Abstract
Survivin is an anti-apoptotic gene that is overexpressed in most human tumors. RNA interference using short interfering RNA (siRNA) can be used to specifically inhibit survivin expression. Tumor cells were treated with a newly designed survivin siRNA, which was modified with 2′-OMe. Cellular survivin mRNA and protein levels were determined by real-time qRT-PCR and Western blot, respectively. Cell cycle and apoptosis were determined by flow cytometry. Cell proliferation was measured by MTT assay. Our data showed that the novel survivin-targeted siRNA could efficiently knockdown the expression of survivin and inhibit cell proliferation. Survivin mRNA was reduced by 95% after 48h treatment with 20nM siRNA. In addition, the siRNA could markedly arrest the cell cycle at the G2/M checkpoint and induce cellular apoptosis in a dose-dependent manner. The percentage of apoptotic cells reached 50% when treated with 40nM siRNA. In conclusion, we have identified a novel chemically modified siRNA against survivin that is highly efficient and delineated its mechanism of action, thus demonstrating a potential therapeutic role for this molecule in cancer. Further evaluation of this siRNA for therapeutic activity is warranted.
Keywords: survivin, RNA interference, cancer, apoptosis, cell cycle checkpoint
1. Introduction
Survivin is a member of the inhibitor of apoptosis (IAP) protein family 1, 2. It inhibits apoptosis and regulates cell division 3-6. Sustained overexpression of survivin has been shown to be cancer specific 7-9. In addition, elevated expression of survivin plays a significant role in the inhibition of apoptosis 10-13.These factors suggest that survivin is a potential therapeutic target 14.
Growth inhibition and apoptosis induction are important mechanisms of cancer therapy 15. RNA interference (RNAi) by small interfering RNA (siRNA) can be used to reduce target gene expression in a sequence specific manner by degradation of the corresponding mRNA 16-19. After uptake by cells, siRNA is loaded into a RNA-induced silencing complex (RISC) 20, 21. The passenger strand is then degraded and the remaining strand (guide strand) binds to a complementary RNA molecule, which is then degraded 22. Gene silencing induced by siRNA is highly efficient and specific to the target gene and therefore has potential application in cancer treatment 23, 24.
In recent years, several siRNA sequences targeting survivin have been reported 25. However, they generally show only moderate activity 26. Unmodified siRNA have issues such as poor stability, off-target effect and immune stimulation 27. Indeed, modifications of the siRNA backbone by chemical groups, such as 2′-O-methyl (OMe) and 2′-fluoro (F), alone or in combination 28, 29, can improve serum stability and reduce off-target effects 30. However, siRNA modification can adversely affect its gene-silencing activity, thus presenting a critical challenge for siRNA drug development 31.
In order to achieve maximum therapeutic effect, it is essential to identify the most active form of drugs. Therefore, several 2′-OMe chemical groups were introduced into a novel survivin siRNA (siRNA-1) and the improvement in potency was evaluated in vitro in the present study.
2. Results and Discussion
2.1. Down-regulation of survivin in human tumor cell lines
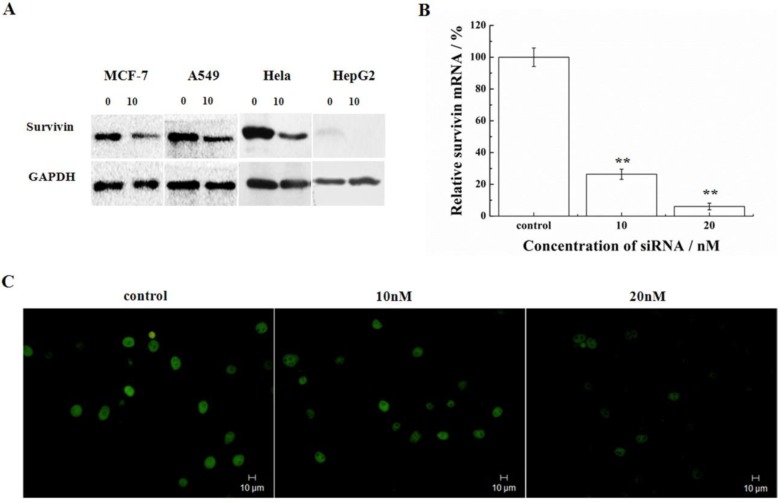
Silencing of survivin expression was examined in a number of cell lines representing different types of tumors (MCF-7, A549, HeLa, and HepG2). Following transfection of cells with 10nM siRNA-1, the protein of survivin was determined by Western blot. HeLa and A549 cells had higher expression of survivin compared with the HepG2 and MCF-7 cells. In these cell lines, the siRNA targeting survivin successfully down-regulated the expression levels of survivin protein after 48h treatment with siRNA-1 (Figure 1A). The mRNA levels of survivin were determined by real-time qRT-PCR at 48h after transfection with different concentrations of siRNA-1 in HeLa cells. As shown in Figure 1B, survivin transcription was reduced by more than 70% at the transcriptional level. At 20nM siRNA, survivin mRNA was reduced by 95%. Analysis by immunofluorescence revealed survivin localization in the nucleus. In cells treated with increasing concentrations of siRNA-1, the fluorescence intensity was gradually diminished (Figure 1C). The cells treated with 20nM siRNA-1 had the weakest fluorescence intensity under a fluorescence microscope. These data suggested concentration-dependent down-regulation of survivin by siRNA-1. In addition, as shown in Figure 1A, the differential expression of survivin in the cells treated by siRNA was cell-line dependent.
