Abstract
The early diagnosis and monitoring of clear-cell Renal Cell Carcinoma (ccRCC), which is the most common renal malignancy, remains challenging. The late diagnosis and lack of tools that can be used to assess the progression of the disease and metastasis significantly influence the chance of survival of ccRCC patients. Molecular biomarkers have been shown to aid the diagnosis and disease monitoring for other cancers, but such markers are not currently available for ccRCC. Recently, plasma and serum circulating nucleic acids, nucleic acids present in urine, and plasma and urine proteins gained interest in the field of cancer biomarker discovery. Here, we describe the applicability of plasma and urine nucleic acids as cancer biomarkers with a particular focus on DNA, small RNA, and protein markers for ccRCC.
Keywords: ccRCC biomarker, cfDNA, CTCs, smallRNA
Introduction
Clear-cell Renal Cell Carcinoma (ccRCC) is the most frequent renal cancer occurring in adults. Together with papillary (10-15% of all renal cell carcinoma cases) and chromophobe carcinoma (5%), renal cell carcinoma comprises 2% of all cancers worldwide 1. ccRCC originates form proximal convoluted tubules in nephrons and is associated with a mutation and/or inactivation of the von Hippel-Lindau (VHL) gene (3p25) and activation of the Raf-MEK-ERK pathway 2. Early-stage ccRCC is frequently discovered accidentally during non-ccRCC related examinations. The disease course is asymptomatic in the majority of the cases, with only 10% of patients displaying the classical triad of symptoms: hematuria, flank pain and weight loss (often accompanied by hypertension and fatigue). In general, patients with ccRCC have the lowest probability of recovery compared with the remaining RCC types 3, and the main curative approach for ccRCC remains complete or partial nephrectomy.
Currently, early ccRCC-specific diagnostic and disease progression tests based on molecular assays are not available, and the presence of recurrent or new tumors is established clinically with X-rays, abdominal ultrasound, and/or abdominal and pectoral CT. Although alterations in various blood variables (such as the erythrocyte sedimentation rate (ESR), platelet count and lactate dehydrogenase) are associated with malignancy, these variables lack specificity and do not aid in the diagnosis of ccRCC 4;5. Similarly, biomarkers for the treatment efficacy of ccRCC are also lacking. In the era of next-generation sequencing, the molecular monitoring of ccRCC progress and the identification of reliable disease and disease progression biomarkers are particularly appealing.
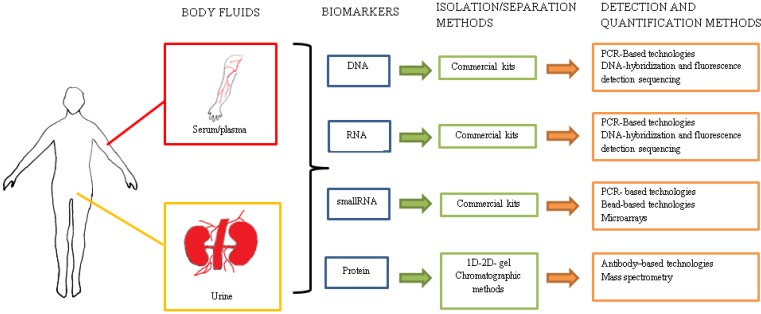
As classical examples, the clinical utility of molecular biomarkers has been shown to be effective with prostate-specific antigen (PSA) and carcinoembryonic antigen (CEA); used in the diagnosis and detection of prostate and colorectal cancers, respectively. Recent efforts concerning the development of less invasive diagnostic methods and the detection of novel disease-markers concentrate on using plasma and urine as target body fluids. The perfect biomarker or biomarker set should be characterized by a high sensitivity, specificity and reproducibility. The test should be relatively non-invasive, and its use must be economically justified. Additionally, the test should enable patient screening, the assessment of the response to treatment and the monitoring of the relapse of the disease. Plasma and serum have been previously shown to be a potential source of promising biomarkers. Both contain a plethora of proteins (including clotting factors in the plasma only), minerals, nucleic acids and hormones 6. Proteomic and molecular approaches have been implemented in the identification of disease-specific proteins and nucleic acids in urine 7 (see Figure 1). The analysis of the urine content should be most applicable to renal diseases because urine consists of a variety of molecules, including naturally occurring polypeptides that are derived directly from kidneys. The concentrations of these compounds originating directly from the kidneys may reflect the pathological state. The undeniable advantage of urine testing is the greater availability and size of fluid volumes used for biomarker isolation and the complete non-invasiveness of the procedure.
Figure 1.
Multiple methods and technologies for the detection and quantification of potential biomarkers isolated from body fluids.
Based on the available literature, we describe the potential applicability of plasma, serum and urine derived nucleic acids as biomarkers in cancer research and provide an overview of recent developments in biomarker identification for ccRCC.
Plasma/serum biomarkers
DNA
Circulating tumor cells (CTCs) are living cells that have shed from the primary tumor and circulate in the bloodstream. The incidence of CTCs is low (approximately 1 per billion of normal blood cells) and depends on the disease stage, thereby complicating the detection of CTC. Despite the technical challenges, Dawson et al. described a method to isolate and quantify CTCs based on an immunomagnetic/fluorescence approach and showed the increase in the number of CTCs in the plasma of women with metastatic breast cancer compared with plasma derived from patients with non-metastatic cancer (≥5 cells per 7.5 ml in 18 of 30 patients) 8. The DNA derived from CTCs can also be used for diagnostic purposes. Vogelstein et al. analyzed the occurrence of circulating tumor DNA (ctDNA) in the plasma from 18 patients with colorectal cancer and showed that the presence of ctDNA decreased by 99% after surgery and that none of the individuals with undetectable ctDNA levels in plasma experienced a recurrence of the disease 9. Based on cited publications, the ctDNA is detected, on average, in 80-99% of the plasma samples derived from cancer patients and can be analyzed using traditional, targeted PCR assays. Additionally, the material has been reported suitable for NGS-based amplicon sequencing 8.
