Abstract
Chromatin insulators are factors involved in higher-order, genome-wide organization of chromatin, and play key roles in regulating transcriptional programs. In this review, we discuss recent studies on the diverse composition of insulator complexes, and on the mechanism by which they establish long-range DNA interactions. Particularly, we describe new biophysical methods that allow for the study of the composition of large molecular complexes, and for defining the factors potentially required to establish long-range DNA contacts.
Keywords: chromatin organization, chromatin insulators, fluorescence correlation spectroscopy, single-molecule, transcription regulation
Eukaryotic chromosomes display several hierarchical levels of organization, from the nanoscale where nucleosomes orderly folds naked double-stranded DNA, to sub-megabase (Mb) scales where the chromatin fiber is condensed into separate, physically-isolated domains (also called topological domains, or TDs). TDs are defined by sub-Mb dense regions of chromatin showing high frequencies of self-interactions.1–5
Chromatin insulators are genetic elements implicated in nuclear organization and transcription regulation in eukaryotes. In Drosophila, 5 insulator families have been identified, that differ by their DNA-binding protein (insulator binding protein, or IBP): Suppressor of Hairy-wing [Su(Hw)],6 boundary element-associated factor (BEAF32),7 Zeste-white 5 (Zw5),8 the GAGA factor (GAF),9 and dCTCF,10 a distant sequence homolog of mammalian CTCF. Recently, chromatin insulators have been shown to play several important roles in the general regulation of transcription and in higher-order chromatin structure. First, IBPs bind thousands of sites genome-wide with a differential distribution, suggesting that different insulators may be involved in the regulation of distinct developmental programs.11-14 Second, insulators regulate transcription of distinct gene ontologies, separate distinct epigenetic chromatin states, and recruit H3K27me3 domains to Polycomb bodies.2,4,11,15 Third, insulators have been typically characterized for their ability to block interactions between enhancers and promoters through the formation of long-range contacts.16-21 Finally, IBPs and co-factors were recently shown to be overwhelmingly over-represented at frontiers between TDs2,4, strongly suggesting that these factors may play an important architectural role in the organization of higher-order chromatin.5
The genome-wide binding profiles of different IBPs often overlap with each other, suggesting that the locus-specific composition of insulator complexes may play a role in their function.5 Most, if not all, insulators share the common Centrosomal Protein 190 (CP190) and/or one of the Mod(mdg4) isoforms as co-factors. CP190 is a protein found only in Drosophila and was originally described for its ability to bind to the centrosome during mitosis.22 CP190 also plays a central role in the insulation function of various IBPs. A large proportion of CP190 binding sites (∼50%) correlate with the presence of BEAF32, and both factors are enriched at borders between TDs.11,13,23 Another factor, Chromator (also known as Chriz/Chro), was also recently found to be overrepresented at those borders shared by BEAF32 and CP1902. Chromator forms a molecular spindle matrix during mitosis with other nuclear-derived factors (Skeletor and Megator),24 localizes to inter-band regions of polytene chromosomes, and plays a role in their structural regulation as well as in transcriptional regulation during interphase.25
Recently, we used novel biophysical approaches to investigate the molecular associations of insulator proteins and the role of different factors in the formation of specific long-range interactions (LRIs).26 First, we revealed that BEAF32 forms a molecular complex with CP190 or Chromator, by performing co-immunoprecipitation (Co-IP) and electrophoretic mobility shift assays (EMSA) on purified proteins or S2 nuclear extracts. BEAF32 interactions with those proteins required the C-terminal domains of CP190 and Chromator. Those complexes were also characterized using Fluorescence Correlation Spectroscopy (FCS). FCS allows for the measurement of the diffusion time of fluorescently-labeled molecules (DNA or protein) moving across a confocal detection volume (Fig. 1a). Upon binding of proteins to fluorescently-labeled DNA fragments, the complex increases in size, which is reflected in an increase in its apparent diffusion time (Fig. 1a-d). Using this technique, we showed that BEAF32 binds specifically to DNA fragments containing its recognition binding site, while neither CP190 nor Chromator were able to form stable complexes under similar conditions. The addition of CP190 or Chromator to preformed BEAF32-DNA complexes led to an increase in the diffusion time, consistent with the binding of CP190/Chromator to BEAF32. Overall, these data indicated that BEAF32 specifically interacts with CP190 and Chromator, but could not inform us on which factors may be required for the formation of LRIs.
