Abstract
Receptor for hyaluronic acid (HA)-mediated motility (RHAMM) is a nonintegral cell surface receptor involved in the aggressive phenotype in a wide spectrum of human malignancies, but the significance of RHAMM in pancreatic ductal adenocarcinoma (PDAC) remains unknown. In this study, we investigated the expression of RHAMM and its clinical relevance in PDAC. RHAMM mRNA expression was examined in 8 PDAC cell lines and in primary pancreatic cancer and adjacent non-tumor tissues from 14 patients using real-time RT-PCR. Western blotting was carried out to analyze the expression of RHAMM protein in PDAC cell lines. We also investigated the expression patterns of RHAMM protein in tissue samples from 70 PDAC patients using immunohistochemistry. The RHAMM mRNA expression was increased in some PDAC cell lines as compared to a non-tumorous pancreatic epithelial cell line HPDE. The RHAMM mRNA expression was significantly higher in PDAC tissues as compared to corresponding non-tumorous pancreatic tissues (P < 0.0001). The RHAMM protein expression was higher in the vast majority of PDAC cell lines relative to the expression in HPDE. The immunohistochemical analysis revealed strong expression of RHAMM in 52 (74%) PDAC tissues. Strong expression of RHAMM was significantly associated with a shorter survival time (P = 0.038). In multivariate analysis, tumor stage (P = 0.039), residual tumor (P = 0.015), and strong RHAMM expression (P = 0.034) were independent factors predicting poor survival. Strong expression of RHAMM may predict poor survival in PDAC patients and may provide prognostic and, possibly, therapeutic value.
Keywords: Receptor for hyaluronic acid-mediated motility, pancreatic ductal adenocarcinoma, prognostic factor, poor survival
Introduction
HA is a large glycosaminoglycan composed of repeating β-1,4-linked D-glucuronic acid and β-1,3-N-acetyl-D-glucosamine disaccharide units that accumulates in the extracellular matrix (ECM) 1. In cancerous tissue, HA is secreted from stromal fibroblasts in response to humoral factors derived from tumor cells 2, promoting tumor transformation, metastasis 3, 4, and cell motility and migration 5-8. In our previous study 9, we demonstrated a relationship between increased expression of HA and poor prognosis in patients with PDAC. Recently, HA receptors have been detected on tumor cells.
One of the HA receptors, RHAMM, was first described by Turley 10; this receptor was originally identified as part of a multimeric complex (HARC) that regulates HA-induced motility of H-ras-transformed fibroblasts 8 and is a peptide in the supernatants from proliferative fibroblasts 5 with many intracellular and extracellular functions. On the cell surface, RHAMM binds with CD44, and HA binding to this complex stimulates downstream signaling that activates RhoA-activated protein kinase and the MAPK/extracellular signal regulated protein kinase (ERK) 1/2 pathway, which results in the expressions of genes that are required for motility and invasion in various cancers 11-13. RHAMM, which is a cancer-associated antigen, alone or in complex with CD44, osteopontin, and integrins is involved in tumorigenesis, progression, invasion and metastasis through interactions with signaling cascades related to ERK1/2, breast cancer 1 (BRCA1), BRCA1-associated RING domain 1, tyrosine and serine/threonine kinases, PDGF receptor, Src and Erk MAP kinases 6, 14-22. Intracellular RHAMM, which is expressed in a cell cycle-dependent manner, has been described as an actin- and microtubule-associated protein that forms a complex with the dynein molecular motor that localizes to the centrosome to maintain spindle integrity 23-26.
In vitro, RHAMM is expressed in many mammalian cell types, including fibroblasts, smooth muscle cells, endothelial cells, macrophages, sperm, nerve cells, and several tumor cell types 27. Clinically, RHAMM expression has been detected in patients with glioma, breast cancer, urinary bladder carcinoma, endometrial carcinoma, PDAC, hepatocellular carcinoma, colorectal cancer, prostate cancer, gastric cancer, lung cancer, and aggressive fibromatosis (i.e., desmoid tumor) 19, 28-37. In this study, we attempted to investigate the mRNA and protein expression levels of RHAMM in PDAC cell lines and tissues.