Because of its stability and relatively easy isolation procedure, circulating cell-free tumor-derived DNA (cfDNA); released by apoptotic and necrotic tumor cells, recently became a main scope of investigations aiming to identify disease-specific biomarkers. Although cfDNA is also detected in healthy individuals, the levels of cfDNA are significantly higher in the plasma of cancer patients, as demonstrated by Frattini et al. in a study of colorectal cancer samples (mean concentrations: 10.3 ng/ml in the plasma of healthy subjects vs. 495.7 ng/ml in the plasma of cancer patients). In that report, the authors observed a decrease in the cfDNA concentrations in the plasma from postoperative colorectal cancer patients after 4 months (with average values of 170.6 ng/ml of cfDNA) 10. Kim et al. compared the cfDNA concentrations in the serum and plasma from patients with gastric cancer and reported significantly higher cfDNA levels in the serum (755.9±1,203.7 ng/ml) than in the plasma (68.7±40.7 ng/ml). Although they were higher in the serum, the cfDNA concentrations were highly variable, which suggests that plasma is a more reliable cfDNA source 11.
Yamada et al. showed that the mutation of K-Ras, found in tissue samples of pancreatic adenocarcinoma, was also detected in the plasma cfDNA from patients with the identical type of cancer, which suggests the potential utility of cfDNA in the detection of mutations that are characteristic for solid tumors 12. Epigenetic changes have been reported to be a frequent adaptation-mechanism observed in tumors, and the increase in epigenetic events (e.g., the silencing of tumor suppressor genes) can be related to cancer progression 13. Although the majority of the epigenetic data is limited to tumor sample analysis, there are publications displaying the utility of cfDNA isolated from plasma as a material suitable for targeted epigenetic assays 14. For example Devos et al. used cfDNA from plasma and reported that 50% of colorectal cancer patients show a high level of hypermethylation in Septin 9 gene; whereas in 90% of healthy patients, this promoter remains unmethylated, mirroring the situation observed in tumors 15;16.
RNA
Circulating RNA from plasma or serum is highly fragmented due to the lack of protective mechanisms preventing RNA from degradation 17. Unfortunately, RNA degradation limits the circulating RNA extraction and analysis to shorter subspecies of RNA: microRNA and long non-coding RNAs (lncRNA). These types of RNA could potentially serve as diagnostic and prognostic cancer biomarkers or could indicate treatment efficiency.
MicroRNAs are non-coding RNAs that are 19-22 nucleotides long and are involved in post-transcriptional gene regulation. They bind to the 3'UTR region of target mRNA and cause the inhibition of transcript translation or mRNA 18. Recent studies have shown that microRNAs enter the circulation as a result of an active release from living cells or as a consequence of apoptotic or necrotic cell death 18. Many miRNAs are abnormally expressed in solid and hematopoietic tumors, and these miRNAs have generated considerable interest as cancer diagnostic tools and indicators of therapy efficacy. Circulating microRNAs were linked to cancer by Lawrie et al., who reported that high levels of miR-155, miR-210, miR21 in serum were characteristic for diffuse B-cell lymphoma 19. Komatsu et al. and Aguilar et al. showed using microarray and Real-Time-based methods that the levels of specific microRNAs were higher in the plasma from patients with esophageal squamous cell carcinoma (ESCC) and the serum from breast cancer patients as compared with the controls. Moreover, in the case of ESCC patients, the miR-25 levels were significantly higher in the plasma from patients with ESCC recurrence and lower in postoperative samples than in the preoperative samples 20;21.
Circulating miRNAs are often stable in plasma and serum because of the incorporation into exosomes and microparticles 22. Except for miRNA, exosomes also carry mRNA and proteins, which are protected from degradation by a bilayer exosomal membrane 23. Exosomes are 30-100 nm membrane vesicles that are secreted by a variety of cells including cancer cells 24. In contrast, microparticles (MPs) are small vesicles (50-1000 nm) released by different types of cell types such as leucocytes, endothelial or platelet cells. MPs are capable of transferring peptides, proteins, lipids, microRNA, mRNA, and DNA from cell-to-cell without direct contact 25. When released into circulation, MPs bind and fuse with cells by using cellular receptors and act like mediators of various processes (e.g. protein expression) such as inflammation 26.
Rabinowits et al. tested the levels of total exosome and miRNA levels in the plasma from lung cancer patients and controls. Their study showed that the mean protein concentration of circulating exosomes was 2.85 mg/ml and 158.6 ng/ml of exosomal miRNA in the plasma from lung adenocarcinoma patients as compared with 0.77 mg/ml of exosomes and 68.1 ng/ml of exosomal miRNA in healthy controls 27. Tanaka et al. tested the expression levels of exosomal miR-21 in patients with ESCC and found levels of exosomal miR-21 were significantly increased in the ESCC group (0.005-0.075 mg/dl, p=0.0009) compared with the levels in plasma exosomes in the control group (0.001-0.037 mg/dl, p=0.0009) 28.
In contrast to microRNA, lncRNAs are approximately 200 nt long, spliced and polyadenylated non-coding RNAs of unknown function 29. lncRNAs have been suggested to take part in almost all aspects of gene regulation, including chromosome dosage-compensation or nuclear and cytoplasmic trafficking 29. Ren et al. reported lncRNAs are stable in both human plasma and serum and tested the expression of lncRNAs in plasma by qRT-PCR. Importantly, their study showed that the plasma and serum lncRNA may be useful biomarkers because of their detectability and stability30.
