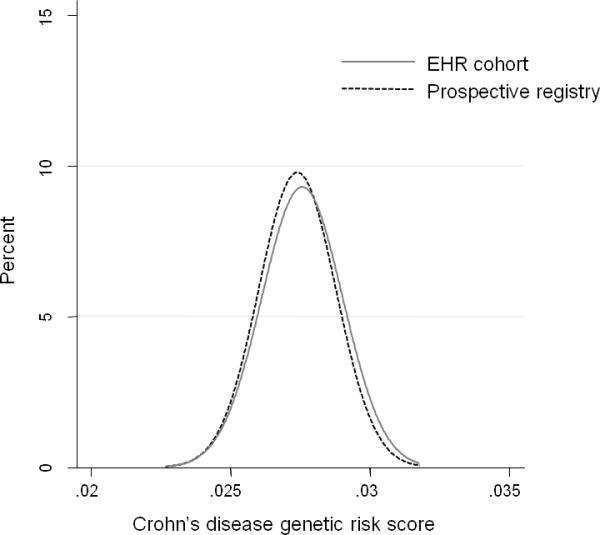
(a) Crohn's disease
Genetic risk score was calculated for 592 CD patients in the EHR cohort and 190 CD patients in the prospective registry.
T-test for comparison of the mean risk scores between the two cohorts = 0.13
(b) Ulcerative colitis
Genetic risk score was calculated for 412 UC patients in the EHR cohort and 111 UC patients in the prospective registry.
T-test for comparison of the mean risk scores between the two cohorts = 0.32
Replication of genotype-phenotype associations using EHR data
Using physician-confirmed phenotypes from our prospective registry, homozygosity at the NOD2 locus was associated with an increased risk for penetrating disease (Odds ratio (OR) 1.69, 95% confidence interval (CI) 1.04 – 2.74) and need for surgery (OR 1.64, 95% CI 0.95 – 2.82) in CD (Supplemental Table 1). The EHR-definitions of disease sub-phenotypes (≥ 1 ICD-9 code for fistulizing disease) and outcomes (≥ 1 ICD-9 code for IBD-related bowel resection) revealed similar strong associations between NOD2 homozygosity for penetrating disease (OR 1.72, 95% CI 1.05 – 2.84) and CD-related surgery (OR 1.67, 95% CI 1.04 – 2.69).
Common genetic variants influence vitamin D levels in IBD
Of the 1,131 patients in the EHR, serum 25(OH)D levels were available in 478 patients. Homozygosity at three of the loci (CYP2R1, NADSYN1, CYP24A1) was associated with lower levels of serum 25(OH)D in comparison to the reference allele (Table 3). The greatest difference in levels between reference alleles and homozygotes for the minor allele was observed for CYP24A1. Individuals with the TT genotype had a serum 25(OH)D level of 30.3ng/mL compared to 24.7 ng/mL in those with the AA genotype (p < 0.001). We then examined the relative contribution of demographic, clinical, and genetic risk factors to vitamin D status in patients with IBD (Table 4). On multivariate analysis, race, season of measurement, type of IBD, and a composite vitamin D genetic risk score were independent predictors of vitamin D status in patients with IBD. Each 1 point increase in the vitamin D genetic risk score was associated with a 1.11 ng/mL decrease in serum 25(OH)D levels (Figure 2). These four common genetic variants explained an additional 3% of the variance in serum 25(OH)D levels in addition to clinical parameters which explained 8%. Black patients had a higher burden of vitamin D risk alleles than whites (4.3 vs. 3.7, p=0.007) but this only partly explained the lower vitamin D levels in blacks. CD patients had a higher burden of vitamin D risk alleles compared to UC (3.9 vs. 3.5, p=0.006). We observed no correlation between either the CD or UC genetic risk score and serum 25(OH)D levels or distribution of vitamin D risk alleles. Repeat measurements of serum 25(OH)D were available in 281 patients among whom 41% of those who were initial deficient normalized their levels on follow-up. We observed no association between the vitamin D genetic risk score or any of the four risk alleles individually and likelihood of normalization of serum 25(OH)D (OR 0.99, 95% CI 0.83 – 1.18).
Table 3.
