Abstract
Background
Disparities in prevalence, HPV status, and mortality rates for head and neck cancer have been described between African Americans (AA) and European Americans (EA).
Methods
We studied the HPV status and gene expression profiles in 56 oropharyngeal/oral cavity tumors and 9 normal tissue samples from EA and AA patients treated in South Carolina between 2010 and 2012.
Results
Overall, 59% of tumors were HPV DNA-positive, but only 48% of those expressed E7 mRNA (HPV-active). The prevalence of HPV-active tumors was 10% in AA and 39% in EA patients. Tumors positive for HPV DNA but negative for HPV mRNA exhibited gene expression profiles distinct from those of both HPV-active and HPV-negative cancers, suggesting that HPV DNA+/RNA− tumors may constitute a unique group.
Conclusions
This study provides a direct assessment of differential expression patterns in HPV-related OPC arising from AA and EA patients, for which there is a paucity of data.
Keywords: HPV, Head and Neck cancer, African American, Cancer Health Disparities, Gene Expression
Introduction
Head and neck cancer (HNC) is the fifth most common malignancy worldwide with an annual mortality rate of 200,0001. About 90% of HNC can be classified as head and neck squamous cell carcinomas (HNSCC), of which approximately 75% are attributed to alcohol and tobacco consumption2.
Approximately 25% of all HNC, and up to 60% of oropharyngeal cancers (OPC) are associated with human papillomavirus (HPV), predominantly HPV163. HPV-associated OPC have better prognosis and a more favorable response to therapy as compared to HPV-negative tumors4. OPC include cancers of the back one-third of the tongue, the soft palate, the side and back walls of the throat, and the tonsils, according to the definition of cancer terms by the National Cancer Institute. HPV-positive cancers exhibit distinct phenotypic features including poor differentiation, scant keratinization and basaloid phenotype, quite different from the keratinizing morphology of HPV-negative squamous cell carcinoma5. Common risk factors of HPV related OPC include number of sexual partners, oral-genital sex, oral-anal sex, smoking, alcohol and marijuana use5. The incidence of OPC increases with more than five sexual partners6 and with human immunodeficiency virus positivity7.
The overall incidence of HNC is on the rise1. This increase is attributed to a surge in HPV-related HNC arising from the oral cavity, oropharynx and pharynx, with reports of increasing incidence worldwide2,3. In the United States, South Carolina (SC) ranks third in oral cancer and OPC mortality, with the most common sites of infection being the tongue, gums and tonsils8.
The high prevalence of HPV-related OPC is determined by a propensity of the virus to infect the discontinuous reticulated epithelium of tonsillar crypts9. High-risk HPV are capable of transforming primary human keratinocytes from either genital10 or oral epithelia in vitro11 and viral oncoproteins play the same role in vivo, by disrupting cell-cycle regulatory pathways leading to progression to anogenital cancer and OPC12. However, the precise mechanisms by which HPV mediates malignant transformation of keratinocytes in the upper digestive tract epithelia are not entirely clear. HPV E7 expression results in overexpression of p16INK4A13, which is commonly used as a clinical surrogate marker for HPV positivity/activity14. However, high p16INK4A alone has insufficient sensitivity and specificity as a biomarker of HPV positivity in different mucosal sub-sites of HNC15. Therefore, increasing emphasis is being placed on the assessment of viral load and viral oncogene expression, resulting in further classification of HPV positive OPC as HPV-active and HPV-inactive16. Differences in risk factors, age of presentation, clinical behavior and gene expression profiles indicate that HPV-positive and HPV-negative tumors develop with different molecular mechanisms and are biologically distinct.
A recent study retrospectively analyzed and confirmed contrasting differences in distribution of disease site, stage, and OS within the oral cavity and oropharyngeal cancers by race17. HPV has been characterized as a risk factor for OPC based on race, life style and sexual behavior, impacting survival outcomes for both African American (AA) and European American (EA) patients.18 According to some reports, the rate of HPV-associated tumors is much lower in AA patients as compared to EA patients in United States19. In general, however, AA males have a higher incidence of HNC than any other racial/gender group, and a mortality rate almost threefold that observed in EA males20. Overall, AA patients tend to present with HPV-negative OPC and have worse prognosis as compared to both HPV-positive and HPV-negative EA patients21.
Despite the unveiling of differential gene expression patterns22, genetic23 and epigenetic profiles24 and more recently the compilation of a mutational landscape25,26 along with preliminary TCGA data27,28 of HPV-related and unrelated HNC, the determinants of the racial disparity in HNC are still relatively unexplored.
This study aimed to further explore possible racial differences in prevalence and mortality rate for oral cancer and OPC between AA and EA South Carolinians, with respect to HPV infection. We determined the frequency of HPV infection, type distribution and HPV status in HNC, by race and gender, in a representative cohort of mostly oral cancer and OPC samples from AA and EA patients treated at the Medical University of South Carolina (MUSC). We also detected differences in OPC survival rates based on the HPV status of the tumors. We then compared the gene expression profiles of HPV-active, -inactive and -negative HNC from these patients. Our results show that AA patients are significantly less likely than EA to present with an HPV-active OPC, and that HPV-inactive OPC have gene expression profiles distinct from those of both HPV-active and HPV-negative cancers, indicating that these tumors may constitute a pathogenetic group of their own. In addition, subtle but significant differences in gene expression can be observed in oral cancer and OPC from AA and EA patients.
Materials and Methods
Tissue samples and extraction of nucleic acids
This study included a total of 65 fresh frozen oral or oropharyngeal tissue samples accrued from AA and EA patients at MUSC, Charleston, SC. About one half of the cancer specimens were consecutive cases accrued directly by the surgeons, the other half derived from the MUSC Hollings Cancer Center Tissue Bank. The sample set included 56 tumor tissue samples, consisting of 38 OPC samples; 16 oral cancers (oral cavity; oral tongue; floor of mouth) 1 hypopharyngeal and 1 maxillary carcinoma. The remaining 9 samples were normal benign uvula tissue samples collected during surgery performed to correct sleep apnea. The study was approved by the MUSC Institutional Review Board (IRB) and USC’s IRB. A written consent for the use of tissue was obtained from patients according to the MUSC Standard Tissue Collection consent, while de-identified tissue samples were provided by the MUSC group to the laboratory at the University of South Carolina School of Medicine. Total RNA and genomic DNA were isolated from fresh frozen tumor samples using miniaturized mortar and pestle systems (CryoGrinder™, CryoCooler™, OPS diagnostics LLC) and TRIzol® Reagent (Ambion®, by Life Technologies). Tissue samples were selected for microarray analysis based upon the quality of the frozen specimen available (mostly tumor cells with scant stroma; absence of necrosis, as judged from H&N stained sections) and the resulting RNA.