Figure 1.
Survivin silencing by siRNA-1 in a number of cell lines. (A) Survivin expression analyzed by Western blot 48h after transfection with siRNA-1. (B) The levels of survivin mRNA determined by real-time qRT-PCR 48 h after transfection in HeLa cells. (C) Survivin expression analyzed by immunofluorescence after transfection with 10nM or 20nM siRNA-1. **Statistically significant at p<0.01.
2.2. Effectiveness of siRNA in MCF-7 cells
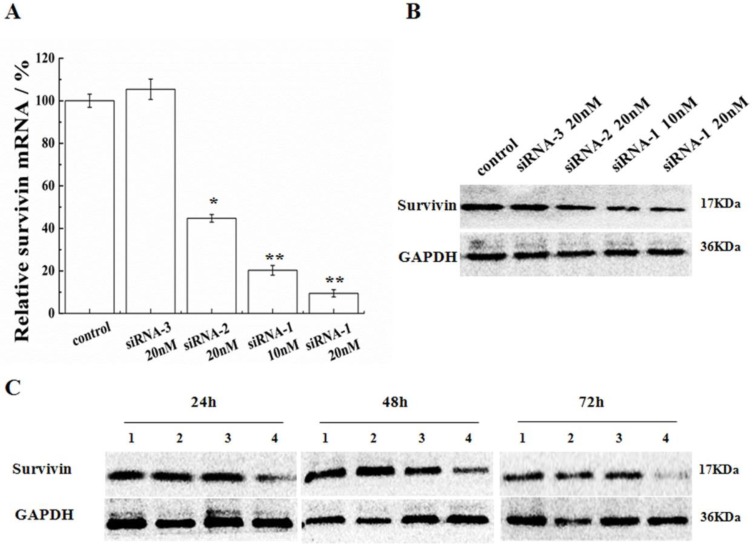
In order to validate the efficiency of siRNA-1 on MCF-7 cells, dosages and durations of treatment were varied. Following transfection by siRNA-1, survivin mRNA and protein expression levels in MCF-7 cells were determined by real-time RT-PCR and Western blot, respectively. As shown in Figure 2A, B, the positive control (siRNA-2) and novel sequence siRNA (siRNA-1) both down-regulated survivin mRNA/protein expression relative to untreated and negative control (siRNA-3) treated cells. With the increasing concentration of siRNA, mRNA and protein levels of survivin were both reduced to a greater extent. At the same dosage, the potency of new siRNA-1 was nearly 1.8 times as high as the positive control, siRNA-2. In addition, protein levels of survivin were analyzed by Western blot at 24, 48 and 72h after transfection (Figure 2C). At 24h after transfection, survivin protein was already reduced. Survivin expression inhibition reached 80% after 72h. In contrast, the inhibitory effect of positive control siRNA-2 was not significant and the siRNA modified by 2′-OMe was more efficient than ordinary siRNA.
Figure 2.
Inhibition of survivin expression in MCF-7 cells. (A) Survivin mRNA levels were analyzed after transfection with siRNAs. Survivin levels were expressed relative to GAPDH. (B) Protein levels of survivin were analyzed by Western blot 48 h after transfection with siRNAs. (C) Protein levels of survivin were analyzed by Western blot at 24, 48 and 72 h after transfection, (1) untreated cells, (2) 20nM siRNA-2, (3) 10nM siRNA-1, (4) 20nM siRNA-1. *Statistically significant with p<0.05, **Statistically significant with p<0.01.