Common genetic variants and serum 25-hydroxy vitamin D levels in patients with inflammatory bowel diseases
| Variant | Serum 25-hydroxy vitamin D levels (in ng/ml), stratified by genotype [Mean (SD)] | |||
|---|---|---|---|---|
| CYP2R1 | AA (n = 51) | AG (n = 201) | GG (n = 223) | P(trend) |
| rs10741657 | 33.2 (14.0) | 29.9 (11.8) | 28.3 (11.9) | 0.010 |
| NADSYN1 | AA (n = 51) | AG (n = 199) | GG (n = 227) | |
| rs2060793 | 33.2 (13.9) | 29.8 (11.8) | 28.4 (11.9) | 0.013 |
| DHCR7 | TT (n = 235) | GT (n = 186) | GG (n = 57) | |
| rs12785878 | 30.4 (12.4) | 28.6 (12.2) | 28.9 (11.1) | 0.197 |
| CYP24A1 | TT (n = 296) | AT (n = 159) | AA (n = 29) | |
| rs6013897 | 30.3 (12.0) | 28.7 (12.4) | 24.7 (11.0) | 0.014 |
CYP – cytochrome; NADSYN1 – Nicotinamide adenine dinucleotide synthetase; DHCR7 – 7-dehydrocholesterol reductase; SD – standard deviation
Table 4.
Multivariate analysis of predictors of low vitamin D in patients with inflammatory bowel diseases
| Predictor | Regression co-efficient | 95% confidence interval (CI) | p-value |
|---|---|---|---|
| Season of measurement | |||
| Spring | Reference | ||
| Summer | 4.16 | 1.23 – 7.08 | 0.005 |
| Fall | −1.24 | −4.56 – 2.06 | 0.46 |
| Winter | −3.87 | −6.66 – −1.07 | 0.007 |
| Race | |||
| White | Reference | ||
| Black | −5.88 | −11.28 – −0.49 | 0.033 |
| Other | 7.26 | −2.25 – 16.78 | 0.13 |
| IBD type | |||
| Ulcerative colitis | Reference | ||
| Crohn's disease | −2.58 | −4.87 – −0.28 | −.028 |
| Sex | |||
| Male | Reference | ||
| Female | 2.09 | −0.14 – 4.32 | 0.066 |
| Age | −0.06 | −0.14 – 0.03 | 0.18 |
| Age at diagnosis | 0.01 | −0.01 – 0.02 | 0.572 |
| IBD-related surgery | |||
| No | Reference | ||
| Yes | −2.58 | −5.43 – 0.16 | 0.064 |
| Smoking | |||
| Never | Reference | ||
| Ever | −0.85 | −2.72 – 1.03 | |
| Body mass index | |||
| 19 – 24.9 | Reference | ||
| < 19 | −4.39 | −09.72 – 0.05 | −.11 |
| 25 – 29.9 | −2.20 | −4..63 – −.23 | 0.076 |
| >30 | −3.04 | −6.77 – 0.68 | −.109 |
| Vitamin D GRS | −1.11 | −1.78 – −0.44 | 0.001 |
IBD – inflammatory bowel diseases; GRS – genetic risk score
Figure 2.
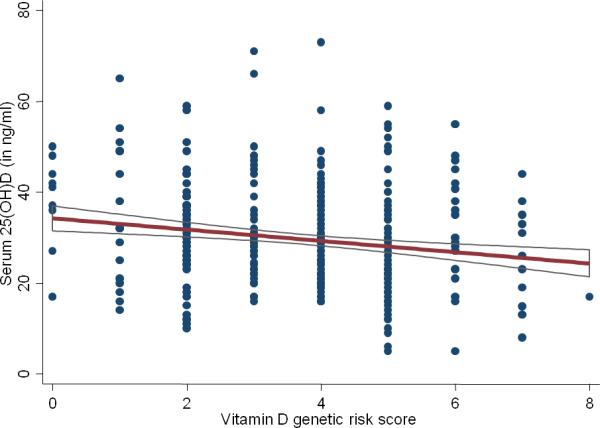
Association between vitamin D genetic risk score and serum 25-hydroxy vitamin D levels in patients with inflammatory bowel diseases
DISCUSSION
There is growing interest in using EHR data for genomic discovery research9, 10. Such use needs to be preceded by establishing the depth and accuracy of EHR-derived data to define genotype-phenotype associations. Utilizing the overlap of patients between a prospectively recruited patient cohort and EHR-derived disease labels, we demonstrate that EHR-cohorts are comparable to prospective registries in terms of genotypic disease burden, while there were some differences in disease phenotypic characteristics. However, we demonstrate that association with disease subphenotypes like penetrating disease and surgery could be replicated as well as novel associations identified.