HPV typing and HPV status
Genomic DNA from oropharyngeal tissue samples was analyzed for the presence of specific HPV types by the INNO-LiPA HPV Genotyping Extra assay (Fujirebio) a multiplex PCR-based assay based upon the reverse line blot hybridization principle. The INNO-LiPA assay targets a 65-bp fragment of the L1 Open Reading Frame to detect and identify 28 different HPV types, including: 18 high-risk HPVs (HR;16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 82); 6 low-risk HPVs (6, 11, 40, 43, 44, 54, 70); and 3 other non-classified HPVs (69, 71, 74). The assay was carried out following the manufacturer’s instructions. Type specific HPV E7 oncogene expression was assessed by RT-qPCR to define the active or inactive status of the virus in the samples, using primers specifically designed to detect E7 from each individual HPV type analyzed. Primer sequences are included in Supplemental Table 1.
mRNA labeling and microarray hybridization
Microarray experiments were performed using the Agilent Technologies (Santa Clara, CA) platform. Total RNA samples were amplified and labeled using Agilent’s Low Input Quick Amp Labeling Kit according to the manufacturer’s recommendations. Briefly, mRNA contained in 200 ng of total RNA was converted into cDNA using a poly-dT primer that also contained the T7 RNA polymerase promoter sequence. Subsequently, T7 RNA polymerase was added to cDNA samples to amplify the original mRNA molecules and to simultaneously incorporate cyanine-3 labeled CTP into the amplification product (cRNA). In the next step, labeled cRNA molecules were purified using Qiagen’s RNeasy Mini Kit (Valencia, CA). After spectrophotometric assessment of dye incorporation and cRNA yield, samples were stored at −80°C until hybridization. Labeled cRNA samples (600 ng) were hybridized to SurePrint G3 Human Gene Expression 8×60K v2 Microarrays at 65°C for 17 h, using Agilent’s Gene Expression Hybridization Kit according to the manufacturer’s recommendations. After washes, arrays were scanned using a High Resolution Agilent DNA Microarray Scanner and images saved in TIFF format.
Data analysis
Data were extracted from images with Feature Extractor Software version 10.7.3.1 (Agilent) where background correction was also performed. Background-corrected data were uploaded into GeneSpring GX version 11.5.1 for analysis. In this process, data were log2 transformed, quantile normalized and base line transformed using the median of all samples. Then, data were filtered by flags in a way that 75% of the samples in at least one of the treatment groups have a “detected” flag. Differentially expressed genes were determined by analysis of the data using the non-parametric Mann-Whitney unpaired test. Cutoff values of 0.02 and 2 were used for p-value and fold-change, respectively. Gene ontology (GO), pathway analysis and hierarchical cluster analysis were also performed with GeneSpring. For the hierarchical cluster analysis, Euclidean similarity metrics and Wards linkage rule were used along with Kruskal-Wallis non-parametric ANOVA. Kaplan-Meier survival curves were generated using GraphPad PRISM version 6.0 (GraphPad, Software, La Jolla, CA). A p-value of 0.05 or less was considered significant.
RT-qPCR
Complementary DNA (cDNA) was synthesized from total RNA with the iScript™ cDNA synthesis Kit (BioRad, Hercules, CA). To achieve uniform template concentration for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays, c-DNA was quantified by Quant-iT™ RiboGreen® RNA Assay (Invitrogen, Grand Island, NY). All primers used in RT-qPCR reactions are listed in Supporting Information Table S1. Primer pairs for TP53 and TGFB2 were as listed in RTPrimerDB (ID2676 and ID1186, respectively). All other primers were designed through Real-Time PCR sciTool (Integrated DNA Technology, Coralville, IA) and Primer-BLAST software (NCBI)29. These custom designed oligonucleotides were analyzed in silico for hairpins, primer dimers and heterodimers through OligoAnalyzer sciTool (Integrated DNA Technology). RT-qPCR assays were performed on iCycler IQ detection system (BioRad) with iScriptTM Sybr® Green Supermix Kit (BioRad). Reactions were performed in duplicate with 200 nm primer mix (2.5 μm stock of forward and reverse primers), 1X SYBR® Green Supermix, 560 pg template c-DNA and nuclease-free water to make up the volume of 25 μl. The cycling conditions for the RT-qPCR assays were as follows: initial denaturation of template at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 10 sec and primer annealing and elongation at the respective temperatures for 1 min. Relative gene expression was analyzed according to recent MIQE guidelines30 using a panel of three reference genes verified by GeNorm/qbase+31. Cumulative normalized relative quantities (CNRQs) were calculated using Biogazelle qbase+ (version 2.5. Biogazelle, Gent, Belgium). A correction factor (CF) was calculated from the inter-run calibrators (IRC). Thus, the final CNRQs were also corrected for inter-run variations. The CNRQ’s were Log transformed for graphical representation and statistical analysis. Individual Log10CNRQs per sample-gene pair were used for multiple comparisons through One Way ANOVA (qbase+ Stat wizard and GraphPad Prism® version 6).
Results
Patient demographics, tumor characteristics and HPV prevalence and activity in OPC samples
We captured most of the incident cases of HNC and sleep apnea (for normal tissue samples) and as many HNC samples as were available to us from the MUSC tissue bank during the duration of the study. The only inclusion criteria were the fact that patients had given consent to the use of samples for research, and that the surgical specimens were sufficient in volume and quality for the study. Clinical characteristics, smoking or alcohol consumption were not significantly different between EA and AA patients included in this study.
The demographic characteristics of tumor and normal samples and their respective HPV status are presented in Supporting Information Tables S2, S3 and S4. Once the OPC samples were analyzed for HPV DNA and RNA, we classified them into three categories based on whether HPV DNA was present and transcribed (HPV-active), HPV DNA was present but not transcribed (HPV-inactive) or HPV DNA was not detected (HPV-negative). The prevalence of HPV, based on the presence of HPV DNA, was 59% overall (Supporting Information, Table S2). However, only a fraction of the samples positive for HPV by INNO-LiPA expressed the HPV E7 oncogene mRNA (Supporting Information, Table S2). Only one of the 9 normal tissue samples was positive for HPV by INNO-LiPA (Supplemental Table S3). HPV typing revealed the presence of HR-HPV types only, and among them HPV16 was present in about 85% of the OPC samples positive by INNO-LiPA (Supplemental Table S4). Other HPV types detected were HPV66, 35, 52 and 58 (Supplemental Table S4).
Age at presentation varied between different groups. As expected, on average, HPV-active oral cancers and OPC presented at younger ages than HPV-negative cancers, especially in males (mean ages in EA males: HPV-active, 52 years; HPV-negative, 71 years; p=0.0003). In addition, HPV-negative cancers in AA patients presented at a younger age than in EA (mean ages of presentation for HPV-negative cancers: AA males, 53 years; EA males, 71 years; p=0.0023). There was no difference between AA and EA patients in the stage of the cancer at presentation, with over 70% of cases presenting at AJCC stage IVa (Supplemental Table S5).