Materials and methods
Cell culture
Eight PDAC cell lines were used: AsPC-1, BxPC-3, Panc-1, Capan-2, and CFPAC1 (ATCC, Manassas, VA, USA); SUIT-2 and KP-2 (JCRB Cell Bank, Osaka, Japan); and NOR-P1 (RIKEN BRC Cell Bank, Tsukuba, Ibaraki, Japan). An immortalized cell line derived from human pancreatic duct, HPDE, was a kind gift from Dr. M.S. Tsao (Univ. of Toronto, Canada). All PDAC cell lines were maintained in RPMI-1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin and HPDE was maintained in HuMedia-KG2 (KURABO, Osaka, Japan) at 37°C in a humidified atmosphere with 5% CO2.
Patients
Archival tissue specimens were collected from 70 PDAC patients who underwent surgical resection between 1982 and 2011 in our department. Clinicopathological characteristics of these patients have been described previously 9. The study was approved by the ethical committee of the university of occupational and environmental health, and written informed consent was obtained from all patients.
Real-time RT- PCR
Total RNA was isolated from all cells and from 14 matched pairs of primary pancreatic tumor and adjacent non-tumor tissues using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Single-stranded cDNA was synthesized from 1.0μg of total RNA. Real-time mRNA expression analysis of RHAMM (Hs00234864_m1) and a housekeeping gene GAPDH (Hs02758991_g1) for control was performed using TaqMan@ Gene Expression Assays and Step One Plus real-time PCR instrument (Applied Biosystems, Foster, CA, USA) according to the manufacture's instruction.
Western blotting
Cultured PDAC and HPDE cells were rinsed once with cold PBS and scrapped in 400µl of PRO-PREP protein extraction solution ( iNtRON Biotechnology, Gyeonggi-do, South Korea) at -20°C for 30 min. After centrifugation at 13000rpm for 5 min at 4°C, the protein concentrations were quantified with a BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, USA). Proteins were then denatured in 6xSDS-PAGE sample buffer solution without reducing reagent (Nacalai Tesque, Kyoto, Japan) for 5 min at 95°C. The same amounts of proteins were subjected to electrophoresis in a 10% Mini-PROTEAN Precast Gel (Bio-Rad, Philadelphia, PA, USA) and transferred onto a poly vinylidene difluoride (PVDF) membrane (ATTO, Tokyo, Japan). The membranes were blocked with 5 % non-fat dried milk in Tris-buffered saline-Tween (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1 % Tween 20) for 1 h at room temperature and incubated with a primary antibody against RHAMM (1:1000; LifeSpan Biosciences, Seattle, WA, USA) or β-actin (1:2000; Sigma-Aldrich, St Louis, MO, USA) overnight at 4°C or for 1 h at room temperature, followed by incubation with an anti-rabbit IgG, horseradish peroxidase-linked species-specific whole antibody (1:1000; GE Healthcare, Little Chalfont, Buckinghamshire, UK) or an anti-mouse IgG, horseradish peroxidase-linked species-specific whole antibody (1:2000; GE Healthcare) for 1 h at room temperature after repeated washing with Tris-buffered saline-Tween. The immunopositive bands were detected by chemiluminescence using the ECL prime western blotting detection reagent (GE Healthcare) and quantified by light-capture with the CS analyzer (ATTO).
Immunohistochemistry and scoring
The PDAC tissues were incubated with anti-RHAMM IgG (Abcam, Cambridge, MA, USA) at a 1:100 dilution for 2 h at room temperature. The sections were incubated for 10 min with biotinylated anti-rabbit antibody for RHAMM according to the manufacture's instruction of Histofine SAB-PO (R) kit (Nichirei Biosciences Inc, Tokyo, Japan). The positive control was archival tissue from a normal human testis. The scoring method, which combined intensity and the percentage of positivity, was previously described 9. The slides were scored by two individual researchers (N.S. and X.C.) on a blinded basis. The extent and intensity measures for each score were combined to identify “weak” expression or “strong” expression.