Figure 1.
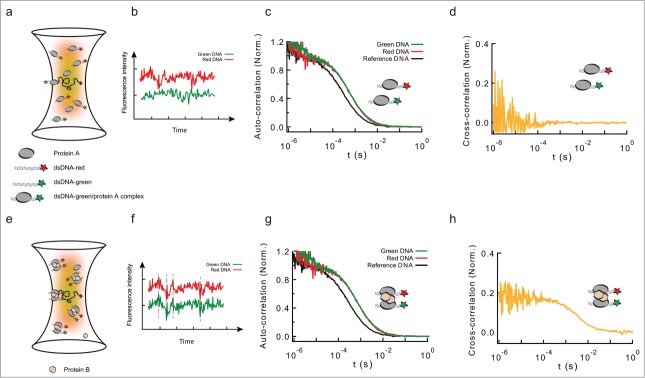
Formation of long-range interactions by insulator proteins studied by PIE-FCCS. (a) Scheme depicting a typical fluorescence cross-correlation fluctuation spectroscopy (FCCS) configuration. Two fluorescently-labeled dsDNA fragments (black ribbons with green or red stars) are specifically bound by a DNA-binding protein (protein A, gray ellipse), and diffuse in and out of an excitation volume (yellow to red gradient). (b) This diffusion produces a time-dependent fluctuation in the fluorescence signals of the green and red-labeled dsDNA fragments (red and green traces). These fluctuations are independent (uncorrelated) if the red and green-labeled dsDNA fragments are not in the same molecular complex. (c) Auto-correlation functions of free dsDNA (no protein, black, could represent either green or red-labeled dsDNA), and of protein-DNA complexes formed by the interaction of protein A with green- and red-labeled DNA (green and red solid lines, respectively). As the shapes of both complexes is the same, the autocorrelation curves are undistinguishable. Binding of protein A leads to a discrete shift in the curve (compare black and red/green curves). (d) Cross-correlation function between the intensity fluctuation signals from protein-DNA complexes formed by the binding of protein A (gray ellipse) to green/red-labeled dsDNA. Absence of cross-correlation is due to the lack of correlated movement of the 2 probes, and indicates that green and red-labeled dsDNA are not in the same molecular complex. (e) Schematic representation of molecular complexes in which protein B (pink hexagon) bridges green- and red-labeled dsDNA fragments pre-bound by protein A. (f) The diffusion of these large complexes in and out of the excitation volume leads to a correlated time-dependent fluctuation in the fluorescence signals of the green and red channels (green and red traces). These fluctuations are correlated at different time-scales. (g) Auto-correlation functions of free dsDNA, and of protein-DNA complexes formed by the interaction of protein B with green- and red-labeled DNA fragments pre-bound by protein A. Even when both dsDNA fragments are part of the same molecular complex, the auto-correlation function displays a single shift in both colors. The size of the shift is larger than in panel (c), due to the increased size of the protein-DNA complex. (h) Cross-correlation function between the intensity fluctuation signals from protein-DNA complexes formed by the formation of a complex involving both green- and red-labeled dsDNA. A positive cross-correlation signal at different time-scales indicates protein-mediated interactions between green and red-dsDNA.
In order to determine which factors may be required to form LRI, we adapted a novel single-molecule assay based on fluorescence cross-correlation spectroscopy (FCCS). FCCS measures the correlated fluorescence intensity fluctuations of 2 spectrally-distinct, fluorescently-labeled molecules to quantitatively determine whether they are in the same molecular complex (Fig. 1a-b). In our assay, we tested for the formation of protein-mediated LRIs by measuring the correlated fluctuations in the 2 channels of 2 different double-stranded (ds) DNA molecules labeled with different fluorophores. Correlated fluctuations was a signature of protein-mediated LRI interactions between 2 dsDNA molecules (Fig. 1e-h). On the contrary, absence of correlation indicated independent diffusion of the probes in the detection volume (Fig. 1a-d). The experimental setup used for our experiments implemented Pulse Interleaved Excitation (PIE) combined with Time Correlated Single Photon Counting (TCSPC) detection, 2 features permitting minimal crosstalk between fluorophores with single-molecule sensitivity.27 Using this approach, we first showed that despite its ability to bind DNA specifically, BEAF32 is not sufficient to mediate LRI in vitro. In contrast, the addition of CP190 or Chromator to pre-formed BEAF32-DNA complexes promoted the formation of intermolecular LRIs. This ability of CP190 and Chromator to establish LRIs in vitro required specific contacts between BEAF32 and their C-terminal domains, consistent with our Co-IP and EMSA results. Importantly, the C-terminal domain of CP190 was not able to establish the molecular contacts required to form specific LRIs, suggesting that this function may be encoded within the N-terminus of CP190, formed by a BTB/POZ and a zinc-finger domain.