Biomarkers in urine
The efficiency of nucleic acid isolation, notably DNA, and their amplification from urine often influenced by the presence of urea, which is a PCR inhibitor 31. The urea in the sample may cause denaturation of polymerase and influence the PCR yields during sample amplification. The yield of nucleic acids, mainly DNA, has been shown to differ depending on the time the sample was collected 7. In the majority of the studies, morning urine was used, although the DNA isolated from such samples has been shown to be more degraded compared with DNA isolated from urine collected later in the day 7. Furthermore, the isolated DNA fragments were longer if later sample was used. On average, isolated DNA fragments range from 150 to 250 bp 7. Additionally, El Bali et al. showed gender-specific differences in DNA yields isolated from urine 7. Urine collected from female patients contained more DNA (36 to 215 ng for 1 ml of sample) compared with 13 to 44 ng of DNA isolated from urine collected from males. The DNA yield was highest from urine samples collected in the afternoon for female patients and in the morning for male patients.
In addition to DNA, urine contains small concentrations of RNA. However, because of the high amount of RNAses, the RNA is highly degraded. Therefore, the majority of studies concentrate on the analysis of microRNA. For example, Bryant et al. found that miR-107 and miR-574-3p were present at significantly higher levels in the urine of men with prostate cancer than in that of controls 32. The microRNA (and mRNA) in the urine is primarily contained in exosomes 33. Nilsson et al. studied exosomal RNA isolated from the urine of patients with prostate cancer. Using nested PCR, the mRNA was analyzed for the fusion TMPRSS2:ERG gene transcripts that are present in high levels in 72% of prostate cancer cases 34. The samples derived from two of the four patients with a high grade prostate cancer (Gleason score) and high PSA levels tested positive for the fusion transcript 35. In contrast, no TMPRSS2:ERG transcripts were detected in samples derived from patients undergoing androgen deprivation therapy (ADT) who displayed a response to ADT treatment 36.
LncRNAs can be isolated from urine. Lee et al. analyzed the total RNA isolated from the urine of patients with prostate cancer and from healthy controls using qPCR. In their study, they identified six lncRNAs of unknown function (AK024556, XLOC_007697, LOC100287482, XLOC_005327, XLOC_008559, and XLOC_009911), which were significantly up-regulated in the urine samples obtained from the prostate cancer patients 37.
Biomarkers in ccRCC - literature overview
Because ccRCC is primarily asymptomatic in the early phase, nearly 40% of all ccRCC patients are diagnosed during the late stage of the disease, when a tumor is already large and/or metastasized. Even when it is diagnosed early and the primary tumor is surgically removed with partial or complete nephrectomy, 30-40% of ccRCC patients experience a relapse in the disease or metastasis. Limited curative options are available because ccRCC is insensitive to chemotherapy and radiotherapy. Therefore, validated and reliable molecular markers that enable the early diagnosis of ccRCC, monitoring of the potential relapse of the disease and indicate treatment efficacy are a major focus in the field of ccRCC research. Here, we summarize the recent reports (2009-2014) describing the identification of potential biomarkers (DNA, RNA, proteins) in the serum and urine from ccRCC patients. A summary of the described markers is displayed in Table 1.
Table 1.
Cancer-associated biomarkers in the plasma, serum and urine from ccRCC patients.
| Biomarker | Source | Detection method | Deregulation | Function | Utility | Source | Ref |
|---|---|---|---|---|---|---|---|
| cfDNA | serum | PCR reaction | upregulated | Unknown | ccRCC diagnosis | Coon JJ et al. | 36 |
| cfDNA | plasma | real-time PCR | upregulated | Unknown | identification of metastatic ccRCC patients during the course of sorafenib therapy | Lee et al. | 37 |
| cfDNA | plasma | real-time PCR | upregulated | Unknown | diagnosis and detection of recurrence after nephrectomy | Gang et al. | 38 |
| miRNA-187 | serum | qPCR | downregulated | Unknown | ccRCC diagnosis | Zhao et al. | 44 |
| miRNA 210 | serum | qPCR | upregulated | role in tumorigenesis | ccRCC detection | Camps et al. | 43 |
| miR‑141 , | serum | qPCR | downregulated | Unknown | ccRCC diagnosis | Cheng et al. | 45 |
| miR‑34a and miR‑21 miR‑224 | serum | qPCR | upregulated | Unknown | ccRCC diagnosis | Cheng et al. | 45 |
| sIL-2R | serum | ELISA immunoassay | upregulated | activation of T cells and regulation immune response | ccRCC diagnosis and detection | Masuda et al. | 46 |
| sB7-H3 | serum | ELISA immunoassay | downregulated | inhibition of T cell activity | ccRCC diagnosis and detection | Masuda et al. | 46 |
| proteasome 20S | serum | sandwich enzyme-linked immunosorbent assay | downregulated | protein degradation | ccRCC diagnosis and prognosis | De Martino et al. | 54 |
| zinc finger proteins | serum | matrix-assisted laser desorption ionization (MALDI) TOF) mass spectrometry (MS) | upregulated | includes a motif which mediates biding to molecules such as protein, DNA and RNA. | ccRCC detection | Yang et al. | 55 |
| RNA-binding proteins (RBPs) | serum | MALDI TOF MS | upregulated | binds to RNA molecules and plays significant role in forming ribonucleoproteins splicing of mRNA, polyadenylation, stabilization, localization and translation | ccRCC detection | Yang et al. | 55 |
| protein TUBB | serum | MALDI TOF MS | upregulated | consisting of microtubules | ccRCC detection | Yang et al. | 55 |
| B7x | serum | ELISA immunoassay | upregulated | downregulation of peripheral immune responses via negative T-cell costimulation | ccRCC diagnosis and detection | Krambeck et al. | 49 |