Statistical analysis
The statistical analyses were performed with SPSS statistical software (version 21.0; SPSS, Inc., Chicago, IL, USA). The data are expressed as the mean ± SD. The differences in the RHAMM mRNA levels among the pancreatic tissues (normal versus cancer) were compared using the Wilcoxon signed rank test. The survival curves were constructed according to the Kaplan-Meier method and were compared using the log-rank test. Cox proportional hazards regression analysis was used to evaluate independent prognostic factors associated with survival. Statistical significance was accepted when P < 0.05. All p values are two-tailed. The experiments performed in this study were repeated three independent times.
Results
RHAMM mRNA expression in cell lines and tissues
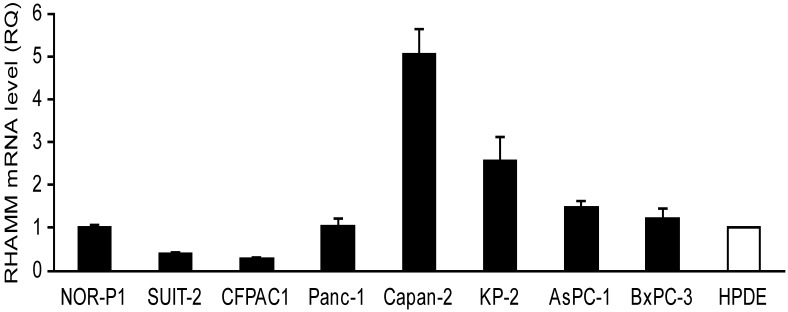
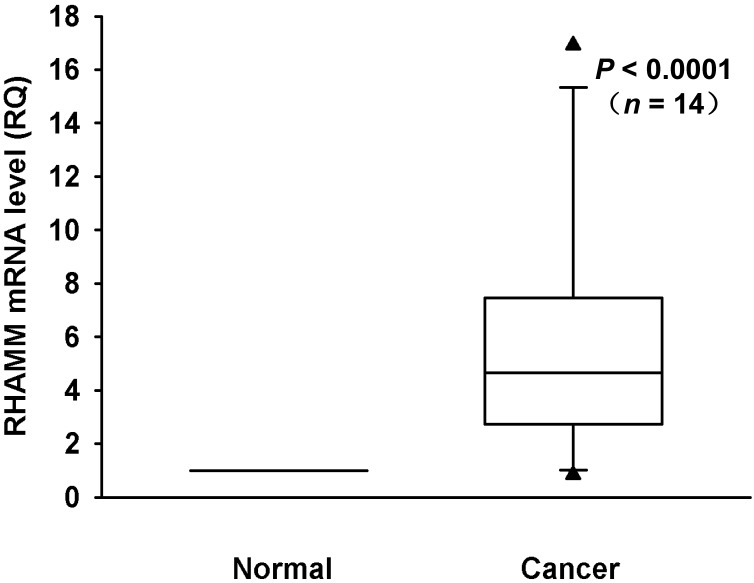
We investigated mRNA expression of RHAMM in a panel of 8 PDAC cell lines and HPDE as a control. The RHAMM mRNA expression was higher in 4 (50%) of 8 PDAC cell lines as compared to HPDE (Fig. 1). We also analyzed the RHAMM mRNA expression levels in tissues from 14 PDAC patients. The RHAMM mRNA expression was significantly higher in PDAC tissues as compared to adjacent non-tumorous pancreatic tissues (median, 4.7-fold; range, 0.8- to 16.9-fold; P < 0.0001) (Fig. 2).
Fig 1.
The receptor for hyaluronic acid-mediated motility (RHAMM) mRNA expression levels in pancreatic ductal adenocarcinoma (PDAC) cell lines and an immortalized cell line derived from human pancreatic duct (HPDE) were examined by real-time RT-PCR. The expression level shown was a relative value when the expression of HPDE was set to 1. Each bar represents the mean ± SD of three replications.
Fig 2.
RHAMM mRNA expression in primary pancreatic cancer and adjacent non-tumor tissues from 14 patients was measured using real-time RT-PCR. RHAMM mRNA expression in most PDAC tissues was higher than in normal pancreatic tissues (median, 4.7-fold; range, 0.8- to 16.9-fold; P < 0.0001; Wilcoxon signed rank test).
