Abstract
Initially identified as a marker of coiled bodies (now Cajal bodies or CBs), the protein coilin was discovered a quarter of century ago. Coilin is now known to scaffold the CB, but its structure and function are poorly understood. Nearly devoid of predicted structural motifs, coilin has numerous reported molecular interactions that must underlie its role in the formation and function of CBs. In this review, we summarize what we have learned in the past 25 years about coilin's structure, post-transcriptional modifications, and interactions with RNA and proteins. We show that genes with homology to human coilin are found in primitive metazoans and comment on differences among model organisms. Coilin's function in Cajal body formation and RNP metabolism will be discussed in the light of these developments.
Keywords: Cajal body, coilin, small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), small Cajal body-specific RNA (scaRNA), small nuclear ribonucleoprotein particle (snRNP), splicing, spliceosome, telomere, histone
Coilin History
Cajal bodies (CBs) were discovered at the beginning of 20th century by Spanish cytologist Ramón y Cajal as structures in neuronal nuclei stained with silver. The bodies were then rediscovered and named coiled bodies in the 1960's due to their specific appearance in electron microscope images; they were later renamed Cajal bodies in honor of their discoverer.1 CB research accelerated with the discovery of autoantibodies present in autoimmune patient sera, which were shown to react with a protein of apparent molecular weight 80 kD (named p80-coilin) that specifically localized to CBs.2 The human protein was characterized and its cDNA cloned.3 The term “coilin” first appeared in a paper published a year earlier, in which Ivan Raska and his co-workers named the protein after coiled bodies and used anti-coilin autoimmune sera to analyze the relationship between CBs and nucleoli.4
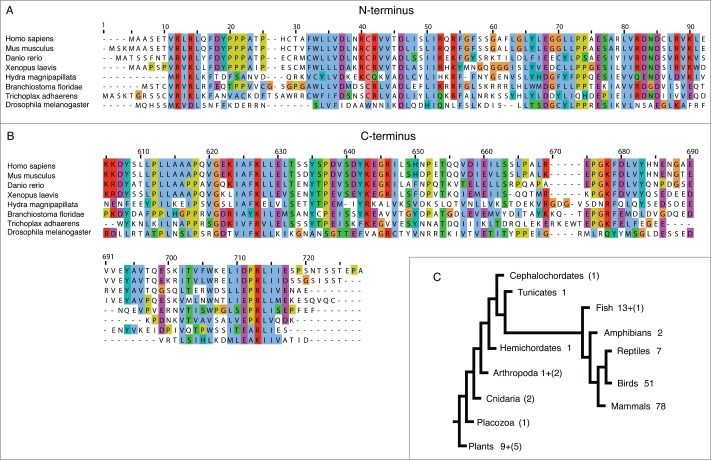
Soon after human coilin was discovered, the Xenopus laevis homologue was identified and shown to specifically localize to sphere organelles within the germinal vesicles of X. laevis oocytes.5 Matera and colleagues cloned mouse, rat and zebrafish coilin genes and revealed the conservation of N- and C-terminal regions across these species (Fig. 1). Coilin's amino acid sequence is not well conserved and is characterized by low complexity and unstructured regions. Hence, it took considerable effort and time to identify genes for coilin in plants6 and fruit flies,7 despite prior evidence for the existence of CBs in these organisms.8,9 Meanwhile, the genomes of a wide variety of organisms have become available. A current search for coilin homologues revealed that predicted proteins with significant sequence homology to human coilin can be found in as primitive an organism as Trichoplax adherens (Fig. 1), which is one of the simplest metazoans. Surprisingly, human coilin is more similar to the predicted Trichoplax coilin than it is to D. melanogaster coilin.
Figure 1.
Coilin is highly conserved among metazoans. Predicted coilin amino acid sequences (RefSeq) for the indicated species were aligned with coilin proteins identified by homology search in Cephalochordates (Lancelet), Cnidarians (Hydra) and Placozoa (Trichoplax). Alignments of conserved (A) N-terminal and (B) C-terminal regions are shown, demonstrating the greatest divergence of Drosophila melanogaster coilin from the other species. (C) Cladogram shows numbers of annotated and/or predicted coilin proteins per clade together with hypothetical coilin proteins found by homology search (numbers in brackets).