To test this hypothesis, we solved the high-resolution structure of the BTB/POZ domain of CP190, which forms strict homo-dimers with a large contact surface. We reasoned that the formation of LRIs may necessitate CP190-CP190 interactions mediated by BTB/POZ. To test this idea, we added CP190-BTB/POZ to pre-formed BEAF32-CP190-BEAF32 complexes in trans. In these experiments, the cross-correlation signal indicative of the formation of LRIs gradually decreased with the concentration of CP190-BTB/POZ, consistent with this domain being responsible for the molecular glue holding distant DNA sites together. Our model for insulator function suggests that BEAF32/dCTCF/Su(HW) provide DNA specificity (first layer proteins) whereas CP190/Chromator are responsible for the physical interactions required for the formation of long-range contacts (second layer).
This biophysical approach shows great potential to dissect the minimal number of factors required for the formation of LRIs in vitro. Importantly, when combined with specific mutations or deletions, this method can be used to determine the molecular mechanism involved in the formation of these contacts: what proteins domains/regions provide DNA-binding specificity, which may be responsible for bridging, or what is the role of protein-protein interactions. The combination of this methodology with structural-based methods will permit, in future, the elucidation of the structural determinants of these mechanisms.
Very recently, the composition of insulator complexes responsible for LRIs was studied genome-wide.28 This study showed the existence of ‘indirect peaks’, defined as sequences enriched in IBPs but devoid of insulator sequences. Indirect peaks were shown to highlight a network of long-range contacts among distinct IBP sites through their common cofactor, CP190. Distinct IBP mutants, in which interactions with CP190 were prevented, revealed that the expression of distant genes associated with indirect peaks was impaired. These results highlight the importance of CP190 in mediating LRIs through recognition of IBPs and further support our model for insulator function in which first layer insulators (BEAF32/dCTCF/Su(HW)) provide DNA specificity while second layer co-factors (CP190/Chromator) provide the physical interactions required for the establishment of long-range contacts.
Despite the finding that CP190 directly interacts with several IBPs (e.g. BEAF32, Su(HW), dCTCF), many of the CP190 binding sites genome-wide were unaccounted for. Over the past year, 4 new proteins were shown to interact with CP190 and to possess insulator function. Ibf1 and Ibf2 were found to localize to insulator bodies, where IBPs of different classes are brought together, and associate with chromatin at CP190-binding sites throughout the genome.29 The novel IBPs Pita and ZIPIC were also found to interact with CP190 and possess a partial enhancer-blocking activity.30 The interaction between CP190 and Pita/ZIPIC is, however, mediated by different CP190 domains: while ZIPIC interacts with the centrosomal targeting domain of CP190, Pita interacts with the BTB/POZ domain of CP190. These studies, together with our own results, suggest that CP190-BTB/POZ may have a multifunctional role in participating of different molecular complexes, as well as in providing the molecular glue required to bridge long-range interactions. The dual roles of this domain in the formation of LRIs and in protein-protein interactions may be important in providing a mechanism of regulation of CP190-dependent LRIs.