2.3. siRNA induced G2/M Cell Cycle Arrest
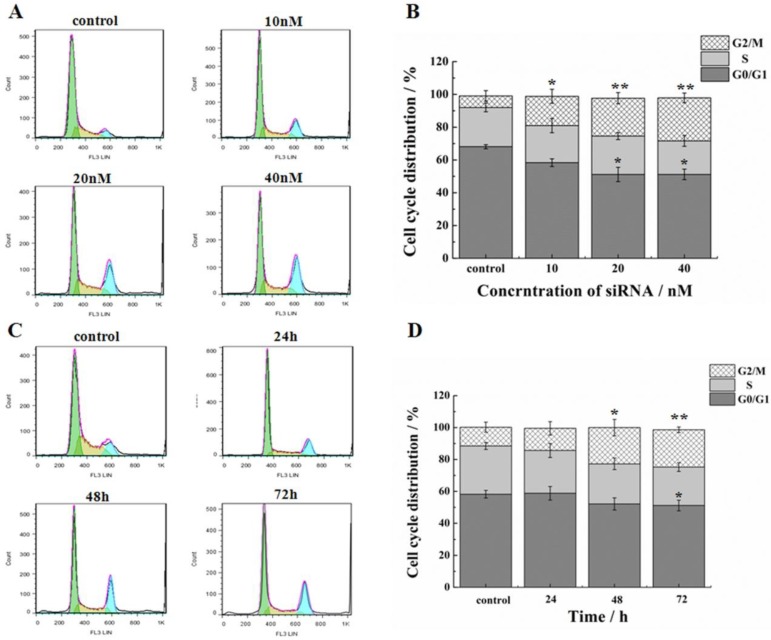
In addition to the suppression of survivin, we evaluated the effect of siRNA on proliferation and progression of cell cycle in HeLa cells. Treatment of cells by siRNA-1 at 48h induced G2/M cell cycle arrest. In addition, the G0/G1 phase ratio decreased significantly (Figure 3A, B). Cell cycle arrest reached the maximum at 48h (Figure 3C, D). At 72 h after transfection, the G2 phase was still blocked. In the meantime, the percentage of cells in the S phase was increased. Based on these data, cell cycle arrest was induced by siRNA-1, mainly at the G2/M phases.
Figure 3.
Effects of down-regulation of survivin on cell cycle. (A) Flow cytometry analysis of the cell cycle of HeLa cells at 48h after transfection with different concentrations of siRNA-1. (B) Cell cycle distribution of HeLa cells at 48h after transfection with different concentrations of siRNA-1. (C) Flow cytometry analysis of the cell cycle of HeLa cells at 24, 48, and 72 h after transfection with 40nM siRNA-1. (D) Cell cycle distribution of HeLa cells at 24, 48, and 72 h following transfection with 40nM siRNA-1. *Statistically significant with p<0.05, **Statistically significant with p<0.01.
2.4. Inhibitory effect on cell proliferation by siRNA
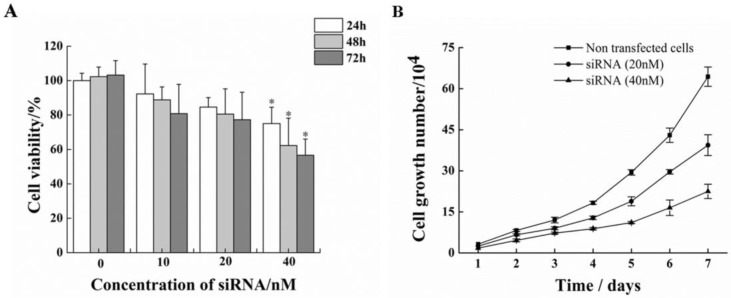
Treatment of HeLa cells with siRNA-1 over 72h caused remarkable reductions in cell proliferation compared to non-treated cells (Figure. 4A). At 72h after transfection, cell proliferation inhibition rate reached to nearly 50%. Furthermore, the effect of siRNA-1 on growth of HeLa cells was investigated (Figure. 4B). The number of surviving cells treated with 40nM siRNA-1 on the fourth day was 50% relative to the control group.