RHAMM protein expression in cell lines
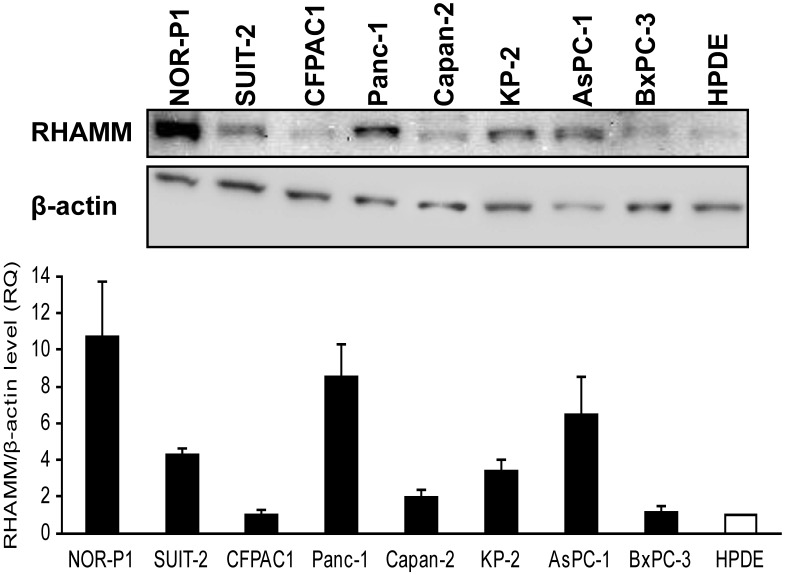
We then used western blotting to examine the protein expression of RHAMM in PDAC cell lines and HPDE. The RHAMM protein expression was higher in the vast majority of PDAC cell lines relative to the expression in HPDE (median, 3.8-fold; range, 1.0- to 10.7-fold) (Fig. 3).
Fig 3.
RHAMM protein expression in PDAC cell lines and HPDE cell line was investigated by western blotting. It was higher in most PDAC cells relative to the expression in HPDE cells (median, 3.8-fold; range, 1.0- to 10.7-fold).
Immunohistochemical analysis of RHAMM in PDAC tissues
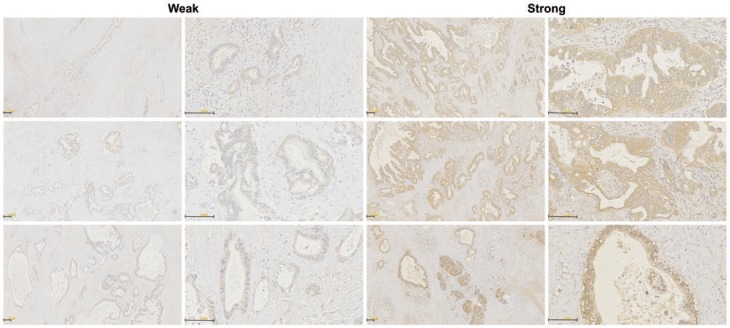
The RHAMM protein expression pattern was investigated in tissue samples from 70 PDAC patients who underwent surgical resection using immunohistochemistry. Staining of RHAMM was found predominantly in the cytomembrane and cytoplasm of tumor cells. Of 70 patients who were evaluable for RHAMM staining, 52 (74%) patients were classified as having strong RHAMM expression, and the remaining 18 (26%) patients were classified as having weak expression, according to the intensity score (Fig. 4).
Fig 4.
Immunohistochemical stainings of RHAMM in PDAC tissues. Weak and strong RHAMM expression patterns were identified, predominantly in tumor cells. The staining patterns were found in the cytomembrane and cytoplasm (original magnification 50× and 200×).