Tonsillar lesions were more common in EA patients (n EA=16, 44%; n AA=3, 15%) and among the HPV-active tumors (n=6, 33%) while lesions of the oral cavity and oropharyngeal sites other than the tonsil were more common in AA patients (n EA=4, 11%; n AA=6, 30%) (Supplemental Table S5). There was no significant difference in anatomic site between HPV-inactive and HPV-negative cancers: the majority of cases of HNC accrued localized to the tonsil, base of tongue or oral tongue in both cases (Supplemental Table S5).
Overall survival (OS) followed a trend similar to that previously reported in HNC, with HPV-negative cancers exhibiting worse OS than HPV-active tumors (Supplemental Figure 1) although, due to the small sample size, this difference did not reach statistical significance. HPV-inactive tumors and HPV-negative lesions had overlapping survival curves.
HPV prevalence in OPC samples from AA and EA patients
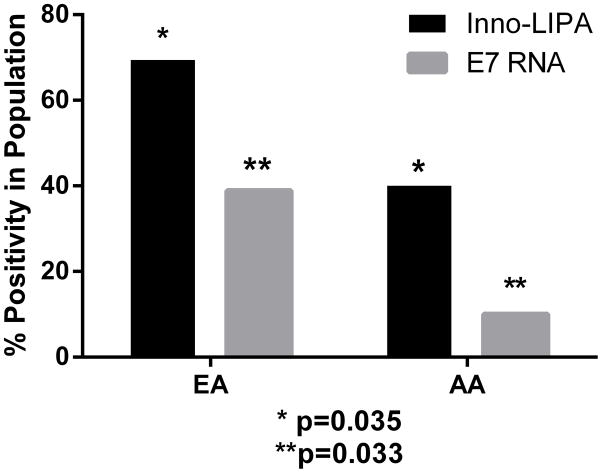
Forty percent of OPC samples from AA patients tested positive for HPV DNA by INNO-LiPA and only 10% of these samples expressed E7 mRNA. In contrast, in OPC samples from EA patients, 69% were positive for HPV DNA, with 39% of those exhibiting HPV-active infection (Figure 1). Therefore, AA patients were more likely to present with HPV-negative tumors as compared to EA patients in this cohort (Figure 1). Despite the small sample size, the observed differences were statistically significant: the odds of having a HPV-positive tumor in EA patients in comparison with AA patients were 3.4, with a 95% confidence interval of 1.08–10.7, p=0.035. The odds of presenting with an HPV-active tumor among EA patients in comparison with AA patients were 5.7, with 95% confidence interval 1.15–28.6, p=0.033 (Figure 1).
Figure 1.
HPV Prevalence in OPC samples from European American (EA) and African American (AA) patients. *Odds ratio for HPV DNA positivity in OPC samples from EA vs AA patients: 3.4; 95% confidence interval: 1.08–10.7; p=0.035. ** Odds ratio for active HPV in OPC from EA vs AA patients: 5.7; 95% confidence interval: 1.15–28.6; p=0.033
Gene expression profiles of HPV-active, HPV-inactive and HPV-negative OPC samples
We next explored gene expression profiles of HPV-active, HPV-inactive and HPV-negative samples. The experimental design of the microarray study is presented in Supplemental Figure 2. Forty RNA samples, (36 tumor and 4 normal tissue samples) of RIN ≥ 6.5 were included in the analysis, conducted on 8×60 K human microarrays (Agilent Technologies).
We first compared HPV-inactive tumors to HPV-negative and HPV–active tumors using Kruskal-Wallis statistics with Benjamini-Hochberg multiple testing correction at fold-change (FC) cutoff 2 and p-value 0.02. The number of differentially-expressed genes (DEGs) with at least 2 FC for the different comparisons were as follows: 92 down/397 up in HPV-active vs HPV-inactive OPC; 123 down/364 up in HPV-active vs HPV-negative OPC; and 32 down/19 up in HPV-inactive vs HPV-negative tumors. Complete lists of these DEGs are presented in Supplemental Tables S6, S7 and S8. A comparison of HPV-negative tumors from AA and EA patients revealed a total of 263 DEGs with 191 down-regulated and 72 up-regulated genes (Supplemental Table S9).
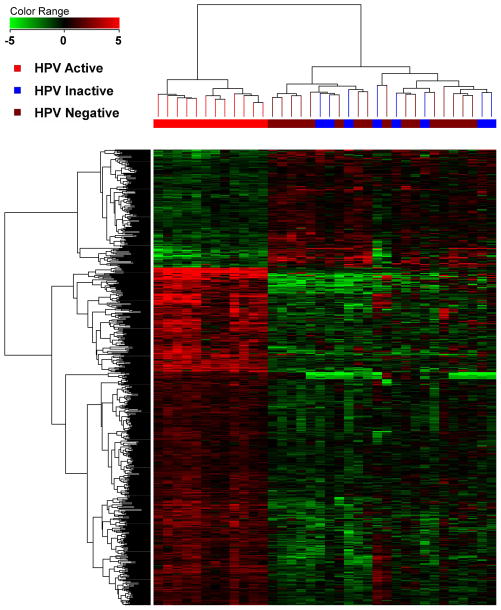
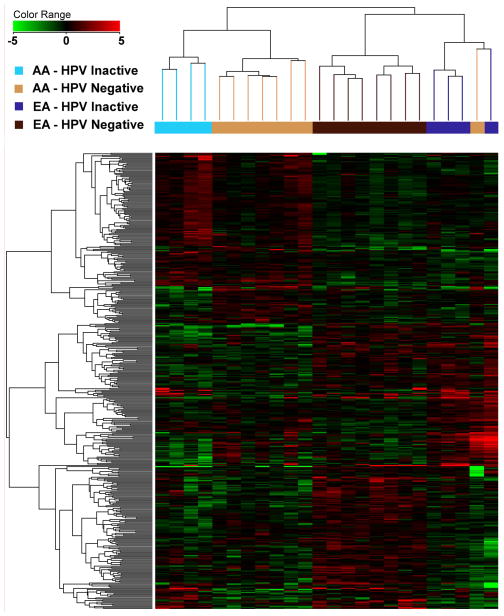
Unsupervised hierarchical clustering of DEGs among different groups of HPV-active, HPV-inactive and HPV-negative samples across the two racial groups allowed for a series of observations: 1) HPV-active tumor samples, irrespective of race, segregated very clearly from the HPV-negative and HPV-inactive tumors (Figure 2A). 2) Within the EA population, HPV-inactive tumors clustered with the HPV-negative tumors, with the only exception of one tumor sample that clustered with the HPV-active tumors (Supplemental Figure 3). When HPV-negative and HPV-inactive tumors were compared to each other, they clearly separated by race and HPV status in an unsupervised cluster analysis, with HPV-inactive and HPV-negative tumors exhibiting distinct and separable gene expression profiles within each racial group (Figure 2B).