| collagen fragments below 1.4 kDa | urine | capillary electrophoresis coupled to mass spectrometry (CE-MS) | downregulated | activation of platelet response, involvement in thrombosis and hemostasis engagement in replacement and restoration of dead skin cells | ccRCC diagnosis and prognosis | Chinello et al. | 57 |
| peptides from inflammatory and immune proteins | urine | CE-MS | downregulated | Unknown | ccRCC diagnosis and prognosis | Frantzi et al. | 51 |
| peptides involved in coagulation and platelet aggregation | urine | CE-MS | upregulated | Unknown | ccRCC diagnosis and prognosis | Frantzi et al. | 51 |
| fragments of the human glycoprotein uromodulin (UMOD/THP) | urine | MALDI TOF MS | upregulated | regulatory protein, prevention of urinary tract from infections and injuries caused by products from tubular fluids. Protection against calcium oxalate crystal formation | ccRCC diagnosis and prognosis | Chinello et al. | 57 |
| fragment of fibrinogen alpha chain (FIBA) |
urine | MALDI TOF MS | upregulated | involvement in hemostasis, platelet aggregation and wound repair | ccRCC diagnosis and prognosis | Frantzi et al. | 51 |
Tumors cell-free DNA (cfDNA) in the plasma and serum
In 2010, Gang et al. postulated that the integrity of necrotic, tumor-derived cfDNA in serum may be different from that released from apoptotic cells (random vs. uniform lengths). To test this hypothesis, the authors performed a PCR on the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene using 4 sets of primers to amplify DNA fragments of different lengths (109-456 bp). Notably, all PCR products were detected using cfDNA isolated from the serum derived from ccRCC patients (n=78), whereas only 109 and 193 bp fragments were amplified using the serum cfDNA of healthy controls (n=42). Although no significant differences were observed when patients were stratified according to gender, age or disease stage, longer cfDNA fragments (397 and 456 bp) were detected at a higher percentage in patients before the surgery than in patients after the removal of the tumor (82% vs. 31% and 82% vs 14%, respectively) 38.
Wan et al. investigated whether a quantitative analysis of plasma cfDNA may aid in post-nephrectomy patient monitoring and provide diagnostic utility in the detection of recurring tumors. The study comprised samples collected from 92 ccRCC patients and 42 controls. The measurements of a 384 bp amplification product were performed using quantitative real-time PCR. Higher cfDNA levels were characteristic for more advanced (G3, G4 Fuhrman grade, and T3 (TNM scale)) and metastatic tumors (6.04± 0.72 ng/ml, p=0.017) compared with localized tumors (5.29 ± 0.53 ng/ml, p=0.017) or controls (0.65 ± 0.29 ng/ml, p < 0.001), which suggested a relationship between the cfDNA levels and the progression of ccRCC 39. The patients were monitored 36 months post-nephrectomy, and in 17 individuals with high cfDNA levels, disease recurrence was diagnosed. The multivariate regression analysis indicated plasma cfDNA levels as an independent predictor of the recurrence-free survival (5.81 ng/ml ± 1.02 ng/ml, 5.23 ± 0.91 ng/ml, p=0.024), which suggested that plasma cfDNA quantification may be useful for recurrence monitoring during patient follow-up 39.
Feng at al. suggested that the plasma cfDNA levels can be indicative of sorafenib treatment efficacy in patients with metastatic ccRCC. The samples were collected from 18 ccRCC patients with metastatic disease who underwent sorafenib treatment at the time of recruitment and at 4-, 8-, 12-, 16- and 24-week time points. The levels of 384 bp amplification products were monitored using real-time PCR. In general, the levels of cfDNA in the plasma from metastatic ccRCC patients were significantly higher than in the plasma from healthy controls (n=18; n=10, respectively) (4.771± 0.404 ng/ml; 0.622±0.288 ng/ml, respectively, p<0.001). Similar to Wan et al., the authors observed a correlation between high cfDNA levels and more advanced and/or metastatic tumors classified with the Fuhrman grade, TNM stage and number of metastatic foci. Receiver operating characteristic (ROC) curves showed that the plasma cfDNA levels could predict disease progression with 100% sensitivity at weeks 8-24 and remission at weeks 4-24. Although the analyzed group is relatively small, the findings suggest that monitoring the levels of the plasma cfDNA may have a predictive value of response to sorafenib therapy 40.
RNA markers in plasma and serum
Zhao et al. investigated whether HIF-1α regulating miR-210 (found highly expressed in many tumors including ccRCC) is detectable in the serum from ccRCC patients and if its expression levels have diagnostic capacity 41;42. First, miR-210 expression was tested in 32 ccRCC tumors and compared with the level in 32 matched non-tumoral renal parenchyma using quantitative real-time PCR. MiR-210 was found to be up-regulated in tumor tissue (64,714.8±19,435.6-fold, p=0.004). This finding was validated in 68 ccRCC sera, in which miR-210 was up-regulated (18.4±8.1-fold) compared with 42 serum samples that were obtained from healthy individuals (1.6±0.5-fold, p<0.0001). Additionally, the miR-210 levels were significantly decreased in 10 samples one week post-nephrectomy (p=0.001). No significant correlation was noted between the miR-210 levels and gender, age, tumor grade and stage (except for the tumor size (p=0.048)) 43. Based on ccRCC tumor deep-sequencing data, Zhao et al. selected miR-187 as a candidate to analyze in the serum from ccRCC patients 44. MiR-187 has been shown to be aberrantly expressed in several types of tumors such as breast and ovarian cancer. MiR-187 functions as a factor down-regulating disabled homolog-2 (Dab2) and, as a consequence, inhibits epithelial-mesenchymal transition and likely cancer cell migration. The authors found a down-regulation of miR-187 in ccRCC tumors (0.42-fold, p<0.01) and confirmed this down-regulation in the plasma from 30 ccRCC patients (0.56, fold, p<0.01).