2.5. siRNA induces cell apoptosis and alters apoptosis-related signaling molecules in HeLa cells
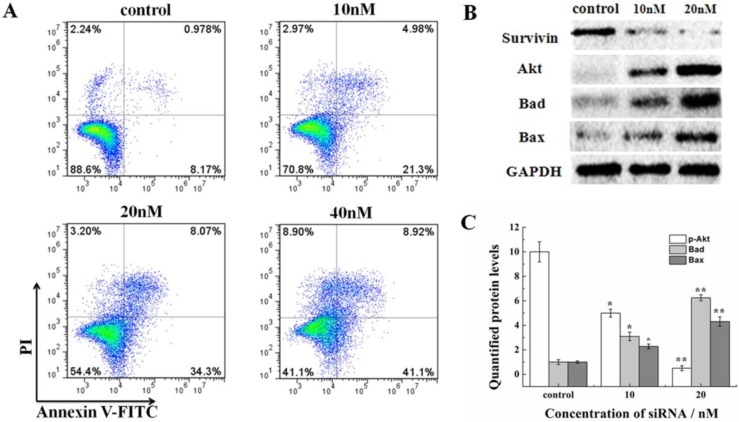
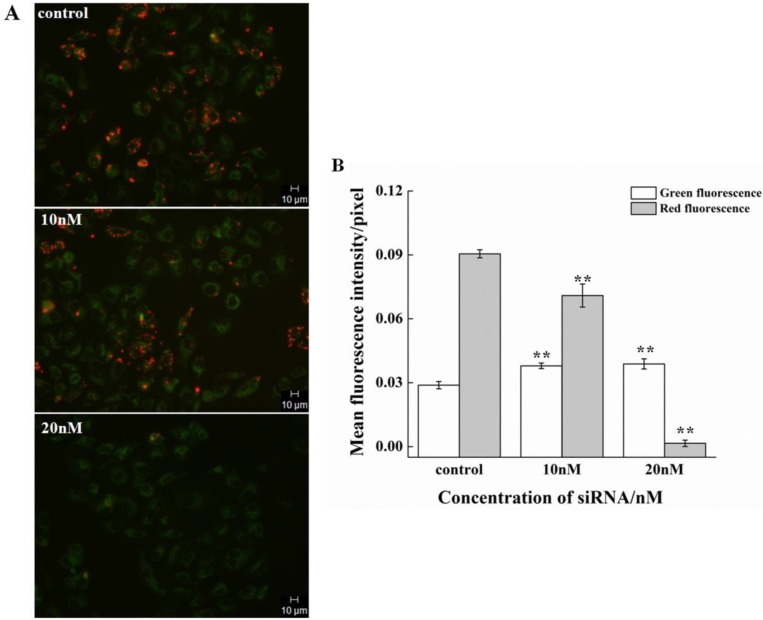
The effect on the cell cycle and proliferation from siRNA-1 was obvious. Cell apoptosis was also studied. Treatment of HeLa cells with different concentrations of siRNA-1 induced different degrees of apoptosis detected by flow cytometry. As shown in Figure 5A, the percentage of apoptotic cells increased with the concentration of siRNA-1. When the concentration of siRNA reached 40nM, the percentage of apoptotic cells was reached 50%. To characterize the molecular mechanism by which survivin alters apoptosis in HeLa cells, we examined the expression of apoptosis-associated proteins in response to survivin. The expression levels of p-Akt, Bax, and Bad were examined using Western blot at 48 h after transfection (Figure. 5B). The protein levels were quantified by densitometry (Figure. 5C). Apoptosis-inducing proteins Bax and Bad were increased in response to the down-regulation of survivin. This showed that siRNA-1 promoted cell apoptosis via Bad and Bax. At the same time, Akt signaling pathway was involved, and the level of p-Akt was similarly down-regulated with a decrease of survivin. In a subsequent experiment, we examined whether siRNA could inhibit the decrease of mitochondrial membrane potential (∆ψm) induced by siRNA (Fig.6A). Under the control conditions, JC-1 emitted high intensity of red fluorescence, and with the increasing concentration of siRNA-1, red fluorescence intensity became weaker compared with control. The data demonstrated that siRNA promoted the dissipation of ∆ψm, which confirmed that cell apoptosis was induced by the Akt signal pathway. In summary, siRNA-1 can affect cell cycle progression, cause G2/M phase arrest and induce tumor cell apoptosis, particularly through the AKT pathway.
Figure 5.
The novel siRNA-1 alters apoptosis-related signaling molecules in HeLa cells. (A) The effect of different concentrations of siRNA-1 on cell apoptosis. (B) The expression levels of the apoptosis signaling proteins Akt, Bax, and Bad examined using Western blot. (C) Quantitation of apoptosis signaling proteins treated by different concentrations of siRNA. *Statistically significant with p<0.05, **Statistically significant with p<0.01.
Figure 6.
The effect of siRNA-1 on ∆ψm at 48h in HeLa cells. (A) HeLa cells exposed to media only (control), 10nM siRNA, or 20nM siRNA. (B) Quantitation of the green and red fluorescence. **Statistically significant with p<0.01.
3. Experimental Section
3.1. siRNA Design
siRNAs were purchased from Guangzhou RiboBio Co. (Guangzhou, China). The sequences used are as follows (Table 1). siRNA purification was conducted by reverse-phase high performance liquid chromatography (HPLC). The second siRNA (siRNA-2), adopted from a previous study, was used as the positive control targeting survivin 26. The third siRNA (siRNA-3) was used as a negative control.