Figure 2.
Unsupervised hierarchical clustering of OPC samples; A: all HPV-active, HPV-inactive and HPV-negative tumors; B: HPV-inactive and HPV-negative cancers from AA and EA patients.
Gene Ontology and Ingenuity Pathway Analysis of microarray results
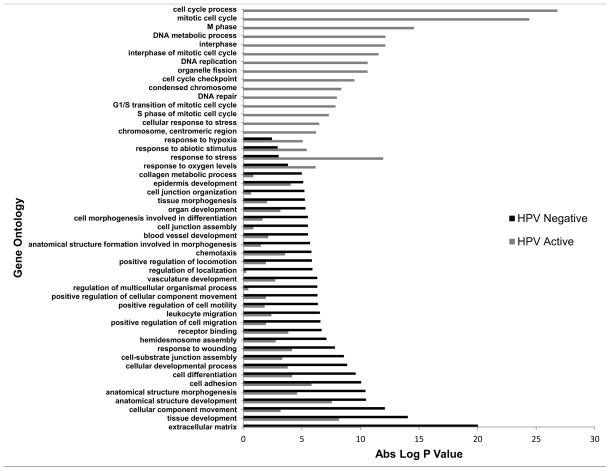
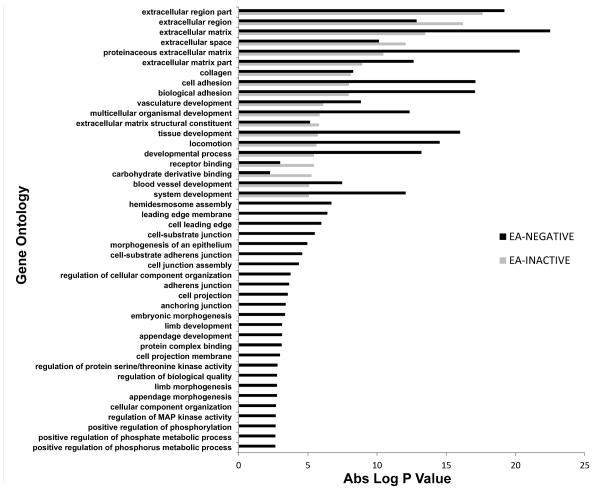
By Gene Ontology (GO) analysis, HPV-active OPC samples show distinct alterations of biological processes including cell proliferation, cell cycle checkpoints and mitosis, with relatively little involvement of pathways of epithelial to mesenchymal transition (EMT), invasion and angiogenesis (see Figure 3 for representative processes; see Supplemental Figure 4 for the entire list). HPV-negative samples presented the opposite picture, with alterations of gene expression pathways indicative of EMT, angiogenesis, chemotaxis and cell motility, all pointing to possible invasive behavior and metastatic potential (Figure 3, Supplemental Figure 4). In addition, a direct comparison of gene ontology biological processes between HPV-inactive and HPV-negative tumors from EA patients revealed that HPV-inactive tumors present with an intermediate profile which resembles, but does not completely overlap that of HPV-negative tumors (Figure 4 and Supplemental Figure 5).
Figure 3.
Comparison of Gene Ontology profiles of HPV-active and HPV-negative tumors
Figure 4.
Comparison of Gene Ontology profiles from HPV-negative and HPV-inactive tumors from EA patients.
The differences in gene expression between AA and EA HPV-negative tumors significantly affected TP63 transcriptional factor networks and cytokine-cytokine receptor interactions among 40 other pathways, as analyzed through the pathway analysis tool of GeneSpring (Cutoff: p value 0.05, Fold Change 2). Additionally, the biomarker tool of Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Inc. 2000–2013) highlighted three major genes (ADAM12, IL1B, MMP14) as plausible markers for differences in HPV-negative OPC tumors between AA and EA patients.
RT-qPCR analysis of selected genes in an expanded sample set
Next, we conducted RT-qPCR analysis of a panel of selected genes. Our goal here was to confirm the differential expression profiles of tumor suppressors genes and oncogenes identified through our microarray analysis which had previously been described to play a role in HPV driven cancers32 (CDKN2A, MCM2, TP53, MAL, KRT15, MMP13, MMP10 and TGFβ2); HPV driven HNSCC22,33 (CDKN2A, MCM2, TP53, MAL, KRT15) and HPV independent HNSCC34 (MET, ADAM12, MMP10, 13, BRCA1, TGFβ2, IL1B, MAL, KRT15). We aimed to investigate the expression of these genes with respect to HPV status and race. Additionally, we categorized our panel of genes into three major categories. (1) Significantly altered genes among HPV-negative, -inactive and -active tumors in all tumors - MCM2, TP53, MAL, KRT15, NOTCH1, CDKN2A. (2) Significantly altered genes among HPV-negative, -inactive and -active tumors within EA population - BRCA1, MAL, CDKN2A, TP53, MMP13, MMP10, TGFβ2, MET, MCM2, KRT15. (3) Lastly, we confirmed the differential expression profile of the three most significantly altered genes obtained from IPA biomarker analysis in AA vs. EA HPV negative tumors - IL1B, ADAM12 and MMP14.