In 2013, the miRNA expression profile in the serum from ccRCC patients was reported to be not fully consistent with the miRNA profile in the tumor 45. Based on the available literature, Cheng et al. selected 8 miRNAs previously reported to be deregulated in ccRCC tumors (miR‑34a, miR‑21, miR‑224, miR‑141, miR‑149, miR‑429, miR‑221 and miR‑211) and tested their expression in both tumors and serum from ccRCC patients. Implementing a qPCR analysis, the authors tested 30 ccRCC tumor tissues (matched with normal tissue samples) and 12 serum samples from patients with ccRCC collected before nephrectomy. MiR‑34a, miR‑21 and miR‑224 were found to be up-regulated in the tumor (6.2-, 22.5- and 27.5-fold, respectively) and serum (84-, 277- and 4-fold, respectively), although not in all samples (n=11, 12 and 9, respectively). miR‑141, miR‑149 and miR‑429 were down regulated in the tumor by 6.3-, 1.7- and 2.4-fold, respectively, although not in all samples (n=26, 21 and 22, respectively). In the serum, only miR-141 was down regulated with a fold change of 5.8x107 (n=12), the difference in the expression of miR-149 and miR-429 between the ccRCC serum and healthy control was not statistically significant. Except for the correlation of the expression of miR‑21 with the disease stage (p<0.05) and the expression of miR-224 with gender (p<0.05), no correlation was observed between the expression of the remaining microRNAs and clinical parameters. 45
Proteomic biomarkers in the serum from ccRCC patients
Because ccRCC is considered an immunogenic tumor, Masuda et al. analyzed the levels of soluble T-cell regulatory molecules (interleukin-2 receptor (sIL-2R), B7-H3 (sB7-H3), and cytotoxic T lymphocyte associated antigen-4 (sCTLA-4) (see Table 1) in the sera from 70 ccRCC patients and 35 healthy controls using ELISA. The serum level of sIL-2R was significantly higher in the patients than in the healthy controls (1080.2 ± 1251.3 pg/ml and 587.4 ± 210.4 pg/ml, respectively; p=0.043). In contrast, the serum levels of sB7-H3 and sCTLA-4 were significantly lower in ccRCC patients than in the controls (p<0.001 and p<0.0001, respectively). The mean levels of sB7-H3 in ccRCC reached 14.5 ± 10.1 ng/ml in the sera from ccRCC patients and 19.3 ± 5.3 ng/ml in samples derived from healthy controls. Similarly, the sCTLA-4 concentration in ccRCC sera was 2.3 ± 0.7 ng/ml on average and 3.0 ± 1.3 ng/ml in the control samples. Although the authors suggested statistically significant differences in sIL-2R sB7-H3 and concentrations in the sera from patients with different disease stages (p< 0.001, p=0.049, respectively), these findings require additional analysis in a larger cohort 46.
Based on previously published data, Thompson et al. analyzed the expression of costimulatory molecule B7x in the serum from ccRCC patients 47. As indicated by experimental evidence, B7x is thought to down-regulate peripheral immune responses via negative T-cell costimulation 48;49. B7x has been reported to be aberrantly expressed in RCC tumors, and its expression correlates with metastatic disease progression and poor survival 50. Using a sandwich ELISA, the authors tested the expression of B7x in the sera from 101 ccRCC patients and 101 healthy, gender-matched controls. Although the increased expression of B7x was detected only in 53 of the 101 ccRCC sera, the expression correlated with tumor thrombus, nodal and distant metastases, and advanced tumor stage. Whether B7x is a ccRCC-specific marker remains unclear because it is not expressed in all patients, its elevated expression has been detected in the blood/serum of ovarian patients and it has not been analyzed in papillary and chromophobe RCC 51.
De Martino et al. tested the potential diagnostic utility of the 20S proteasome levels as a ccRCC serum biomarker. Proteasome 20S has been previously described as a potential biomarker of myeloproliferative solid tumors, breast cancer, and ovarian cancer (among other types) 52;53. Here, 20S proteasome serum levels were measured in 113 ccRCC and 15 control samples using a sandwich enzyme-linked immunosorbent assay. The 20S levels were increased in ccRCC sera (median 4.66 mg/ml vs. 1.52 mg/ml in controls; p<0.0001) and were found to be increased in patients with ccRCC-associated symptoms (p=0.0008), with distant metastases (p=0.0011), higher Fuhrman grades (p=0.0247), and necrotic tumors (p=0.0462). Additionally, a significant correlation was noted between the 20S levels and the tumor size (r=0.27, p=0.004). Moreover, patients with high 20S proteasome serum levels displayed a 9.41-fold increased risk of death from ccRCC compared with patients who had low 20S proteasome serum levels 54.