Table 1.
siRNAs sequences
| siRNA-1 | sense | mGCA GGU UCC UmUA UCU GUCA dTdT |
| antisense | UGA mCAG AmUA AGG AAC CUGmCdTdT | |
| siRNA-2 | sense | GGC UGG CUU CAU CCA CUGC dTdT |
| antisense | GCA GUG GAU GAA GCC AGCC dTdT | |
| siRNA-3 | sense | UUC UCC GAA CGU GUC ACG UTT |
| antisense | ACG UGA CAC GUU CGG AGA ATT |
m, represents as a single 2′-OMe modification was made at the position of the siRNA strand.
3.2. Cell lines and cell culture
Cell lines were purchased from the American Type Culture Collection (ATCC; Rockville, MD). Cells were grown in DMEM or RPMI-1640 medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics/antimycotics (Sigma-Aldrich, St. Louis, MO) at 37˚C in a humidified atmosphere containing 5% CO2.
3.3. Cell Transfection
Cells (1.4~1.6×105cells/well) were seeded in 6-well plates and incubated at 37˚Cuntilthey reached60-70%confluency. Before transfection, culture medium was removed and replaced with fresh serum-free Opti-MEMI (Gibco) medium. Tumor cells were transfected with siRNA-1 or controls using RNAiMAX (Invitrogen, Grand Island, NY). The culture supernatant was replaced after 4h and fresh medium containing 10% FBS was added. The cells were harvested and evaluated after 48h.
3.4. Real-time RT-PCR for determination of survivin mRNA
Survivin gene expression was determined by real-time RT-PCR. Total RNA was extracted from the transfected cells using TRIZOL reagent (TaKaRa, Dalian, China). Complementary deoxyribonucleic acids (cDNAs) were synthesized by reverse transcription from 1μg of total RNA. Forward and reverse primers used were as follows: 5'-CAGTGTTTCTTCTGCTTCAAGG-3' and 5'-CTTATTGTTGGTTTCCTTTGCAT-3' (Sangon Biotech, China). SYBR Green PCR kit (TaKaRa, Dalian, China) was used for the PCR reaction and each sample was analyzed in triplicate. The relative expression of each mRNA was detected and normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as described 32.
3.5. Western blot analysis for expression of survivin protein
Western blot analysis was performed following standard methods 33. After a period of incubation, cells were washed twice with PBS. Total protein was extracted on ice by RIPA (Sigma-Aldrich, St. Louis, MO). Protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit (Bio-rad, Hercules, CA). Electrophoretic analysis of 30μl protein from each sample was performed by SDS-PAGE. The proteins are transferred to a PVDF membrane after the separation. The detection of target protein was carried out using a secondary antibody (sheep anti-rabbit antibody, 1:10000 dilutions, Pierce, Rockford, IL, USA) following the primary antibody (rabbit anti-human survivin antibody, 1:1000 dilutions, Cell Signaling Technology, Inc.). Binding was detected using an enhanced chemiluminescence (ECL) kit (GE Healthcare, United Kingdom). The results were analyzed by gel imaging and analysis system (Upland, CA, USA).
3.6. Visualization of survivin expression by fluorescence microscopy
The expression of survivin in cancer cells was detected using immunofluorescence 34. Coverslips were placed into a 12 well plate and cells were seeded (1×104) into the coverslips/well. The cells were incubated overnight at 37˚C. Tumor cells were transfected with different concentrations of siRNA-1. After 48h, medium was removed from wells. The cells were washed with warm PBS and fixed in 4% paraformaldehyde/PBS for 15min. Cells were permeabilized by TritonX-100 and then treated with primary survivin antibody and FITC-conjugated donkey anti-goat antibody. Then, mounting solution was added. The cells were visualized on a fluorescence microscope in the dark.
3.7. Flow Cytometry analysis of cell cycle and apoptosis
Cell cycle and apoptosis were determined by flow cytometry 35. Cells (2×105cells/well) were plated onto 6-well plates overnight and then replaced with serum free medium for cell cycle synchronization. Then the cells were treated with 10, 20, 40nM siRNA complex.
The cells were harvested and fixed in 75% methanol at 4℃ overnight. The cells were washed twice with cold PBS and stained with PI (0.5mg/ml RNase, and 0.1mg/ml PI in PBS) for 30 min at room temperature. The stained cells were characterized by a flow cytometer flow cytometer (Beckman Coulter Corp., Tokyo, Japan), and DNA content was analyzed by FlowJo software.
After 48h, the cells were harvested and washed twice with PBS. The cells were stained using Annexin V-FITC/PI kit (KeyGEN Biotech, China), following manufacturer's protocol.