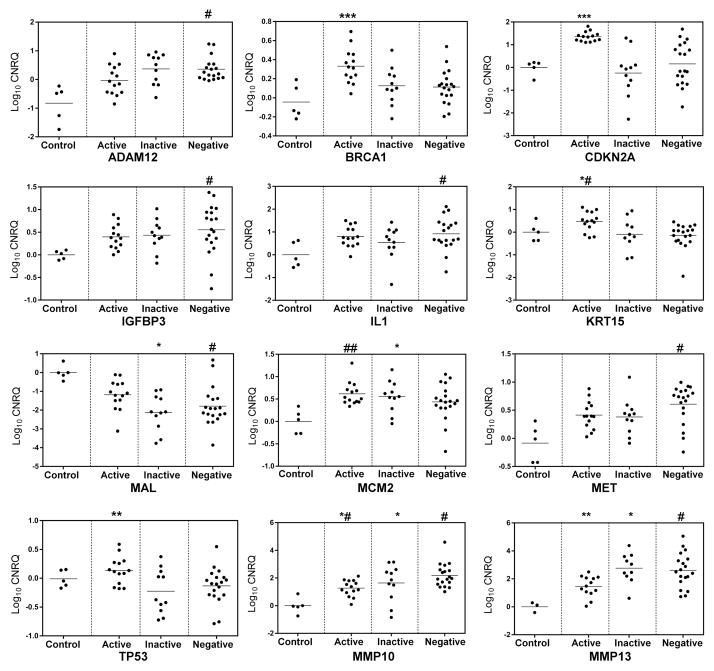
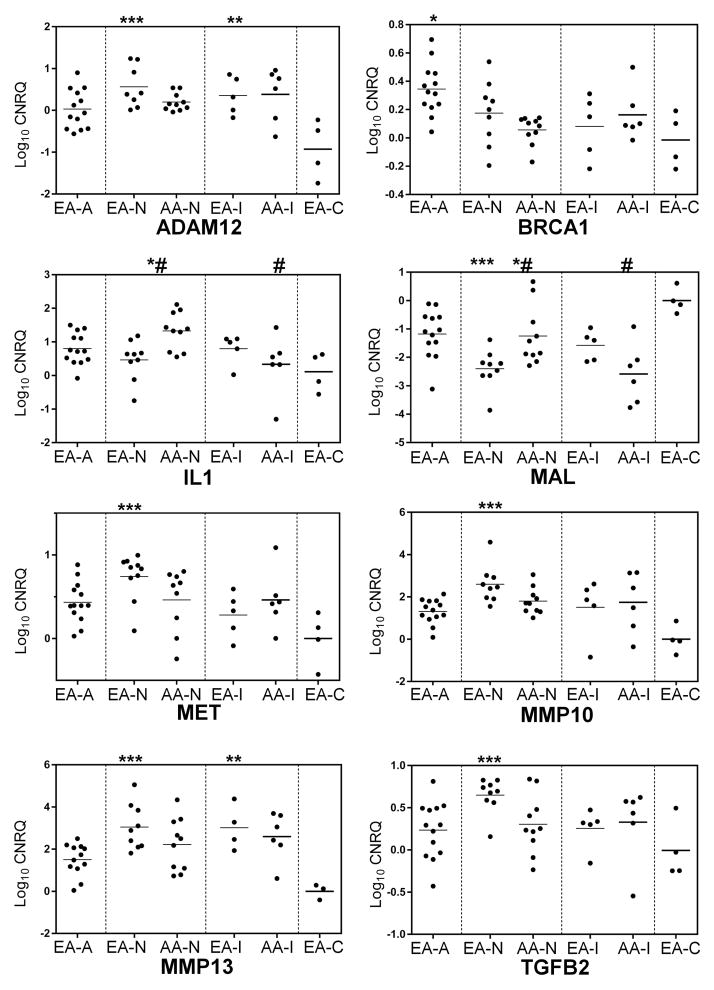
RT-qPCR assays confirmed the expression profiles of the selected DEGs (Figure 5) within HPV-active, HPV-inactive and HPV-negative tumors in the 36 tumor samples used for microarray analysis and in an additional set of 13 samples, for a total of 49 samples, including 5 normal and 44 tumor samples. The expression data for genes that were significantly differentially expressed in this sample set are shown in Figure 5 and Figure 6. As expected, HPV-active cancers expressed significantly higher levels of CDKN2A than HPV-negative tumors (Figure 5). We detected overexpression of BRCA1 and KRT15 in HPV-active in comparison with HPV-negative tumors; MMP10 expression was lower in HPV-active tumors and there was a significant difference in the expression of MMP13 between HPV-inactive and HPV-active tumors (Figure 5); TP53 was overexpressed in HPV-active vs. HPV-inactive tumors, and c-MET and MCM2 were overexpressed in all tumor samples with respect to controls, with a trend for the highest levels of c-MET expression in HPV-negative tumors (Figure 5). A similar behavior was observed for ADAM12, IL1, TGF-β2 and IGFBP3 (Figure 5). HPV-active tumors had the lowest levels of MAL as compared to HPV-negative and HPV-inactive tumors, while all the tumors showed down-regulation of MAL as compared to controls (Figure 5). Figure 6 shows the results of RT-qPCR for each of the selected DEGs, separated by HPV status and by race. c-MET was overexpressed in HPV-negative tumors in comparison with HPV-active tumors from EA patients (Figure 6). HPV-inactive and HPV-negative tumors in AA patients exhibited lower levels of c-MET RNA (Figure 6). A similar trend was observed for TGF-β2, with the highest levels of expression in HPV-negative tumors from EA patients (Figure 6). TP53 mRNA levels were high in HPV-active tumors from EA patients and similar to normal tissue in all other racial and HPV-status categories (Figure 6). Similarly, we observed significant up-regulation of RNA for the BRCA1 tumor suppressor gene in HPV-active tumors from EA patients, as compared to the HPV-negative and inactive tumors from both racial groups. In addition, expression levels of IL1B and MAL were significantly lower in HPV-negative tumors from EA patients than in HPV-negative tumors from AA patients (Figure 6).
Figure 5.
RT-qPCR validation of a panel of differentially expressed genes with significant inter-group variations. Log10 transformed cumulative normalized relative quantities (CNRQs) calculated through qbasePLUS are plotted by HPV status. Significant relationships determined by multiple testing are as follows: (***) Active/Control, Active/Inactive and Active/Negative; (**) Active/Inactive; (*) Inactive/Control; (*#) Active/Negative; (#) Negative/Control; (##) Active/Control
Figure 6.
Differential expression of selected genes in tissue samples from EA and AA patients. EA-A: European American Active, EA-N: European American Negative, EA-I European American Inactive, EA-C: European American Control. AA-N: African American Negative, AA-I: African American Inactive, (*) EA-/EA-C; (**) EA-I/EA-C; (***) EA-N/EA-C, (#) AA-N/AA-I; (*#) EA-N/AA-N.
Overall, the RT-qPCR results confirmed the microarray data as well as the IPA biomarker profile, and pointed to differences in gene expression between HPV-active and HPV-negative tumors within each racial group. These data also suggest more subtle differences between HPV-negative and HPV-inactive tumors, and between tumors derived from EA and AA patients.
Discussion
HPV is an established independent risk factor for the development of OPC’s and its detection in SCC of the oral cavity has come of age with the advent of RNA based assays to test HPV activity. In surgical pathology, immunohistochemistry for p16INK4a is a well-known surrogate marker for HPV expression in cervical cancers14 and HNC4. However, the sensitivity and specificity of this protein as a biomarker for HPV-driven HNC is highly debated15. This biomarker’s reliability for HPV-driven OPC increases in combination with DNA/RNA based assays for viral oncogenes35. A recent study classified OPC tumors into HPV-active (DNA+RNA+), -inactive (DNA+RNA−) and -negative (DNA−RNA−) based on multiplex nested PCR for HPV E6 DNA and E6I/E1^E4 RNA24.