A few prior studies have used EHR-derived cohorts to examine genetic basis of disease, though most restricted analysis to associations with disease status rather than deeper EHR-derived phenotypes16-18, 26. In an elegant study, Kurreeman et al. compared the effect sizes of 28 rheumatoid arthritis (RA) risk alleles in 1,515 EHR-derived RA cases and 1,480 EHR-controls with GWAS data from 5,539 autoantibody positive RA cases13. They demonstrated similar association for nearly all risk alleles in their EHR-cohort and a similar distribution of genetic risk burden. Ritchie et al. also similarly demonstrated that many disease-related associations could be replicated using EHR-derived cohorts including four loci for CD18. However, few studies have examined the robustness of EHR-derived deeper disease phenotypes to discover genetic associations. Indeed, that is a major strength of such EHR-linked cohorts as they can readily utilize a spectrum of data available from clinical care that is not captured in prospective registries.
While initially recognized for its role in bone and mineral metabolism, there is growing interest in the immunologic role of vitamin D in IBD29-31. Vitamin D deficiency is common and associated with increased risk of surgery32, 34, 43, 44; normalization of vitamin D levels reduces such risk and prevents relapses34, 36. However, factors predisposing to the high frequency of vitamin D deficiency in IBD patients have been inconsistently defined. Several prior studies have demonstrated the effect of common genetic variants on vitamin D levels39, 40. Since the initial landmark publications, the associations between serum 25(OH)D levels and polymorphisms in NADSYN1, group-specific component vitamin D binding protein (GC), CYP2R1 and CYP24A1, and DHCR7 have been replicated in different cohorts and ethnic populations. However, few studies have examined their contribution in disease states, particularly autoimmune disease and none have previously examined their role in IBD. Using serum 25(OH)D levels obtained as part of routine clinical care, we demonstrate that previously described common genetic variants independently contribute to vitamin D deficiency in IBD patients, and could partly explain the higher proportion of low vitamin D levels in Blacks and in those with CD. Such genetic risk loci explain an additional 3% of variance in circulating vitamin D levels in IBD (25% of the total explained variance). Adding in demographic and disease related factors, up to 11% of the variance in vitamin D levels in IBD patients can be explained suggesting that the major contribution to serum vitamin D levels remains unknown. We did not identify an association between the genetic risk alleles and normalization of vitamin D levels; however there was considerable heterogeneity in intervals between the two measures of vitamin D and doses of supplementation and the number of patients in this analysis was small precluding stastistically robust comparisons. Two prior randomized controlled trials have shown polymorphisms in the CYP2R1, CYP24A1, and VDR loci may influence response to low- and high- dose supplementation45, 46. Given the potential therapeutic role of vitamin D in IBD, future studies examining such associations in IBD are warranted.
There are several implications to our findings. Our findings demonstrate the utility of EHR-linked genetic cohorts for replicating genetic associations with clinical parameters and genotype-phenotype correlations. The wealth of data in the EHR obtained as part of routine clinical care including a spectrum of clinical, laboratory, and free text data makes this an invaluable resource for future studies9, 16, 19, 20, 25. Furthermore, such phenotypes can be derived more efficiently and cost-effectively than prospective registries which require added personnel support for recruitment and data management20. An added value to EHR-derived cohorts is the ability to phenotype patients efficiently, accurately, and repeatedly at different points in time, particularly as such longitudinal follow-up is often missing in cohorts from large genetic consortia. Such longitudinal follow-up can also be used to determine genetic determinants of treatment response (or adverse events of therapy)9, 19, 25, 28 including, as in our study, determining effects of genetic polymorphisms on normalization of vitamin D levels. As laboratory values including vitamin D levels are not routinely available in prospective registries or require to be measured for research, linking prospective registries to EHR allows us to efficiently perform analyses using readily available data. Additionally, EHR-based algorithms may be portable to diverse health systems with preserved accuracy, though some refinement may be necessary at each site12. However, prior to widespread application of such methods in distinct EHRs, additional validation studies are required for other diseases.