Coilin protein domains
Evolutionary conservation at the amino acid level is highest within coilin's N- and C-termini. The middle portion is not conserved and low complexity region and RG box found in human coilin appear to be specific for vertebrates. The first N-terminal 92-amino acid domain was shown to self-interact and to be essential for proper targeting of coilin to CBs.10-12 The central part contains 2 nuclear localization signals and a putative nucleolar localization sequence,12 which might explain coilin's affinity toward the nucleolus, which was noticed already 25 years ago.4 The conserved C-terminal domain folds into a Tudor domain like structure.13 Several Tudor domains were found to bind methylated amino acids but direct tests did not show any interaction of coilin Tudor domain with monomethyl-lysine, trimethyl-lysine and dimethyl-arginine.13 However, the C-terminal domain was shown to interact with Sm proteins, which contain symmetrically dimethylated arginines.14-16 Coilin interaction is stronger when Sm proteins are purified from eukaryotic cells rather than bacterially expressed, suggesting that posttranscriptional modifications might play a role in Sm protein-coilin interactions.15,16 Thus, the precise molecular mechanism of coilin-Sm protein binding is still unknown.
The search for coilin function
Coilin loss of function has been analyzed in Arabidopsis thaliana, Drosophila melanogaster, Danio rerio and Mus musculus. It was initially disappointing that coilin gene disruptions in plants and flies did not detectably affect viability or fertility.6,7 Yet coilin depletion has strong effects on the viability of vertebrate embryos. In mouse, gene disruption leads to a dramatic loss of homozygote pups in matings of heterozygotes, and coilin−/− mice are significantly less fertile.17,18 In zebrafish embryos, coilin depletion through morpholino injection was lethal within the first 24 hours of development and was accompanied by reduced levels of snRNPs and spliced mRNAs.19 Fish embryos were rescued upon injection of spliceosomal small nuclear ribonucleoprotein particles (snRNPs), which suggests that coilin's essential function in embryos is to promote macromolecular assembly of snRNPs.19 This conclusion agrees with early findings that snRNAs and snRNP proteins are concentrated in CBs.20,21
Spliceosomal snRNPs are assembled and recycled in CBs.22-28 Mathematical modeling and measurement of snRNP kinetics in CBs suggest that concentration of snRNPs in the CB increases snRNP assembly rate by a factor of 10.29,30 Similarly, co-localization of snRNAs and scaRNAs, which guide snRNA modifications, in CBs might enhance snRNA modification efficiency. However, in D. melanogaster, scaRNAs function in the absence of coilin, showing that concentration of snRNA and scaRNAs in CBs is not essential in flies.31,32 Recent work showed that coilin directly binds snoRNAs33 and that snoRNAs shuttle via CBs before they reach their nucleolar destination.33,34 Therefore it is tempting to speculate that snoRNP assembly, like snRNP assembly, takes place in CBs and that coilin plays an important role as well.
Based on these observations, we suggest that coilin promotes RNP biogenesis by acting as a chaperone of nuclear small non-coding RNAs. This might not be essential in differentiated cells or cells lines cultured in vitro, but may become critical in rapidly developing embryos. A clear example is zebrafish, where embryos depleted of coilin and lacking CBs are unable to complete embryogenesis, display splicing defects and reduced numbers of mature snRNPs.19,35 Because zebrafish embryos require splicing of zygotically transcribed pre-mRNAs,36 these combined data lead to the interpretation that coilin supports the high embryonic demand for splicing by promoting rapid snRNP assembly in CBs. This explanation could easily also apply to mouse embryos. We suggest that the viability of a small number of homozygous coilin−/− mice reflects the chance that a sufficient number of embryonic cells survive on maternal stores of snRNPs. In other systems, like plants or insects, differences in mechanisms of development might determine whether coilin's function is essential or not. For example, insects are characterized by a syncytial blastoderm that may confer a survival advantage for embryos. If snRNP assembly is slower in D. melanogaster colin null mutants, perhaps blastoderm nuclei are able to share the snRNP deficit among all of the cells of the blastula during the critical period when rapid splicing is needed. An additional or auxiliary possibility is that more cell death is tolerated in insect embryos in general. Given that conserved coilin domains in D. melanogaster diverged significantly from vertebrate coilin (Fig. 1), it is also possible that flies have evolved a mechanism for snRNP assembly that does not depend on coilin as strongly as in vertebrates.