Our approach of classifying tumors based on HPV status involved INNO-LiPA based HPV DNA detection and determination of E7 expression through RT-qPCR. Thus, tumors positive for both INNO-LiPA and E7 RT-qPCR were classified as HPV-active, INNO-LiPA positive but E7 RT-qPCR negative were classified as HPV-inactive and those which tested negative for both were classified as HPV-negative. Amongst the tumor samples which tested negative for HPV DNA by INNO-LiPA, one sample expressed HPV16 E7 mRNA and was hence categorized as HPV-active. We presume that the negative DNA result in this sample may be due to loss of the L1 ORF, perhaps a consequence of integration events. Our results are in line with conclusions drawn from other studies, indicating that HPV status of HNC cannot be defined solely by the presence of HPV DNA, rather a combination of several different assays is needed.15,36 The current consensus is that the presence of HPV DNA, in the absence of HPV gene expression, does not indicate a HPV-driven pathogenesis in OPC21,24.
The prevalence of HPV in our set of 56 OPC’s & oral cavity tumors was in agreement with previously reported data: AA patients exhibited more HPV-negative tumors as compared to EA patients21,37–39. 2While prevalence of HPV by DNA measures was about twice in EA as compared to AA patients, the prevalence of active HPV by RNA was over four times greater in EA than in AA patients. The observed differences in HPV prevalence might derive from differences in lifestyle, sexual practices and perhaps also genetic factors that render EA patients more susceptible to HPV infection in the oropharynx. These differences deserve further investigation.
Data are now available from landmark studies conducted to investigate the role of HPV in HNSCC with respect to gene expression patterns22, genetic and epigenetic profiles23,24, mutational landscape25,26 and more recently a comprehensive study on a small set of samples targeting the four ‘omics’: methylation, transcription, miRNA and genomic based analysis40. The molecular mechanisms governing HPV-mediated OPC are better understood in view of these studies. The novelty of our results lies in the differential expression patterns of OPC arising in AA and EA patients. Our analysis revealed subtle differences in gene expression between cancers from AA and EA patients, and between HPV-negative and HPV-inactive cancers. These differences will need to be further explored and corroborated in larger sample sets, as the small number of samples available to us is a limitation of this study. However, even in this small sample set, our observations provide a rationale for further investigation. For example, HPV in HPV-inactive cancers is usually dismissed as a “passenger” that has no bearing on the pathogenesis of the cancer. We propose that HPV-inactive tumors might have originated as HPV-active, and may have lost dependence on HPV during disease progression. This hypothesis is supported by our observations of subtle differences in gene expression between HPV-inactive and HPV-negative cancers, and by the reported observation that HPV-positive HNC tend to be HPV-active at initial presentation, whereas HPV-positive recurring HNC are more often inactive.21 If proven correct, this hypothesis has the potential to dramatically change the way we view the role of HPV not only in HNC but at other sites, and would also have implications for the potential of HPV vaccines in the prevention of HNC and other cancers. The idea that HPV-transformed cells may become independent of the continued expression of E6/E7 for growth at certain cancer sites is novel and deserves further investigation. Silencing of HPV sequences may occur by methylation in cells that have acquired mutations that can “replace” E6/E7 function. HPV methylation has been investigated in a variety of cancers and cancer cell lines (see41 for a recent review) however the role of HPV methylation and other mechanisms leading to a possible progression from HPV-dependence to HPV-independence in HNC remains to be determined.
Recent studies have highlighted the relevance of matrix metalloproteinases (MMP’s)42 and a disintegrin and metalloproteinase domain-containing protein 12 (ADAM12)43 in the tumor progression of HPV negative OPC’s. Our microarray studies detected about 25-fold up-regulation for MMP10 and 23-fold for MMP13 in HPV-negative tumors as compared to the HPV-active tumors in samples from EA patients. Differential expression profiles of MMP10 and MMP13 have been reported as potential biomarkers of disease progression in OPC44. In HPV-negative tumors, we observed up-regulation of ADAM12, MMP10 and MMP13, through microarray and RT-qPCR, in EA patients as compared to AA patients. We further confirmed the differential expression of three biomarkers (ADAM12, IL1B, MMP14), as given by IPA, through RT-qPCR in OPC samples from AA and EA patients.
Interleukin 1-beta (IL1B) has been reported to convert normal periodontal ligament fibroblasts to tumor associated fibroblasts and further cause EMT in SCC-25 co-cultures45. IL1B was recently proposed as a biomarker for oral squamous cell carcinoma46. Overexpression of IL1B was observed in HPV-negative tumors in AA patients compared to EA.
Myelin and lymphocyte-associated protein (MAL) is found to be down-regulated in HNC47 and a similar trend was observed in our OPC samples, with maximum down-regulation in HPV-negative tumors from EA patients, as compared to HPV-negative tumors from AA patients and HPV-active OPC. While the TP63 network and transcriptional targets of ΔNp63 isoforms were well represented in HPV-negative tumors from both racial groups, differential gene expression analysis revealed an emphasis on protein degradation profiles for OPC arising from EA patients (MMP10, MMP13, ADAM12) as compared to tumor suppressor and cytokine-based proliferation in AA patients (TGFβ2, IL1).
Another important biomarker considered in our analysis was the c-MET oncogene. The c-MET/HGF pathway has been well studied in HNC with c-MET overexpression reported in 80% of cases48. Recently, c-MET has been proposed as a potential cancer stem cell marker49. However, c-MET’s role with respect to HPV-positive OPC hasn’t yet been reported. Our study indicates differential expression patterns of c-MET, with highest c-MET levels in HPV-negative tumors as compared to HPV-inactive and HPV-active tumors. Additionally, HPV-negative tumors arising from EA patients expressed higher levels of c-MET when compared to those from AA patients. This difference confirms the EMT and proliferative nature of HPV-negative tumors, and also indicates that HPV-negative tumors from EA patients may be driven by c-MET to a larger extent than those in AA patients.
In conclusion, we observed a profile of differentially expressed biomarkers (ADAM12hi, MMP10hi, MMP13hi, METhi, IL1low, MALlow and TGFβ2hi) for HPV-negative tumors arising from EA patients as compared to those from AA patients. These biomarkers deserve further investigation as possible markers of invasive, EMT-based tumor phenotypes amenable to differential therapeutic interventions.
An important limitation of this study is the relatively small number of tumors and normal specimens that were available to us: this limited the power of some of our analyses and prevented us from directly examining HPV-active tumors from AA patients, performing a full comparison of tumor and normal specimens from EA and AA patients, or segregating samples based upon exact tissue site of origin. Nevertheless, we believe that our results warrant follow-up, as they point to the possibility that HPV-inactive tumors may begin as HPV-active lesions, and that in the course of tumor progression mutational events and/or epigenetic changes may redirect the tumor development process along lines more similar to those of HPV-negative tumors. If this is true, this finding will have major implications for the prevention of OPC, and also for the interpretation of conflicting reports on the presence of HPV at a variety of tumors sites, where the possibility of cancers beginning as HPV-active and then escaping the need for continued expression of E6/E7 for growth may also exist. Last, but not least, possible differences in tumor progression patterns between EA and AA patients also deserve further investigation.