Correlation between RHAMM expression and prognosis in PDAC patients
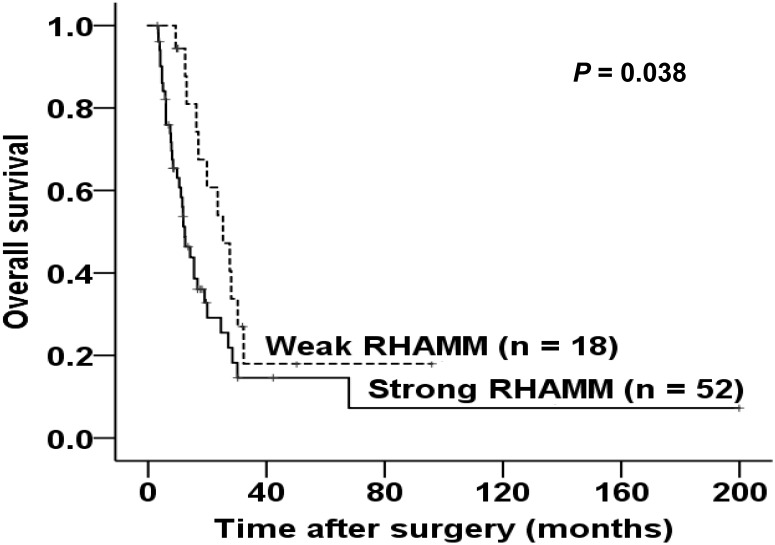
We explored the correlations between the RHAMM expression pattern and patient prognosis by comparing the survival of patients between the strong and weak expression groups. As shown in Fig. 5, strong expression of RHAMM was significantly associated with a shorter survival time after surgery (log rank = 4.284, P = 0.038).
Fig 5.
Kaplan-Meier survival curve of patients according to the expression patterns of RHAMM. Strong expression of RHAMM was significantly associated with poor survival (log rank = 4.284, P = 0.038).
Finally, we used cox proportional hazard model to analyze the prognostic significance of RHAMM expression. Univariate analysis showed tumor stage (P = 0.009), residual tumor (P = 0.003), and RHAMM expression (P = 0.043) correlated significantly with survival. Multivariate analysis revealed tumor stage (P = 0.039), residual tumor (P = 0.015), and RHAMM expression (P = 0.034) were independent factors, which affected survival (Table 1).
Table 1.
Cox multivariate analysis of clinicopathological parameters for survival
| Variables | Univariate P value |
Multivariate HR (95 % CI) |
P value |
|---|---|---|---|
| UICC stage | 0.009 | 1.66 (1.03 - 2.68) | 0.039 |
| Tumor diameter | 0.911 | ||
| Histological grade | 0.694 | ||
| Age (> 75 years) | 0.157 | ||
| Gender (M) | 0.302 | ||
| Chemotherapy (+) | 0.249 | ||
| Residual tumor (+) | 0.003 | 2.32 (1.18 - 4.59) | 0.015 |
| Lymph node metastasis (+) | 0.438 | ||
| Lymphatic invasion (+) | 0.261 | ||
| Vessel invasion (+) | 0.713 | ||
| Neural invasion (+) | 0.135 | ||
| RHAMM expression (strong) | 0.043 | 2.07 (1.06 - 4.07) | 0.034 |
HR hazard ratio, CI confidence interval, UICC union for international cancer control, RHAMM receptor for hyaluronic acid-mediated motility
Discussion
Several studies have identified that RHAMM overexpression is associated with the invasion, progression and poor prognosis of breast cancer 20, 29, 38; with the histological grade, invasion and metastasis of endometrial carcinoma 19; with the invasion of pancreatic adenocarcinoma 39; and with adverse prognostic factors in colorectal and gastric cancer 33, 35. Abetamann V et al. indicated that RHAMM contributes to the invasion of pancreatic adenocarcinoma by investigating RHAMM mRNA expression in 10 cell lines. These authors could not investigate RHAMM mRNA expression in PDAC tissues and RHAMM protein expression in PDAC cell lines and did not clarify the clinical effects of the RHAMM expression or the prognostic value for PDAC. Our study is the first report to determine the correlation between the expression pattern of RHAMM and the survival of patients with PDAC.
We used immunohistochemistry to identify RHAMM protein expression in the cytomembrane and cytoplasm in PDAC cells, although expression was not observed in the nucleus, which was similar to the pattern observed in other tumor types. Strong RHAMM expression has been reported in 65% of oral squamous cell cancers 40, 40% of colorectal cancers 41 and 28% of gastric cancers 35. Thus, expression may depend on the cancer type.