We acknowledge several limitations to our study. Since all patients included in this present study were enrollees in our prospective registry, they may have greater severity of disease by virtue of seeking care at a specialized IBD center. Second, there is heterogeneity in the depth and specificity of data present in the EHR. While our algorithms were tailored to improve accuracy of phenotyping, not all phenotypic associations may be replicable in an EHR. Third, patients who had serum vitamin D measured may be different from those without levels. Only a few patients had repeat levels available; the heterogeneity in supplementation and small numbers resulted in insufficient power to examine effect of genotype on supplementation. As in any observational study, one cannot exclude the effect of unmeasured confounders. EHR-linked data is also susceptible to the effect of changes in coding practices over time. However, as our algorithm development, training, and validation was on random sets, we do not expect temporal impact our findings. Compared to prospective registries, EHR-data is also susceptible to missing or sparse data that may impact capture by disease-defining algorithms. Finally, the number of patients with available genotype data and vitamin D levels was low. A post-hoc power calculation revealed a statistical power of 73% in detecting an OR of 2 across tertiles of vitamin D genetic risk score, and 87% for an OR of 2.25.
In conclusion, we demonstrate than an EHR-derived cohort can be a powerful tool to examine genotype-phenotype associations in IBD. We demonstrate comparability of genetic burden between the two groups and replicated associations between the NOD2 locus and complicated CD. We also demonstrate that common genetic variants explain an additional 3% of the variance of serum vitamin D levels in IBD patients and future studies should examine the effect of such variants on response to supplementation and patient outcomes.
Supplementary Material
Acknowledgments
Sources of Funding: The study was supported by NIH U54-LM008748. A.N.A is supported by funding from the US National Institutes of Health (K23 DK097142). K.P.L. is supported by NIH K08 AR060257 and the Harold and Duval Bowen Fund. E.W.K is supported by grants from the NIH (K24 AR052403, P60 AR047782, R01 AR049880). This work is supported by the National Institutes of Health (NIH) (P30 DK043351) to the Center for Study of Inflammatory Bowel Diseases.
Footnotes
Financial conflicts of interest: None
REFERENCES
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kappelman MD, Rifas-Shiman SL, Porter CQ, et al. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907–13. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–8. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 4.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler-Milstein J, DesRoches CM, Furukawa MF, et al. More Than Half of US Hospitals Have At Least A Basic EHR, But Stage 2 Criteria Remain Challenging For Most. Health Aff (Millwood) 2014;33:1664–71. doi: 10.1377/hlthaff.2014.0453. [DOI] [PubMed] [Google Scholar]
- 8.DesRoches CM, Charles D, Furukawa MF, et al. Adoption of electronic health records grows rapidly, but fewer than half of US hospitals had at least a basic system in 2012. Health Aff (Millwood) 2013;32:1478–85. doi: 10.1377/hlthaff.2013.0308. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 11.Ananthakrishnan AN, Cai T, Savova G, et al. Improving Case Definition of Crohn's Disease and Ulcerative Colitis in Electronic Medical Records Using Natural Language Processing: A Novel Informatics Approach. Inflamm Bowel Dis. 2013;19:1411–20. doi: 10.1097/MIB.0b013e31828133fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19:e162–9. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet. 2011;88:57–69. doi: 10.1016/j.ajhg.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS One. 2013;8:e78927. doi: 10.1371/journal.pone.0078927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford DC, Crosslin DR, Tromp G, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–72. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birdwell KA, Grady B, Choi L, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet Genomics. 2012;22:32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6:234cm3. doi: 10.1126/scitranslmed.3008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis MF, Sriram S, Bush WS, et al. Automated extraction of clinical traits of multiple sclerosis in electronic medical records. J Am Med Inform Assoc. 