Supplementary Material
Overall survival of patients with HPV-active oral cancer or OPC compare to HPV-negative and HPV-inactive cancers
Supplemental Figure 2. Experimental design of microarray studies
Supplemental Figure 3. Unsupervised hierarchical clustering of HPV active, inactive and negative oral cancer and OPC samples from European American patients
Supplemental Figure 4. A comparison of Gene Ontology profile from HPV active and HPV negative tumors
Supplemental Figure 5. A comparison of Gene Ontology profile from HPV negative and HPV inactive tumors from European American (EA) patients
Supplemental Table S1. Sequence of primers used in RT-qPCR
Supplemental Table S2. Demographical characteristics of OPC patients and HPV status of tumor samples
Supplemental Table S3. Characteristics of normal tissue samples. (*) Positive for HPV16 by INNO-LiPA; could not be tested for HPV activity due to low RIN
Supplemental Table S4. HPV type distribution in tumor samples
Supplemental Table S5. Demographics & baseline characteristics of patients whose tumor samples were used in microarray analysis
Supplemental Table S6. List of DEGs between HPV-active and HPV–inactive tumors
Supplemental Table S7. List of DEGs between HPV-active and HPV-negative tumors
Supplemental Table S8. List of DEGs between HPV-inactive and HPV-negative tumors
Supplemental Table S9. List of DEGs between HPV-negative tumors from AA and EA patients
Acknowledgments
This work was funded by grant 1P20MD001770 from the National Institutes of Health. Gene expression analysis by microarrays was conducted at the South Carolina College of Pharmacy/USC DNA Microarray Facility, which receives partial support from the Bioinformatics Core of SC INBRE (P20GM103499). The sponsors had no role in the design or execution of the experiments described in this paper, or in the interpretation of the results.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Curado MP, Boyle P. Epidemiology of head and neck squamous cell carcinoma not related to tobacco or alcohol. Curr Opin Oncol. 2013;25(3):229–234. doi: 10.1097/CCO.0b013e32835ff48c. [DOI] [PubMed] [Google Scholar]
- 3.Joseph AW, D’Souza G. Epidemiology of human papillomavirus-related head and neck cancer. Otolaryngol Clin North Am. 2012;45(4):739–764. doi: 10.1016/j.otc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus--associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24(5):736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 5.Cardesa A, Nadal A. Carcinoma of the head and neck in the HPV era. Acta Dermatovenerol Alp Panonica Adriat. 2011;20(3):161–173. http://www.ncbi.nlm.nih.gov/pubmed/22131117. [PubMed] [Google Scholar]
- 6.Gillison ML, D’Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 7.Beachler DC, D’souza G. Oral human papillomavirus infection and head and neck cancers in HIV-infected individuals. Curr Opin Oncol. 2013 doi: 10.1097/CCO.0b013e32836242b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen KL, Horner M-JD, Reed SG, et al. Head and neck cancer disparities in South Carolina: descriptive epidemiology, early detection, and special programs. [Accessed August 20, 2013];J S C Med Assoc. 2006 102(7):192–200. http://www.ncbi.nlm.nih.gov/pubmed/17319230. [PubMed] [Google Scholar]
- 9.Andersen A, Sølling AS, Ovesen T, Rusan M. The interplay between HPV and host immunity in head and neck squamous cell carcinoma. Acta Otorhinolaryngol Ital. 2013 doi: 10.1002/ijc.28411. [DOI] [PubMed] [Google Scholar]
- 10.Werness BA, Münger K, Howley PM. Role of the human papillomavirus oncoproteins in transformation and carcinogenic progression. [Accessed September 17, 2013];Important Adv Oncol. 1991 :3–18. http://www.ncbi.nlm.nih.gov/pubmed/1651285. [PubMed]
- 11.Al-Bakkal G, Ficarra G, McNeill K, Eversole LR, Sterrantino G, Birek C. Human papilloma virus type 16 E6 gene expression in oral exophytic epithelial lesions as detected by in situ rtPCR. [Accessed September 17, 2013];Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999 87(2):197–208. doi: 10.1016/s1079-2104(99)70273-8. http://www.ncbi.nlm.nih.gov/pubmed/10052376. [DOI] [PubMed] [Google Scholar]
- 12.Campisi G, Panzarella V, Giuliani M, et al. Human papillomavirus: its identity and controversial role in oral oncogenesis, premalignant and malignant lesions (review) [Accessed August 19, 2013];Int J Oncol. 2007 30(4):813–823. http://www.ncbi.nlm.nih.gov/pubmed/17332919. [PubMed] [Google Scholar]
- 13.Li Y, Nichols MA, Shay JW, Xiong Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. [Accessed September 17, 2013];Cancer Res. 1994 54(23):6078–6082. http://www.ncbi.nlm.nih.gov/pubmed/7954450. [PubMed] [Google Scholar]
- 14.Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. [Accessed September 17, 2013];Int J Cancer. 2001 92(2):276–284. doi: 10.1002/ijc.1174. http://www.ncbi.nlm.nih.gov/pubmed/11291057. [DOI] [PubMed] [Google Scholar]
- 15.Wilson DD, Rahimi AS, Saylor DK, et al. P16 Not a Prognostic Marker for Hypopharyngeal Squamous Cell Carcinoma. Arch Otolaryngol Head Neck Surg. 2012;138(6):556–561. doi: 10.1001/archoto.2012.950. [DOI] [PubMed] [Google Scholar]
- 16.Deng Z, Hasegawa M, Kiyuna A, et al. Viral load, physical status, and E6/E7 mRNA expression of human papillomavirus in head and neck squamous cell carcinoma. Head Neck. 2013;35(6):800–808. doi: 10.1002/hed.23034. [DOI] [PubMed] [Google Scholar]
- 17.Zandberg DP, Liu S, Goloubeva O, et al. Oropharyngeal cancer (OPC) drives racial outcome disparities in squamous cell carcinoma of the head and neck (HNSCC): Ten year experience at the university of maryland greenebaum cancer center (UMGCC) Head Neck. 2014 doi: 10.1002/hed.23933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brawley OW. Oropharyngeal cancer, race, and the human papillomavirus. Cancer Prev Res (Phila) 2009;2(9):769–772. doi: 10.1158/1940-6207.CAPR-09-0150. [DOI] [PubMed] [Google Scholar]
- 19.Cole L, Polfus L, Peters ES. Examining the incidence of human papillomavirus-associated head and neck cancers by race and ethnicity in the U.S., 1995–2005. PLoS One. 2012;7(3):e32657. doi: 10.1371/journal.pone.0032657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murdock JM, Gluckman JL. African-American and white head and neck carcinoma patients in a university medical center setting. Are treatments provided and are outcomes similar or disparate? [Accessed September 24, 2013];Cancer. 2001 91(1 Suppl):279–283. doi: 10.1002/1097-0142(20010101)91:1+<279::aid-cncr19>3.0.co;2-x. http://www.ncbi.nlm.nih.gov/pubmed/11148594. [DOI] [PubMed] [Google Scholar]