Yang et al. implemented magnetic beads-based weak cation-exchange chromatography in combination with matrix-assisted laser desorption ionization (MALDI) time of flight (TOF) mass spectrometry (MS) to identify differentially expressed peptides and proteins in the serum from 58 ccRCC patients, 40 additional samples of paired pre- and post-operative ccRCC patients and 64 samples from healthy volunteers. All serum samples were analyzed in triplicate to identify reliable serum peptidome profiles. Cancer patients were accurately distinguished from healthy individuals, and preoperative patients were distinguished from postoperative ccRCC patients using 18 discriminating m/z (mass to charge ratio) peaks with an 82.5-92.7% sensitivity and a specificity above 83.8-98.3%, depending on the classification model used (genetic algorithm (GA), supervised neural networks (SNN), and the quick classifier (QC)). Three peptides (regions of RNA-binding protein 6 (RBP6), tubulin beta chain (TUBB), and zinc finger protein 3 (ZFP3)) were found to be specific for ccRCC sera (see Table 1) 55.
ccRCC biomarkers in urine
Studies of ccRCC urine biomarkers are based mainly on proteomic approaches that implement advanced, although not identical, methods for peptide identification. Additionally, diverse sample pre-fractionation methods and/or chromatographic separations can lead to the identification of different, often non-overlapping, markers. Therefore, the data covering urinary peptidome are non-uniform but likely complementary and are primarily to be emphasized as proof-of concept experiments.
High-resolution capillary electrophoresis coupled to mass spectrometry (CE-MS) was implemented by Frantzi et al. to identify urinary peptides that could serve as a ccRCC diagnostic and a disease screening tool. The discovery set consisted of 40 ccRCC and 68 non-diseased control urine samples, and the peptide profiles with a molecular range from 0.8 to 20 kDa were compared. Disease-specific peptide models were generated using the Support Vector Machine-based (SVM-based) software, and 86 peptides were identified with a statistically significant distribution in the two groups. The peptide-based classifier enabled the differentiation of ccRCC from the control with an area under the curve (AUC) of 0.92 (95% CI, 0.84 to 0.97; p = 0.0001). A ROC probability cut-off of 0.38 resulted in the correct classification of 25 of the 30 ccRCC patients (80% sensitivity [95% CI, 65% to 94%]) and 39 of the 46 normal controls (87% specificity [95% CI, 74% to 95%]). Additionally, the authors tested the peptide set's potential to discriminate ccRCC from closely related cancer types and non-malignant renal and systemic diseases such as diabetic nephropathy (n=195), focal segmental glomerulosclerosis (n=54), membranous glomerulonephritis (n=65), systemic lupus erythematosus (n=46), IgA nephropathy (n=126), vasculitis (n=121), cardiovascular disease (n=33) and bladder cancer (n=219). At 80% sensitivity and a 0.38 cut-off value, the peptide set clearly discriminates between the RCC and most of the remaining diseases, as shown in the model classification scores. A considerable degree of overlap (~25%) was observed between vasculitis (64% specificity) and bladder cancer (76% specificity), which suggested the existence of common processes leading to the generation of such markers (e.g., vascular damage) 51.
Sequences for 40 of the 86 ccRCC specific peptides were identified and were mainly represented by collagen fragments. These sequences were also abundantly found in healthy controls, as described previously 56. Notably, collagen fragments below 1.4 kDa were consistently down-regulated in ccRCC patients, likely reflecting changes in the extracellular matrix (ECM) turnover that results in a specific degradation profile. This profile was mirrored by the presence of altered-length products in the urine. Additionally, the ccRCC peptide marker panel contained fragments of serum proteins (fibrinogen chains, immunoglobulin Fc regions and hemoglobin subunits) and fragments of proteins that are likely expressed in the kidney (e.g., Na/K-transporting ATPase subunit alfa, retinitis pigmentosa GTPase regulator, VPS10 domain-containing receptor SorCS3 and the endothelial adhesion molecule CD99 antigen-like protein 2), represented by the C-terminal fragment of the deleted region in the malignant brain tumors 1 (DMBT1) protein. In general, this analysis shows that peptide levels from inflammatory and immune proteins decreased but that those involved in coagulation and platelet aggregation increased. Combining these 40 sequenced biomarkers enabled the classification of the validation set (40 ccRCC samples) with a sensitivity of 77% and a specificity of 85%; the AUC was 0.87 (95% CI, 0.77 to 0.94; P = 0.0001) and showed that the unidentified biomarkers (included in the 86 biomarker model) enhanced the performance of the classification.
An extensive study of urinary protein profiles using material from 118 ccRCC (73 men and 45 women) and 137 healthy subjects (81 men and 56 women) was performed by Chinello et al. The analyzed group was extended with 35 non-ccRCC samples (16 benign renal masses and 19 malignant non-ccRCC; 22 men and 13 women) 57. The proteins were isolated using a pre-purification method (magnetic beads-based procedure) in combination with a MALDI-TOF analysis. In total, 60% of the collected samples comprised the training group; the remaining 40% were used for the independent evaluation of the models. The authors were able to discriminate ccRCC patients from healthy subjects using twelve peptides (with a SVM algorithm). The discriminating capability was confirmed in the validation cohort with an AUC of 0.96. The identity of the seven (of twelve) signals included in the clusters were obtained by MALDI-TOF/TOF and nLC-ESI-MS/MS analysis. These signals differed from those identified by Frantzi et al. Surprisingly, none of the ions identified (fragments of the human glycoprotein uromodulin (UMOD/THP, m/z 1670 and 2216)), meprin- a zinc-dependent metalloproteinases abundantly expressed in the apical membranes of renal proximal tubules (MEP1A, m/z 1727), osteopontin (m/z 2528), a fragment of the extracellular domain of SeCreted and TransMembrane protein 1 (SCTM1, m/z 1116) were ccRCC specific (as shown in the literature). Notably, the ccRCC discriminant signal observed at the m/z of 2192 was assigned to two possible candidates: a probable G-protein coupled receptor 162 (GP162) and a sequence derived from a Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (KPB1)51. Whereas GP162 is an orphan receptor assigned to the G protein coupled receptors (GPCRs) family involved in signal transmission and associated with cancer-correlated processes such as cell proliferation, angiogenesis, tumor progression and development, KPB1 is a key regulatory enzyme of glycogen metabolism with no established or direct link to cancer.