After incubation for 20 min at room temperature in the dark, cell apoptosis was immediately detected on a flow cytometer as described 36, 37.
3.8. MTT Assay
Cell viability was measured by MTT assay 38. Cells (1×104cells/well) were grown in 96-well plates and incubated overnight. Various concentrations of siRNA (0-50nM) and RNAiMAX complexes were added to the cells after the replacement of culture medium with serum free medium. The medium was removed after 4 h and the cells were cultured in fresh medium for a period of time. MTT assay was performed to evaluate cell viability. Briefly, 20μl MTT stock solution (5mg/ml in PBS) was added and incubated for 4 h at 37˚C. The medium was then removed. After that, 150μl /well DMSO was added to dissolve formazan crystals that formed and the absorbance was measured at 540 nm using an automatic microplate reader (Biotek, VT, USA) 39.
3.9. Cell proliferation
Cells (1×104cells/well) were grown in 24-well plates and incubated overnight. Different concentrations of siRNA complexes were added and after 4 h incubation, the cells were cultured in fresh medium. Cells from three wells were counted with trypan blue staining method in each group every day. The survival rate of the cells was observed for a total of seven days.
3.10 Mitochondrial membrane potential analysis
JC-1 (5, 5´, 6, 6´-tetrachloro-1, 1´, 3, 3´-tetraethylbenzimidazol carbocyanine iodide, Sigma-Aldrich), a fluorescent probe, was used to measure alterations in ∆ψm. Healthy cells with a high ∆ψm exhibit red fluorescence. Meanwhile, apoptotic or unhealthy cells with a low ∆ψm exhibit green fluorescence (19). HeLa cells were seeded onto dishes at a density of 1x105cells/well. Subsequent to pretreatment with different concentrations of siRNA, cells were incubated with JC-1 at a final concentration of 2 µM at 37˚C for 15 min in the dark. Cells were washed with PBS and changes in mitochondrial fluorescence were analyzed on a fluorescent microscope.
3.11. Statistical Analysis
The data were analyzed using SPSS 16.0 software (IBM, Armonk, New York). The difference between two independent samples was analyzed by Student's t test. A statistically significant difference was considered to be present at p<0.05.
4. Conclusions
Because survivin is an essential component of most types of cancers 40-42, it can serve as a biomarker for a number of malignancies 43-45. Although targeting survivin by siRNA has shown some promise in cancer, the potency of siRNA needs further improvement. In order to achieve maximum therapeutic effect, however, it is essential to identify the most active form of siRNA drugs. In this respect, rational design of the siRNA itself must precede the design of the delivery vehicle.
We have shown that a novel siRNA sequence we designed (siRNA-1), which was modified by several 2′-OMe chemical groups in both strands, is much better than the positive control sequence reported previously. In addition, the suppression of siRNA was cell-line dependent. Meanwhile, siRNA-1 exerted a significant effect on cell proliferation and cell cycle by enhancing the G2/M phase arrest. The contribution of siRNA to cell proliferation, inhibition, and cell apoptosis were also regulated and controlled by signaling pathways. For instance, Bad and Bax expression increased in the cancer cells treated with siRNA, along with induction of apoptosis. Moreover, the Akt-mediated signal pathway may be involved in apoptosis induction. In response to apoptotic stimuli, survivin is trafficked from the mitochondria to the cytosol where it can inhibit apoptosis. Similarly the decreasing ∆ψm is one of the important hallmarks of cellular apoptosis 46, 47. The results of this study suggest that the novel siRNA sequence has high potency and warrants further evaluation as a therapeutic agent. Clinical translation of siRNA requires both optimization of the siRNA design and development of an efficient delivery vehicle. Our future work will focus on assessing the in vivo activities of the novel survivin siRNA.
Author Contributions
Yuhuan Li and Da Liu performed the laboratory experiments. Yuhuan Li drafted the manuscript; Lesheng Teng and Robert. J Lee conceived and designed most of the studies and revised the manuscript; Yong Cai and Jing Xie participated in study design and the manuscript revision; Yulin Zhou and Yujing Li contributed research materials and were involved in helpful discussions.
Figure 4.
Inhibitory effect on cell proliferation by siRNA-1 targeting survivin. (A) MTT assay analysis of viability of HeLa cells following 24-72 h of treatment with different doses of siRNA-1. (B) Assessment of HeLa cell growth after transfection with different concentrations of siRNA over seven days. *Statistically significant with p<0.05.