RHAMM mediates the Ras and TGF-b signaling pathways and is associated with poor prognosis in a variety of tumorous entities 20, 38, 42. RHAMM expression in PDAC was found to significantly inversely correlate with survival in this study. This finding is in agreement with findings for other types of tumors such as breast, colon, and gastric cancer. Additionally, we found that the RHAMM mRNA levels were elevated in pancreatic tumor tissues, regardless of the tumor grade, compared with the levels in normal pancreatic tissues. Our studies suggest that RHAMM could be used in clinical settings as a novel prognostic marker in patients with PDAC.
Frequent overexpression of RHAMM has led to the proposal that it could be a target for PDAC therapy. By targeting its extracellular functions, the biological study of RHAMM in tumor progression has led to novel tumor therapies. Pre-clinical, phase I and II trials are currently investigating the ability of a RHAMM-derived peptide vaccine to promote immune recognition and destruction of tumors by activated T cells, thus targeting RHAMM as a novel cancer-associated antigen 43-45. RHAMM has been used to block the extracellular function of RHAMM on the surface of cancer cells as a promising target for antibody therapy 34. Thus, these findings suggest that RHAMM may serve as a prognostic marker and as an attractive target for PDAC therapy.
Acknowledgments
We thank Ms. Ueda for her technical assistance and Ms. Ying Jiang (Ph.D., research associate, department of health development, institute of industrial ecological sciences, university of occupational and environmental health, Japan) for her help with the statistical analysis.
Abbreviations
- HA
hyaluronic acid
- RHAMM
receptor for hyaluronic acid-mediated motility
- PDAC
pancreatic ductal adenocarcinoma
- ECM
extracellular matrix
- ERK
extracellular signal regulated protein kinase
- BRCA1
breast cancer 1
- BCA
bicinchoninic acid
- PVDF
poly vinylidene difluoride.
References
- 1.Meyer K. Chemical structure of hyaluronic acid. Fed Proc. 1958;17:1075–7. [PubMed] [Google Scholar]
- 2.Yeo TK, Nagy JA, Yeo KT, Dvorak HF, Toole BP. Increased hyaluronan at sites of attachment to mesentery by CD44-positive mouse ovarian and breast tumor cells. Am J Pathol. 1996;148:1733–40. [PMC free article] [PubMed] [Google Scholar]
- 3.Hall CL, Yang B, Yang X, Zhang S, Turley M, Samuel S. et al. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell. 1995;82:19–26. doi: 10.1016/0092-8674(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 4.Bartolazzi A, Peach R, Aruffo A, Stamenkovic I. Interaction between CD44 and hyaluronate is directly implicated in the regulation of tumor development. J Exp Med. 1994;180:53–66. doi: 10.1084/jem.180.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D. et al. Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility. J Cell Biol. 1992;117:1343–50. doi: 10.1083/jcb.117.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall CL, Wang C, Lange LA, Turley EA. Hyaluronan and the hyaluronan receptor RHAMM promote focal adhesion turnover and transient tyrosine kinase activity. J Cell Biol. 1994;126:575–88. doi: 10.1083/jcb.126.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–13. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 8.Turley EA, Austen L, Vandeligt K, Clary C. Hyaluronan and a cell-associated hyaluronan binding protein regulate the locomotion of ras-transformed cells. J Cell Biol. 1991;112:1041–7. doi: 10.1083/jcb.112.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng XB, Sato N, Kohi S, Yamaguchi K. Prognostic impact of hyaluronan and its regulators in pancreatic ductal adenocarcinoma. PLoS One. 2013;8:e80765. doi: 10.1371/journal.pone.0080765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turley EA. Purification of a hyaluronate-binding protein fraction that modifies cell social behavior. Biochem Biophys Res Commun. 1982;108:1016–24. doi: 10.1016/0006-291x(82)92101-5. [DOI] [PubMed] [Google Scholar]