Interestingly, a recent study shed new insight into the possible actors involved in the role of CP190-dependent insulators in the opening of heterochromatin.31 This study showed that CP190 binding to dCTCF or to other IBPs mediate recruitment of the nucleosome remodeling factor NURF and the complex dREAM to insulator sites.31 NURF remodels chromatin by promoting nucleosome sliding or ejection and was shown to promote transcriptional activation or repression of target genes. It is thus a good candidate to mediate the nucleosomal depletion necessary for chromatin opening at CP190-dependent insulators. dCTCF or CP190, together with unknown factors, might prepare the epigenetic landscape such that dREAM or NURF target specific chromatin modifications at insulator sites.31
A second potential pathway for heterochromatin opening may involve Z4 and Chromator. A subset of CP190 binding sites correlate with the binding pattern of Putzig/Z4 and Chromator,31 while Z4 associates with the Chromator complex to recruit the kinase JIL-1, which is key in defining de-condensed domains of larval polytene chromosomes.32-34 Importantly, JIL-1 participates in a complex histone modification network that characterizes active, de-condensed chromatin, and is thought to reinforce the status of active chromatin through the phosphorylation of histone H3 at serine 10 (H3S10). Taken together, these and our data suggest that BEAF32, in complex with CP190, may be responsible for the recruitment of the Chromator/JIL-1 complex to active chromatin domains to prevent heterochromatin spreading and/or in chromatin opening. This mechanism would be consistent with the observation that BEAF32 localizes primarily to de-condensed chromatin regions in polytene chromosomes, is implicated in the regulation of active genes and frequently delimits the boundaries of chromatin silencing. Interestingly, a very recent study showed that the targeting of CP190 to dCTCF binding sites within a condensed chromatin locus leads to large-scale unfolding of the local chromatin structure.35 dCTCF was insufficient to cause locus-specific chromatin de-condensation, but was required to recruit CP190 (and probably other factors essential for chromatin opening) to provide genomic specificity. But, what specific roles do insulators play in genome-wide chromatin organization?
Recent studies have shown that insulator proteins are over-represented at barriers between TDs,2,4 leading to the suggestion that insulators may play an important architectural role in the organization of higher-order chromatin.5,36 The mechanisms by which insulators may delimit genomic interactions between proximal TDs is unknown, but several distinct models have been put forward (Fig. 2): (1) proximal TDs may be brought together in a rosetta-like structure;5 (2) the 2 barriers of a single TD may be joined together by insulator-mediated interactions; (3) the borders of distant TDs could be brought physically together despite being genomically distant. The specific action of insulators may depend on the chromatin context or genomic locus, so that these mechanisms may simultaneously co-exist within the cell. Further work will be required to specifically test these models to more precisely define the role of insulators as architectural factors.36
Figure 2.
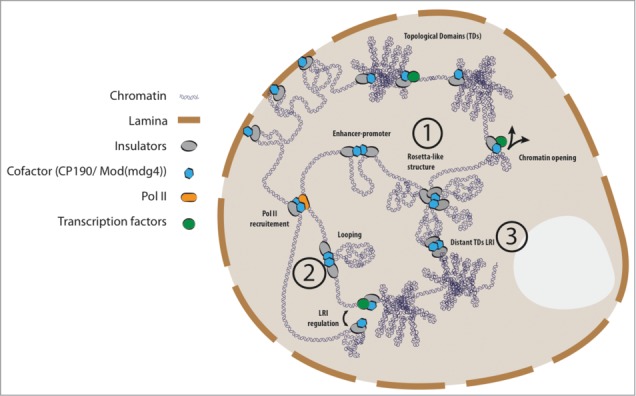
Schematic representation of models for the putative roles of chromatin insulators in nuclear organization. Insulator proteins have been ascribed many different functions, schematically represented in the figure (enhancer-blocking, isolation of TDs, chromatin opening, attachment of heterochromatin to lamina, recruitment of RNAPII). See annotations in main text for possible mechanisms by which insulators may organize higher-order chromatin (models 1–3).
The model proposing distinct roles for IBPs (BEAF32, dCTCF, etc) and co-factors (CP190/Mod(mdg4)) combined with the preferential, genome-wide localization of insulator proteins on barriers between TDs suggest a role for this multi-layer organization in the establishment of transcriptional states throughout the cell cycle. First layer proteins remain bound to chromatin at all stages of the cell cycle.7,37 In contrast, both CP190 and Chromator seem to be bound to chromatin specifically during interphase, and display a dramatic cellular re-localization during mitosis: CP190 strongly binds to centrosomes while Chromator co-localizes with the spindle matrix.22,25 Thus, the dissociation and cellular redistribution of second layer insulator proteins during cell division could be responsible for the massive remodeling of chromosome architecture occurring during mitosis, and for the re-establishment of higher-order contacts at the onset of interphase.38 In contrast, first layer insulator proteins may act as anchor points for the re-establishment of higher-order interactions after mitosis, and for the maintenance of the transcriptional identity of TDs. Thus, this model suggest distinct roles for insulator binding proteins and co-factors in actively re-shaping the organization of chromatin into TDs during the cell cycle. This model is consistent with recent genome-wide data suggesting that, overall, first layer insulator proteins remain bound to their binding sites during mitosis, whereas second layer insulator proteins tend to show a large change in binding patterns,37,39
Interestingly, recent Hi-C studies on cells synchronized in G1 or metaphase showed that topological domain organization describes the organization of chromatin in G1 chromosomes, but during metaphase this organization is dramatically disturbed.38 Future single-cell Hi-C40 and microscopy methods may enlighten us on the specific roles of the different insulator factors in the remodeling of topological domain structures throughout cell division and on the specific roles played by these important proteins in regulating transcription.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by funding from the European Research Council under the 7th Framework Program (FP7/2007–2013) to M.N (ERC grant agreement 260787).