Perspectives
Currently, no clinically validated biomarkers are available to aid the diagnosis, disease monitoring and treatment efficacy in ccRCC. The most advanced approaches to ccRCC biomarker discovery from plasma and urine are represented by proteomic studies. Although promising, the data generated using different methodologies are non-overlapping and require extensive validation, first in larger cohorts and then with diverse technologies to define a reliable protein marker pool. Up to date nucleic acid biomarkers were mainly a subject of a targeted and low throughput analysis without concurrent conformation in additional cohorts and populations. Hopefully, with the recent technological advances, notably next-generation sequencing, we will be able to define nucleic acid biomarkers quantitatively and qualitatively in the plasma and serum and boost the research aiming to identify reliable ccRCC biomarkers.
Acknowledgments
This publication was supported by the KNOW RNA Research Centre in Poznan (No. 01/KNOW2/2014).
References
- 1.Rydzanicz M, Wrzesiński T, Bluyssen H a R, Wesoły J. Genomics and epigenomics of clear cell renal cell carcinoma: Recent developments and potential applications. Cancer Lett. 2013;341:111–126. doi: 10.1016/j.canlet.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Ojo D, Kapoor A, Hill B, Lin X, Wong N, Pinthus J. RKIP is commonly downregulated in clear cell renal cell carcinoma (ccRCC) Cancer Cell Microenviron. 2015:1–5. [Google Scholar]
- 3.Beck SDW. Effect of Papillary and Chromophobe Cell Type on Disease-Free Survival After Nephrectomy for Renal Cell Carcinoma. Ann Surg Oncol. 2003;11:71–77. doi: 10.1007/BF02524349. [DOI] [PubMed] [Google Scholar]
- 4.Lam JS, Breda A, Belldegrun AS, Figlin R a. Evolving principles of surgical management and prognostic factors for outcome in renal cell carcinoma. J Clin Oncol. 2006;24:5565–5575. doi: 10.1200/JCO.2006.08.1794. [DOI] [PubMed] [Google Scholar]
- 5.Ljungberg B. Prognostic markers in renal cell carcinoma. Curr Opin Urol. 2007;17:303–308. doi: 10.1097/MOU.0b013e328277f180. [DOI] [PubMed] [Google Scholar]
- 6.Ehresman DJ, Froehlich JW, Olsen GW, Chang SC, Butenhoff JL. Comparison of human whole blood, plasma, and serum matrices for the determination of perfluorooctanesulfonate (PFOS), perfluorooctanoate (PFOA), and other fluorochemicals. Environ Res. 2007;103:176–184. doi: 10.1016/j.envres.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Bali L El, Diman A, Bernard A, Roosens NHC, De Keersmaecker SCJ. Comparative Study of Seven Commercial Kits for Human DNA Extraction from Urine Samples Suitable for DNA Biomarker-Based Public Health Studies. J Biomol Tech. 2014:jbt.14–2504. doi: 10.7171/jbt.14-2504-002. -002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F. et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 9.Diehl F, Schmidt K, Choti M a, Romans K, Li M, Thornton K. et al. NIH Public Access. NIH public access. 2010;14:985–990. [Google Scholar]
- 10.Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G. et al. Quantitative analysis of plasma DNA in colorectal cancer patients: A novel prognostic tool. Ann N Y Acad Sci. 2006;1075:185–190. doi: 10.1196/annals.1368.025. [DOI] [PubMed] [Google Scholar]
- 11.Kim YS. Quantitative analysis of cell-free DNA in the plasma of gastric cancer patients. Oncol Lett. 2012:921–926. doi: 10.3892/ol.2012.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada T. et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998 Jun;4(6):1527–32. [PubMed] [Google Scholar]
- 13.Baylin S. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2:21231. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 14.Bock C. Epigenetic biomarker development. Epigenomics. 2009;1:99–110. doi: 10.2217/epi.09.6. [DOI] [PubMed] [Google Scholar]
- 15.DeVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J. et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–1346. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 16.Wasserkort R, Kalmar A, Valcz G, Spisak S, Krispin M, Toth K. et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer. 2013;13:398. doi: 10.1186/1471-2407-13-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerkovnik P, Perhavec A, Zgajnar J, Novakovic S. Optimization of an RNA isolation procedure from plasma samples. Int J Mol Med. 2007;20:293–300. [PubMed] [Google Scholar]
- 18.Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V. Challenges in Using Circulating miRNAs as Cancer Biomarkers. 2014. [DOI] [PMC free article] [PubMed]
- 19.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K. et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W. et al. Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer. 2014;111:1614–1624. doi: 10.1038/bjc.2014.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mar-Aguilar F, Mendoza-Ramírez J a, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P. et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittmann J, Jäck HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta - Rev Cancer. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Keller S, Ridinger J, Rupp A-K, Janssen JWG, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barteneva NS, Fasler-Kan E, Bernimoulin M, Stern JNH, Ponomarev ED, Duckett L. et al. Circulating microparticles: square the circle. BMC Cell Biol. 2013;14:23. doi: 10.1186/1471-2121-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N. et al. Microparticles: Major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633–644. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M. et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 29.Maruyama R, Suzuki H. Long noncoding rna involvement in cancer. BMB Rep. 2012;45:604–611. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren S, Wang F, Shen J, Sun Y, Sun Y, Xu W. et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Huggett JF, Novak T, Garson J a, Green C, Morris-Jones SD, Miller RF. et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008;1:70. doi: 10.1186/1756-0500-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant RJ, Pawlowski T, Catto JWF, Marsden G, Vessella RL, Rhees B. et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–74. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2013;86:1–12. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 34.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken J a. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 35.Ploussard G, Haese A, Van Poppel H, Marberger M, Stenzl A, Mulders PF a. et al. The prostate cancer gene 3 (PCA3) urine test in men with previous negative biopsies: Does free-to-total prostate-specific antigen ratio influence the performance of the PCA3 score in predicting positive biopsies? BJU Int. 2010;106:1143–1147. doi: 10.1111/j.1464-410X.2010.09286.x. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson J, Skog J, Nordstrand a, Baranov V, Mincheva-Nilsson L, Breakefield XO. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee B, Mazar J, Aftab MN, Qi F, Shelley J, Li J-L. et al. Long Noncoding RNAs as Putative Biomarkers for Prostate Cancer Detection. J Mol Diagnostics. 2014;16:615–626. doi: 10.1016/j.jmoldx.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gang F, Guorong L, An Z, Anne G-P, Christian G, Jacques T. Prediction of clear cell renal cell carcinoma by integrity of cell-free DNA in serum. Urology. 2010;75:262–265. doi: 10.1016/j.urology.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 39.Wan J, Zhu L, Jiang Z, Cheng K. Monitoring of plasma cell-free DNA in predicting postoperative recurrence of clear cell renal cell carcinoma. Urol Int. 2013;91:273–278. doi: 10.1159/000351409. [DOI] [PubMed] [Google Scholar]
- 40.Feng G, Ye X, Fang F, Pu C, Huang H, Li G. Quantification of plasma cell-free DNA in predicting therapeutic efficacy of sorafenib on metastatic clear cell renal cell carcinoma. Dis Markers. 2013;34:105–111. doi: 10.3233/DMA-120950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartels CL, Tsongalis GJ. Mini-reviews micrornas:novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- 42.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H. et al. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 43.Zhao A, Li G, Péoc'h M, Genin C, Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94:115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y. et al. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438:439–444. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 45.Cheng T, Wang L, Li Y, Huang C, Zeng L, Yang J. Differential microRNA expression in renal cell carcinoma. Oncol Lett. 2013;6:769–776. doi: 10.3892/ol.2013.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda A, Arai K, Nishihara D, Mizuno T, Yuki H, Kambara T. et al. Clinical Significance of Serum Soluble T Cell Regulatory Molecules in Clear Cell Renal Cell Carcinoma. Biomed Res Int. 2014;2014:1–6. doi: 10.1155/2014/396064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zang X, Loke P, Kim J, Murphy K, Waitz R, Allison JP. B7x: a widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci U S A. 2003;100:10388–10392. doi: 10.1073/pnas.1434299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zang X, Allison JP. The B7 family and cancer therapy: Costimulation and coinhibition. Clin Cancer Res. 2007;13:5271–5279. doi: 10.1158/1078-0432.CCR-07-1030. [DOI] [PubMed] [Google Scholar]
- 49.Krambeck AE, Thompson RH, Dong H, Lohse CM, Park ES, Kuntz SM. et al. B7-H4 expression in renal cell carcinoma and tumor vasculature: associations with cancer progression and survival. Proc Natl Acad Sci U S A. 2006;103:10391–10396. doi: 10.1073/pnas.0600937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocellin S, Keilholz U, Rossi CR, Nitti D. Circulating tumor cells: The 'leukemic phase' of solid cancers. Trends Mol Med. 2006;12:130–139. doi: 10.1016/j.molmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Frantzi M, Metzger J, Banks RE, Husi H, Klein J, Dakna M. et al. Discovery and validation of urinary biomarkers for detection of renal cell carcinoma. J Proteomics. 2014;98:44–58. doi: 10.1016/j.jprot.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Lavabre-Bertrand T, Henry L, Carillo S, Guiraud I, Ouali A, Dutaud D. et al. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer. 2001;92:2493–2500. doi: 10.1002/1097-0142(20011115)92:10<2493::aid-cncr1599>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 53.Hoffmann O, Heubner M, Anlasik T, Winterhalter M, Dahlmann B, Kasimir-Bauer S. et al. Circulating 20S proteasome in patients with non-metastasized breast cancer. Anticancer Res. 2011;31:2197–2201. [PubMed] [Google Scholar]
- 54.De Martino M, Hoetzenecker K, Ankersmit HJ, Roth G a, Haitel a, Waldert M. et al. Serum 20S proteasome is elevated in patients with renal cell carcinoma and associated with poor prognosis. Br J Cancer. 2012;106:904–8. doi: 10.1038/bjc.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J, Yang J, Gao Y, Zhao L, Liu L, Qin Y. et al. Identification of Potential Serum Proteomic Biomarkers for Clear Cell Renal Cell Carcinoma. PLoS One. 2014;9:e111364. doi: 10.1371/journal.pone.0111364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coon JJ, Zürbig P, Dakna M, Dominiczak AF, Fliser D, Frommberger M, Discovery and Disease Diagnostics. Proteomics; 2010. p. 2. doi:10.1002/prca.200800024.CE-MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chinello C, Cazzaniga M, De Sio G, Smith AJ, Gianazza E, Grasso A. et al. Urinary Signatures of Renal Cell Carcinoma Investigated by Peptidomic Approaches. PLoS One. 2014;9:e106684. doi: 10.1371/journal.pone.0106684. [DOI] [PMC free article] [PubMed] [Google Scholar]