- 11.Itoh K, Yoshioka K, Akedo H, Uehata M, Ishizaki T, Narumiya S. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat Med. 1999;5:221–5. doi: 10.1038/5587. [DOI] [PubMed] [Google Scholar]
- 12.Somlyo AV, Phelps C, Dipierro C, Eto M, Read P, Barrett M. et al. Rho kinase and matrix metalloproteinase inhibitors cooperate to inhibit angiogenesis and growth of human prostate cancer xenotransplants. FASEB J. 2003;17:223–34. doi: 10.1096/fj.02-0655com. [DOI] [PubMed] [Google Scholar]
- 13.Bourguignon LY, Singleton PA, Zhu H, Diedrich F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J Biol Chem. 2003;278:29420–34. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 14.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G. et al. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–35. doi: 10.1016/s0301-472x(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 15.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr Opin Cell Biol. 1994;6:726–33. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 16.Tolg C, Hamilton SR, Nakrieko KA, Kooshesh F, Walton P, McCarthy JB. et al. Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J Cell Biol. 2006;175:1017–28. doi: 10.1083/jcb.200511027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telmer PG, Tolg C, McCarthy JB, Turley EA. How does a protein with dual mitotic spindle and extracellular matrix receptor functions affect tumor susceptibility and progression? Commun Integr Biol. 2011;4:182–5. doi: 10.4161/cib.4.2.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiner J, Bullinger L, Guinn BA, Dohner H, Schmitt M. Leukemia-associated antigens are critical for the proliferation of acute myeloid leukemia cells. Clin Cancer Res. 2008;14:7161–6. doi: 10.1158/1078-0432.CCR-08-1102. [DOI] [PubMed] [Google Scholar]
- 19.Rein DT, Roehrig K, Schondorf T, Lazar A, Fleisch M, Niederacher D. et al. Expression of the hyaluronan receptor RHAMM in endometrial carcinomas suggests a role in tumour progression and metastasis. J Cancer Res Clin Oncol. 2003;129:161–4. doi: 10.1007/s00432-003-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Thor AD, Moore DH 2nd, Zhao Y, Kerschmann R, Stern R. et al. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res. 1998;4:567–76. [PubMed] [Google Scholar]
- 21.Greiner J, Ringhoffer M, Taniguchi M, Li L, Schmitt A, Shiku H. et al. mRNA expression of leukemia-associated antigens in patients with acute myeloid leukemia for the development of specific immunotherapies. Int J Cancer. 2004;108:704–11. doi: 10.1002/ijc.11623. [DOI] [PubMed] [Google Scholar]
- 22.Turley EA, Noble PW, Bourguignon LY. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–92. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 23.Godar S, Weinberg RA. Filling the mosaic of p53 actions: p53 represses RHAMM expression. Cell Cycle. 2008;7:3479. doi: 10.4161/cc.7.22.7320. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell CA, Keats JJ, Crainie M, Sun X, Yen T, Shibuya E. et al. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell. 2003;14:2262–76. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assmann V, Jenkinson D, Marshall JF, Hart IR. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J Cell Sci. 1999;112( Pt 22):3943–54. doi: 10.1242/jcs.112.22.3943. [DOI] [PubMed] [Google Scholar]
- 26.Sohr S, Engeland K. RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53. Cell Cycle. 2008;7:3448–60. doi: 10.4161/cc.7.21.7014. [DOI] [PubMed] [Google Scholar]
- 27.Zaman A, Cui Z, Foley JP, Zhao H, Grimm PC, Delisser HM. et al. Expression and role of the hyaluronan receptor RHAMM in inflammation after bleomycin injury. Am J Respir Cell Mol Biol. 2005;33:447–54. doi: 10.1165/rcmb.2004-0333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akiyama Y, Jung S, Salhia B, Lee S, Hubbard S, Taylor M. et al. Hyaluronate receptors mediating glioma cell migration and proliferation. J Neurooncol. 2001;53:115–27. doi: 10.1023/a:1012297132047. [DOI] [PubMed] [Google Scholar]
- 29.Assmann V, Gillett CE, Poulsom R, Ryder K, Hart IR, Hanby AM. The pattern of expression of the microtubule-binding protein RHAMM/IHABP in mammary carcinoma suggests a role in the invasive behaviour of tumour cells. J Pathol. 2001;195:191–6. doi: 10.1002/path.941. [DOI] [PubMed] [Google Scholar]