2013;20:e334–40. doi: 10.1136/amiajnl-2013-001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–21. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao KP, Kurreeman F, Li G, et al. Associations of autoantibodies, autoimmune risk alleles, and clinical diagnoses from the electronic medical records in rheumatoid arthritis cases and non-rheumatoid arthritis controls. Arthritis Rheum. 2013;65:571–81. doi: 10.1002/art.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin Transl Sci. 2012;5:394–9. doi: 10.1111/j.1752-8062.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 2011;89:379–86. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantorna MT, Mahon BD. Mounting evidence for vitamin D as an environmental factor affecting autoimmune disease prevalence. Exp Biol Med (Maywood) 2004;229:1136–42. doi: 10.1177/153537020422901108. [DOI] [PubMed] [Google Scholar]
- 30.Cantorna MT, Zhu Y, Froicu M, et al. Vitamin D status, 1,25-dihydroxyvitamin D3, and the immune system. Am J Clin Nutr. 2004;80:1717S–20S. doi: 10.1093/ajcn/80.6.1717S. [DOI] [PubMed] [Google Scholar]
- 31.Cantorna MT, Mahon BD. D-hormone and the immune system. J Rheumatol Suppl. 2005;76:11–20. [PubMed] [Google Scholar]
- 32.Ananthakrishnan AN, Khalili H, Higuchi LM, et al. Higher predicted vitamin d status is associated with reduced risk of Crohn's disease. Gastroenterology. 2012;142:482–9. doi: 10.1053/j.gastro.2011.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramagopalan SV, Heger A, Berlanga AJ, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–60. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ananthakrishnan AN, Cagan A, Gainer VS, et al. Normalization of plasma 25-hydroxy vitamin D is associated with reduced risk of surgery in Crohn's disease. Inflamm Bowel Dis. 2013;19:1921–7. doi: 10.1097/MIB.0b013e3182902ad9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantorna MT, Munsick C, Bemiss C, et al. 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease. J Nutr. 2000;130:2648–52. doi: 10.1093/jn/130.11.2648. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen SP, Agnholt J, Glerup H, et al. Clinical trial: vitamin D3 treatment in Crohn's disease - a randomized double-blind placebo-controlled study. Aliment Pharmacol Ther. 2010;32:377–83. doi: 10.1111/j.1365-2036.2010.04355.x. [DOI] [PubMed] [Google Scholar]
- 37.Mouli VP, Ananthakrishnan AN. Review article: vitamin D and inflammatory bowel diseases. Aliment Pharmacol Ther. 2014;39:125–36. doi: 10.1111/apt.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farraye FA, Nimitphong H, Stucchi A, et al. Use of a novel vitamin D bioavailability test demonstrates that vitamin D absorption is decreased in patients with quiescent Crohn's disease. Inflamm Bowel Dis. 2011;17:2116–21. doi: 10.1002/ibd.21595. [DOI] [PubMed] [Google Scholar]
- 39.Ahn J, Yu K, Stolzenberg-Solomon R, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19:2739–45. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang TJ, Zhang F, Richards JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–8. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ananthakrishnan AN, Huang H, Nguyen DD, et al. Differential effect of genetic burden on disease phenotypes in Crohn's disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol. 2014;109:395–400. doi: 10.1038/ajg.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adler J, Rangwalla SC, Dwamena BA, et al. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am J Gastroenterol. 2011;106:699–712. doi: 10.1038/ajg.2011.19. [DOI] [PubMed] [Google Scholar]
- 43.Froicu M, Weaver V, Wynn TA, et al. A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol. 2003;17:2386–92. doi: 10.1210/me.2003-0281. [DOI] [PubMed] [Google Scholar]
- 44.Garg M, Lubel JS, Sparrow MP, et al. Review article: vitamin D and inflammatory bowel disease - established concepts and future directions. Aliment Pharmacol Ther. 2012;36:324–44. doi: 10.1111/j.1365-2036.2012.05181.x. [DOI] [PubMed] [Google Scholar]
- 45.Barry EL, Rees JR, Peacock JL, et al. Genetic Variants in CYP2R1, CYP24A1, and VDR Modify the Efficacy of Vitamin D3 Supplementation for Increasing Serum 25-Hydroxyvitamin D Levels in a Randomized Controlled Trial. J Clin Endocrinol Metab. 2014;99:E2133–7. doi: 10.1210/jc.2014-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse M, Tran B, Armstrong BK, et al. Environmental, personal, and genetic determinants of response to vitamin D supplementation in older adults. J Clin Endocrinol Metab. 2014;99:E1332–40. doi: 10.1210/jc.2013-4101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