- 21.Weinberger PM, Merkley MA, Khichi SS, et al. Human papillomavirus-active head and neck cancer and ethnic health disparities. Laryngoscope. 2010;120(8):1531–1537. doi: 10.1002/lary.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slebos RJC, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(3 Pt 1):701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 23.Smeets SJ, Braakhuis BJM, Abbas S, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25(17):2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 24.Kostareli E, Holzinger D, Bogatyrova O, et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J Clin Invest. 2013;123(6):2488–2501. doi: 10.1172/JCI67010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoury JD, Tannir NM, Williams MD, et al. Landscape of DNA Virus Associations across Human Malignant Cancers: Analysis of 3,775 Cases Using RNA-Seq. J Virol. 2013;87(16):8916–8926. doi: 10.1128/JVI.00340-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13(1):134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustin Sa, Benes V, Garson Ja, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 31.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyeon D, Newton Ma, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlecht NF, Burk RD, Adrien L, et al. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213(3):283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 34.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5(5):489–500. doi: 10.1016/s1535-6108(04)00112-6. http://www.ncbi.nlm.nih.gov/pubmed/15144956. [DOI] [PubMed] [Google Scholar]
- 35.Schache AG, Liloglou T, Risk JM, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17(19):6262–6271. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holzinger D, Flechtenmacher C, Henfling N, et al. Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int J Cancer. 2013;133(6):1389–1399. doi: 10.1002/ijc.28142. [DOI] [PubMed] [Google Scholar]
- 37.Settle K, Posner MR, Schumaker LM, et al. Racial survival disparity in head and neck cancer results from low prevalence of human papillomavirus infection in black oropharyngeal cancer patients. Cancer Prev Res (Phila) 2009;2(9):776–781. doi: 10.1158/1940-6207.CAPR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chernock RD, Zhang Q, El-Mofty SK, Thorstad WL, Lewis JS. Human papillomavirus-related squamous cell carcinoma of the oropharynx: a comparative study in whites and African Americans. Arch Otolaryngol Head Neck Surg. 2011;137(2):163–169. doi: 10.1001/archoto.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worsham MJ, Stephen JK, Chen KM, et al. Improved survival with HPV among African Americans with oropharyngeal cancer. Clin Cancer Res. 2013;19(9):2486–2492. doi: 10.1158/1078-0432.CCR-12-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung aC, Job S, Ledrappier S, et al. A Poor Prognosis Subtype of HNSCC Is Consistently Observed across Methylome, Transcriptome, and miRNome Analysis. Clin Cancer Res. 2013;19(15):4174–4184. doi: 10.1158/1078-0432.CCR-12-3690. [DOI] [PubMed] [Google Scholar]
- 41.Johannsen E, Lambert PF. Epigenetics of human papillomaviruses. Virology. 2013:1–8. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koontongkaew S. The tumor microenvironment contribution to development, growth, invasion and metastasis of head and neck squamous cell carcinomas. J Cancer. 2013;4(1):66–83. doi: 10.7150/jca.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao VH, Kandel A, Lynch D, et al. A positive feedback loop between HER2 and ADAM12 in human head and neck cancer cells increases migration and invasion. Oncogene. 2012;31(23):2888–2898. doi: 10.1038/onc.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lallemant B, Evrard A, Combescure C, et al. Reference gene selection for head and neck squamous cell carcinoma gene expression studies. BMC Mol Biol. 2009;10:78. doi: 10.1186/1471-2199-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudás J, Fullár A, Bitsche M, et al. Tumor-produced, active interleukin-1β regulates gene expression in carcinoma-associated fibroblasts. Exp Cell Res. 2011;317(15):2222–2229. doi: 10.1016/j.yexcr.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamatani T, Shiogama S, Yoshihama Y, Kondo S, Shirota T, Shintani S. Interleukin-1 beta in unstimulated whole saliva is a potential biomarker for oral squamous cell carcinoma. [Accessed September 28, 2013];Cytokine. 2013 doi: 10.1016/j.cyto.2013.08.011. http://www.sciencedirect.com/science/article/pii/S1043466613006741. [DOI] [PubMed]
- 47.Cao W, Zhang Z-Y, Xu Q, et al. Epigenetic silencing of MAL, a putative tumor suppressor gene, can contribute to human epithelium cell carcinoma. Mol Cancer. 2010;9(1):296. doi: 10.1186/1476-4598-9-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burtness B, Bauman JE, Galloway T. Novel targets in HPV-negative head and neck cancer: overcoming resistance to EGFR inhibition. Lancet Oncol. 2013;14(8):e302–e309. doi: 10.1016/S1470-2045(13)70085-8. [DOI] [PubMed] [Google Scholar]
- 49.Sun S, Wang Z. Head neck squamous cell carcinoma c-Met+ cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer. 2011;129(10):2337–2348. doi: 10.1002/ijc.25927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overall survival of patients with HPV-active oral cancer or OPC compare to HPV-negative and HPV-inactive cancers
Supplemental Figure 2. Experimental design of microarray studies
Supplemental Figure 3. Unsupervised hierarchical clustering of HPV active, inactive and negative oral cancer and OPC samples from European American patients
Supplemental Figure 4. A comparison of Gene Ontology profile from HPV active and HPV negative tumors
Supplemental Figure 5. A comparison of Gene Ontology profile from HPV negative and HPV inactive tumors from European American (EA) patients
Supplemental Table S1. Sequence of primers used in RT-qPCR
Supplemental Table S2. Demographical characteristics of OPC patients and HPV status of tumor samples
Supplemental Table S3. Characteristics of normal tissue samples. (*) Positive for HPV16 by INNO-LiPA; could not be tested for HPV activity due to low RIN
Supplemental Table S4. HPV type distribution in tumor samples
Supplemental Table S5. Demographics & baseline characteristics of patients whose tumor samples were used in microarray analysis
Supplemental Table S6. List of DEGs between HPV-active and HPV–inactive tumors
Supplemental Table S7. List of DEGs between HPV-active and HPV-negative tumors
Supplemental Table S8. List of DEGs between HPV-inactive and HPV-negative tumors
Supplemental Table S9. List of DEGs between HPV-negative tumors from AA and EA patients