- 30.Kong QY, Liu J, Chen XY, Wang XW, Sun Y, Li H. Differential expression patterns of hyaluronan receptors CD44 and RHAMM in transitional cell carcinomas of urinary bladder. Oncol Rep. 2003;10:51–5. [PubMed] [Google Scholar]
- 31.Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B. et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–22. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CW, Su JY, Tsou AP, Chau GY, Liu HL, Chen CH. et al. Integrative genomics based identification of potential human hepatocarcinogenesis-associated cell cycle regulators: RHAMM as an example. Biochem Biophys Res Commun. 2005;330:489–97. doi: 10.1016/j.bbrc.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Zlobec I, Baker K, Terracciano LM, Lugli A. RHAMM, p21 combined phenotype identifies microsatellite instability-high colorectal cancers with a highly adverse prognosis. Clin Cancer Res. 2008;14:3798–806. doi: 10.1158/1078-0432.CCR-07-5103. [DOI] [PubMed] [Google Scholar]
- 34.Gust KM, Hofer MD, Perner SR, Kim R, Chinnaiyan AM, Varambally S. et al. RHAMM (CD168) is overexpressed at the protein level and may constitute an immunogenic antigen in advanced prostate cancer disease. Neoplasia. 2009;11:956–63. doi: 10.1593/neo.09694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishigami S, Ueno S, Nishizono Y, Matsumoto M, Kurahara H, Arigami T. et al. Prognostic impact of CD168 expression in gastric cancer. BMC Cancer. 2011;11:106. doi: 10.1186/1471-2407-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolg C, Poon R, Fodde R, Turley EA, Alman BA. Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor) Oncogene. 2003;22:6873–82. doi: 10.1038/sj.onc.1206811. [DOI] [PubMed] [Google Scholar]
- 37.Teder P, Bergh J, Heldin P. Functional hyaluronan receptors are expressed on a squamous cell lung carcinoma cell line but not on other lung carcinoma cell lines. Cancer Res. 1995;55:3908–14. [PubMed] [Google Scholar]
- 38.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C. et al. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–80. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abetamann V, Kern HF, Elsasser HP. Differential expression of the hyaluronan receptors CD44 and RHAMM in human pancreatic cancer cells. Clin Cancer Res. 1996;2:1607–18. [PubMed] [Google Scholar]
- 40.Yamano Y, Uzawa K, Shinozuka K, Fushimi K, Ishigami T, Nomura H. et al. Hyaluronan-mediated motility: a target in oral squamous cell carcinoma. Int J Oncol. 2008;32:1001–9. [PubMed] [Google Scholar]
- 41.Zlobec I, Baker K, Terracciano L, Peter S, Degen L, Beglinger C. et al. Two-marker protein profile predicts poor prognosis in patients with early rectal cancer. Br J Cancer. 2008;99:1712–7. doi: 10.1038/sj.bjc.6604729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxwell CA, Rasmussen E, Zhan F, Keats JJ, Adamia S, Strachan E. et al. RHAMM expression and isoform balance predict aggressive disease and poor survival in multiple myeloma. Blood. 2004;104:1151–8. doi: 10.1182/blood-2003-11-4079. [DOI] [PubMed] [Google Scholar]
- 43.Greiner J, Schmitt A, Giannopoulos K, Rojewski MT, Gotz M, Funk I. et al. High-dose RHAMM-R3 peptide vaccination for patients with acute myeloid leukemia, myelodysplastic syndrome and multiple myeloma. Haematologica. 2010;95:1191–7. doi: 10.3324/haematol.2009.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt M, Schmitt A, Rojewski MT, Chen J, Giannopoulos K, Fei F. et al. RHAMM-R3 peptide vaccination in patients with acute myeloid leukemia, myelodysplastic syndrome, and multiple myeloma elicits immunologic and clinical responses. Blood. 2008;111:1357–65. doi: 10.1182/blood-2007-07-099366. [DOI] [PubMed] [Google Scholar]
- 45.Tabarkiewicz J, Giannopoulos K. Definition of a target for immunotherapy and results of the first Peptide vaccination study in chronic lymphocytic leukemia. Transplant Proc. 2010;42:3293–6. doi: 10.1016/j.transproceed.2010.07.022. [DOI] [PubMed] [Google Scholar]