References
- 1. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376-80; PMID:22495300; http://dx.doi.org/ 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sexton T, Yaffe E, Kenigsberg E, Bantignies FDR, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the drosophila genome. Cell 2012; 148:458-72; PMID:22265598; http://dx.doi.org/ 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 3. Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012; 485:381-5; PMID:22495304; http://dx.doi.org/ 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hou C, Li L, Qin Z, Corces V. Gene density, transcription, and insulators contribute to the partition of the drosophila genome into physical domains. Mol Cell 2012; 48:471-84; PMID:23041285; http://dx.doi.org/ 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Bortle K, Nichols MH, Li L, Ong C-T, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol 2014; 15:R82; PMID:24981874; http://dx.doi.org/ 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geyer PK, Corces VG. DNA position-specific repression of transcription by a drosophila zinc finger protein. Genes Dev 1992; 6:1865-73; PMID:1327958; http://dx.doi.org/ 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- 7. Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 1995; 81:879-89; PMID:7781065; http://dx.doi.org/ 10.1016/0092-8674(95)90008-X [DOI] [PubMed] [Google Scholar]
- 8. Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev 1999; 13:2098-107; PMID:10465787; http://dx.doi.org/ 10.1101/gad.13.16.2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maeda RK, Karch F. Making connections: boundaries and insulators in drosophila. Curr Opin Genet Dev 2007; 17:394-9; PMID:17904351; http://dx.doi.org/ 10.1016/j.gde.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from drosophila to humans and confers enhancer blocking of the fab-8 insulator. EMBO Rep 2005; 6:165-70; PMID:15678159; http://dx.doi.org/ 10.1038/sj.embor.7400334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bushey AM, Ramos E, Corces VG. Three subclasses of a drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 2009; 23:1338-50; PMID:19443682; http://dx.doi.org/ 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 2009; 28:877-88; PMID:19229299; http://dx.doi.org/ 10.1038/emboj.2009.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the drosophila genome. PLoS Genet 2010; 6:e1000814; PMID:20084099; http://dx.doi.org/ 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a drosophila insulator. Mol Cell 2011; 44:29-38; PMID:21981916; http://dx.doi.org/ 10.1016/j.molcel.2011.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Kas E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol 2008; 6:2896-910; PMID:19108610; http://dx.doi.org/ 10.1371/journal.pbio.0060327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 2003; 17:664-75; PMID:12629048; http://dx.doi.org/ 10.1101/gad.1052003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dean A. In the loop: long range chromatin interactions and gene regulation. Brief Funct Genomics 2011; 10:3-10; PMID:21258045; http://dx.doi.org/ 10.1093/bfgp/elq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA 2011; 108:9566-71; PMID:21606361; http://dx.doi.org/ 10.1073/pnas.1019391108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 2006; 7:703-13; PMID:16909129; http://dx.doi.org/ 10.1038/nrg1925 [DOI] [PubMed] [Google Scholar]
- 20. Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, Georgiev P. Orientation-dependent interaction between drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res 2008; 36:7019-28; PMID:18987002; http://dx.doi.org/ 10.1093/nar/gkn781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev 2007; 17:400-7; PMID:17913488; http://dx.doi.org/ 10.1016/j.gde.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oegema K, Whitfield WG, Alberts B. The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol 1995; 131:1261-73; PMID:8522588; http://dx.doi.org/ 10.1083/jcb.131.5.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res 2012; 22:2176-87; PMID:22722341; http://dx.doi.org/ 10.1101/gr.136788.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rath U, Ding Y, Deng H, Qi H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. The chromodomain protein, chromator, interacts with JIL-1 kinase and regulates the structure of drosophila polytene chromosomes. J Cell Sci 2006; 119:2332-41; PMID:16723739; http://dx.doi.org/ 10.1242/jcs.02960 [DOI] [PubMed] [Google Scholar]