References
- 1.Srinivasula SM, Ashwell JD. IAPs: what's in a name? Mol Cell. 2008;30:123–35. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohi T, Okada K, Xia F. et al. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279:34087–90. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 3.Mita AC, Mita MM, Nawrocki ST. et al. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Biddle D, Hanks AN. et al. Activation of dual apoptotic pathways in human melanocytes and protection by survivin. J Invest Dermatol. 2006;126:2247–56. doi: 10.1038/sj.jid.5700381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colnaghi R, Wheatley SP. Liaisons between Survivin and Plk1 during Cell Division and Cell Death. J Biol Chem. 2010;285:22592–604. doi: 10.1074/jbc.M109.065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zang XP, Pento JT. SiRNA inhibition of ER-alpha expression reduces KGF-induced proliferation of breast cancer cells. Anticancer Res. 2008;28:2733–5. [PubMed] [Google Scholar]
- 7.Waligorska-Stachura J, Andrusiewicz M, Sawicka-Gutaj N, Survivin Delta Ex3 Overexpression in Thyroid Malignancies. Plos One; 2014. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng W, Meng FK, Liu ZM. et al. Bortezomib-based chemotherapy regimens can improve response in newly diagnosed multiple myeloma patients with bcl-2 and survivin overexpression. Int J Clin Exp Patho. 2014;7:4239–46. [PMC free article] [PubMed] [Google Scholar]
- 9.Poomsawat S, Punyasingh J, Vejchapipat P. Overexpression of Survivin and Caspase 3 in Oral Carcinogenesis. Appl Immunohisto M M. 2014;22:65–71. doi: 10.1097/PAI.0b013e31828a0d0c. [DOI] [PubMed] [Google Scholar]
- 10.Tazo Y, Hara A, Onda T. et al. Bifunctional roles of survivin-Delta Ex3 and survivin-2B for susceptibility to apoptosis in endometrial carcinomas. J Cancer Res Clin. 2014;140:2027–37. doi: 10.1007/s00432-014-1762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YB, Gao XH, Deeb D. et al. Ubiquitin-proteasomal degradation of antiapoptotic survivin facilitates induction of apoptosis in prostate cancer cells by pristimerin. Int J Oncol. 2014;45:1735–41. doi: 10.3892/ijo.2014.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohi T, Beltrami E, Wall NR. et al. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114:1117–27. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbot DC, Blackhall FH, Kowalski D, A randomized open-label phase II study evaluating antitumor activity of the survivin antisense oligonucleotide LY2181308 (LY) in combination with docetaxel (DO) for second-line treatment of patients with non-small cell lung cancer (NSCLC) using change in tumor size (CTS) J Clin Oncol; 2013. p. 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobahat M, Narendran A, Riabowol K. Survivin as a Preferential Target for Cancer Therapy. Int J Mol Sci. 2014;15:2494–516. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendruschk S, Wiedemuth R, Aigner A. et al. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro Oncol. 2011;13:1074–89. doi: 10.1093/neuonc/nor098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang ZG, Yu B, Zhu J. et al. A microfluidic method to synthesize transferrin-lipid nanoparticles loaded with siRNA LOR-1284 for therapy of acute myeloid leukemia. Nanoscale. 2014;6:9742–51. doi: 10.1039/c4nr01510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boukany PE, Morss A, Liao WC. et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–54. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 19.Wen Y, Meng WS. Recent In Vivo Evidences of Particle-Based Delivery of Small-Interfering RNA (siRNA) into Solid Tumors. J Pharm Innov. 2014;9:158–73. doi: 10.1007/s12247-014-9183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou CG, Yang ZG, Teng LS. Nanomedicine based on Nucleic Acids: Pharmacokinetic and Pharmacodynamic Perspectives. Curr Pharm Biotechno. 2014;15:829–38. doi: 10.2174/1389201015666141020155620. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, Wang XM, Zhou CG. et al. Insight into Mechanisms of Cellular Uptake of Lipid Nanoparticles and Intracellular Release of Small RNAs. Pharm Res-Dordr. 2014;31:2685–95. doi: 10.1007/s11095-014-1366-7. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir SM, Harborth J, Lendeckel W. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Devi GR. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006;13:819–29. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- 24.Seth S, Matsui Y, Fosnaugh K. et al. RNAi-based Therapeutics Targeting Survivin and PLK1 for Treatment of Bladder Cancer. Mol Ther. 2011;19:928–35. doi: 10.1038/mt.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XJ, Zhang XY, Li XY. et al. The Role of Survivin in Podocyte Injury Induced by Puromycin Aminonucleoside. Int J Mol Sci. 2014;15:6657–73. doi: 10.3390/ijms15046657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharmaceut. 2006;3:579–88. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 27.Lee SJ, Son S, Yhee JY. et al. Structural modification of siRNA for efficient gene silencing. Biotechnol Adv. 2013;31:491–503. doi: 10.1016/j.biotechadv.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Prakash TP, Allerson CR, Dande P. et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J Med Chem. 2005;48:4247–53. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 29.Kraynack BA, Baker BF. Small interfering RNAs containing full 2 '-O-methylribonucleotide-modified sense strands display argonaute2/eIF2C2-dependent activity. Rna. 2006;12:163–76. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng J, Zhang L, Zhang J. et al. Single modification at position 14 of siRNA strand abolishes its gene-silencing activity by decreasing both RISC loading and target degradation. FASEB J. 2013;27:4017–26. doi: 10.1096/fj.13-228668. [DOI] [PubMed] [Google Scholar]
- 32.Chakrabarti S, Wu XF, Yang ZG. et al. MOG1 Rescues Defective Trafficking of Na(v)1.5 Mutations in Brugada Syndrome and Sick Sinus Syndrome. Circ-Arrhythmia Elec. 2013;6:392–401. doi: 10.1161/CIRCEP.111.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang ZG, Sun W, Hu KL. Adenosine A(1) receptors selectively target protein kinase C isoforms to the caveolin-rich plasma membrane in cardiac myocytes. Bba-Mol Cell Res. 2009;1793:1868–75. doi: 10.1016/j.bbamcr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Yang ZG, Sun W, Hu KL. Molecular mechanism underlying adenosine receptor-mediated mitochondrial targeting of protein kinase C. Bba-Mol Cell Res. 2012;1823:950–8. doi: 10.1016/j.bbamcr.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YP, Kong QH, Huang Y. et al. Inhibition of c-FLIP by RNAi enhances sensitivity of the human osteogenic sarcoma cell line U2OS to TRAIL-induced apoptosis. APJCP. 2015;16:2251–6. doi: 10.7314/apjcp.2015.16.6.2251. [DOI] [PubMed] [Google Scholar]
- 36.Xie J, Teng LS, Yang ZG, A Polyethylenimine-Linoleic Acid Conjugate for Antisense Oligonucleotide Delivery. Biomed Res Int; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin LL, Batra S, Douda DN. et al. CXCL1 Contributes to Host Defense in Polymicrobial Sepsis via Modulating T Cell and Neutrophil Functions. J Immunol. 2014;193:3549–58. doi: 10.4049/jimmunol.1401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder BR, Ghare MI, Bhattacharya C. et al. The disaccharide moiety of bleomycin facilitates uptake by cancer cells. J Am Chem Soc. 2014;136:13641–56. doi: 10.1021/ja507255g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang XM, Huang XM, Yang ZG. et al. Targeted Delivery of Tumor Suppressor microRNA-1 by Transferrin- Conjugated Lipopolyplex Nanoparticles to Patient-Derived Glioblastoma Stem Cells. Curr Pharm Biotechno. 2014;15:839–46. doi: 10.2174/1389201015666141031105234. [DOI] [PubMed] [Google Scholar]
- 40.Xing Z, Conway EM, Kang C. et al. Essential role of survivin, an inhibitor of apoptosis protein, in T cell development, maturation, and homeostasis. J Exp Med. 2004;199:69–80. doi: 10.1084/jem.20031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roca H, Varsos ZS, Mizutani K. et al. CCL2, survivin and autophagy: new links with implications in human cancer. Autophagy. 2008;4:969–71. doi: 10.4161/auto.6822. [DOI] [PubMed] [Google Scholar]
- 42.Small S, Keerthivasan G, Huang Z. et al. Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia. 2010;24:1920–6. doi: 10.1038/leu.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikeguchi M, Kaibara N. survivin messenger RNA expression is a good prognostic biomarker for oesophageal carcinoma. Br J Cancer. 2002;87:883–7. doi: 10.1038/sj.bjc.6600546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu B, Gonzalez A, Massion PP. et al. Nuclear survivin as a biomarker for non-small-cell lung cancer. British journal of cancer. 2004;91:537–40. doi: 10.1038/sj.bjc.6602027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan DJ, Rexhepaj E, O'Brien SL. et al. Altered cytoplasmic-to-nuclear ratio of survivin is a prognostic indicator in breast cancer. Clin Cancer Res. 2008;14:2681–9. doi: 10.1158/1078-0432.CCR-07-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dohi T, Beltrami E, Wall NR. et al. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114:1117–27. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Liu J, Liu X. et al. Panax notoginseng saponins inhibit ischemia-induced apoptosis by activating PI3K/Akt pathway in cardiomyocytes. J Ethnopharmacol. 2011;137:263–70. doi: 10.1016/j.jep.2011.05.011. [DOI] [PubMed] [Google Scholar]