- 25. Rath U, Wang D, Ding Y, Xu YZ, Qi H, Blacketer MJ, Girton J, Johansen J, Johansen KM. Chromator, a novel and essential chromodomain protein interacts directly with the putative spindle matrix protein skeletor. J Cell Biochem 2004; 93:1033-47; PMID:15389869; http://dx.doi.org/ 10.1002/jcb.20243 [DOI] [PubMed] [Google Scholar]
- 26. Vogelmann J, Le Gall A, Dejardin S, Allemand F, Gamot A, Labesse G, Cuvier O, Nègre N, Cohen-Gonsaud M, Margeat E, et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the drosophila genome. PLoS Genet 2014; 10:e1004544; PMID:25165871; http://dx.doi.org/ 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olofsson L, Margeat E. Pulsed interleaved excitation fluorescence spectroscopy with a supercontinuum source. Opt Express 2013; 21:3370-8; PMID:23481797; http://dx.doi.org/ 10.1364/OE.21.003370 [DOI] [PubMed] [Google Scholar]
- 28. Liang J, Lacroix L, Gamot A, Cuddapah S, Queille S, Lhoumaud P, Lepetit P, Martin PGP, Vogelmann J, Court F, et al. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and pol II pausing. Mol Cell 2014; 53:672-81; PMID:24486021; http://dx.doi.org/ 10.1016/j.molcel.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuartero S, Fresán U, Reina O, Planet E, Espinàs ML. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J 2014; 33:637-47; PMID:24502977; http://dx.doi.org/ 10.1002/embj.201386001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maksimenko O, Bartkuhn M, Stakhov V, Herold M, Zolotarev N, Jox T, Buxa MK, Kirsch R, Bonchuk A, Fedotova A, et al. Two new insulator proteins, pita and ZIPIC, target CP190 to chromatin. Genome Res 2014; 25:89-99; PMID:25342723; http://dx.doi.org/ 10.1101/gr.174169.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bohla D, Herold M, Panzer I, Buxa MK, Ali T, Demmers J, Krüger M, Scharfe M, Jarek M, Bartkuhn M, et al. A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS ONE 2014; 9:e107765; PMID:25247414; http://dx.doi.org/ 10.1371/journal.pone.0107765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang C, Cai W, Li Y, Deng H, Bao X, Girton J, Johansen J, Johansen KM. The epigenetic H3S10 phosphorylation mark is required for counteracting heterochromatic spreading and gene silencing in drosophila melanogaster. J Cell Sci 2011; 124:4309-17; PMID:22247192; http://dx.doi.org/ 10.1242/jcs.092585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Regnard C, Straub T, Mitterweger A, Dahlsveen IK, Fabian V, Becker PB. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet 2011; 7:e1001327; PMID:21423663; http://dx.doi.org/ 10.1371/journal.pgen.1001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y, Cai W, Wang C, Yao C, Bao X, Deng H, Girton J, Johansen J, Johansen KM. Domain requirements of the JIL-1 tandem kinase for histone H3 Serine 10 phosphorylation and chromatin remodeling in vivo. J Biol Chem 2013; 288:19441-9; PMID:23723094; http://dx.doi.org/ 10.1074/jbc.M113.464271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ahanger SH, Günther K, Weth O, Bartkuhn M, Bhonde RR, Shouche YS, Renkawitz R. Ectopically tethered CP190 induces large-scale chromatin decondensation. Sci Rep 2014; 4:3917; PMID:24472778; http://dx.doi.org/ 10.1038/srep03917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gómez-Díaz E, Corces VG. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol 2014; 24(11); PMID:25218583; http://dx.doi.org/ 10.1016/j.tcb.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Sung E, Donlin-Asp PG, Corces VG. A subset of drosophila myc sites remain associated with mitotic chromosomes colocalized with insulator proteins. Nat Commun 2013; 4:1464; PMID:23403565; http://dx.doi.org/ 10.1038/ncomms2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science 2013; 342:948-53; PMID:24200812; http://dx.doi.org/ 10.1126/science.1236083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gurudatta BV, Yang J, Van Bortle K, Donlin-Asp PG, Corces VG. Dynamic changes in the genomic localization of DNA replication-related element binding factor during the cell cycle. Cell Cycle 2013; 12:1605-15; PMID:23624840; http://dx.doi.org/ 10.4161/cc.24742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013; 502:59-64; PMID:24067610; http://dx.doi.org/ 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]