Additional data connecting coilin, CBs and snRNPs were added this year when Novotny et al. showed that incomplete or defective snRNPs are anchored to coilin and CBs. This finding suggests that coilin is part of a quality control mechanism that proofreads final snRNP assembly.37 Again, this function might not be essential under normal conditions but becomes important when snRNP assembly is perturbed or when transcription and splicing rates are high, producing a large quantity of mono-snRNPs that require recycling and reassembly. Coilin-dependent concentration of mono-snRNPs in CBs would increase their assembly rate and at the same time prevent incomplete snRNPs from entering splicing reaction.
Coilin and CBs were suggested to be important for telomerase assembly and telomere maintenance. Early experiments detected telomerase RNA in CBs, and CBs associate with telomerase during S-phase.38-42 Interestingly, telomerase RNA localization to CBs seems to be human-specific because telomerase RNA was not found in mouse CBs.43 Telomerase RNA is retained in CBs via protein WRAP53, which binds the CAB box sequence found in telomerase RNA and many scaRNAs.39,44-47 WRAP53 interacts with coilin, providing the mechanistic link between telomerase and coilin.48,49 Telomerase RNA localization to CBs was suggested to be important for telomerase function and mislocalization of telomerase RNA from CBs correlates with reduced telomerase activity.45,50,51 In addition, coilin depletion by siRNA inhibits association of telomerase with telomeres.52,53 However, the role of coilin or CBs in telomerase assembly is unclear. Loss of coilin could be overcome by overexpression of telomerase,52 and human cancer cells lacking coilin do not exhibit any inhibition of telomerase activity or defects in telomere lengths.54 Why did coilin depletion by siRNA, which is never complete, reveal defects in telomere maintenance, when a genetic coilin knockout did not show any telomere shortening? While RNAi mediated knockdowns are transient and cells are assayed 2-3 days after RNAi treatment, knockout requires prolonged period of clone selection. Perhaps only clones that successfully adapted to coilin deletion survive. The situation is similar to mouse, where some coilin knockout embryos die during embryogenesis while surviving pups are viable despite reduce size and fertility.18
The interaction between coilin and WRAP53 could also explain why coilin is found at sites of DNA damage, and CBs are sensitive to UV irradiation.55,56 While coilin function during DNA repair is unclear, WRAP53 was recently shown to be critical for DNA double-strand break repair.57 Independent work found coilin associated with DNA repair factor Ku.58-60
Coilin could be also involved in higher order chromatin organization albeit molecular details of this function remain elusive. Genes encoding snRNAs, snoRNAs and histone mRNAs were found closely associated with CBs in cytological assays.61-67 Coilin association with snRNA and histone genes was recently re-examined by genome-wide ChIP-Seq, which detected coilin peaks at most of these genes.33 CBs contain pre-U2 snRNA68 and contact between CBs and chromatin is maintained by active snRNA gene transcription.61,69 CBs may be assembled on or transported toward active snRNA genes.70 This transcription-dependent contact between snRNA genes and CBs was recently confirmed by the finding that coilin ChIP signals on snRNA genes is cell cycle specific, with the highest signal in S-phase when snRNA genes are actively transcribed.33 Similarly, coilin binding to U7 snRNA brings histone genes to a close proximity of CBs.33,71 Taken together, these findings suggest that coilin brings several gene loci together within the 3D space of the nucleus and thereby function in global organization of chromatin.
Toward Cajal body formation
Coilin is an indispensable structural component of CBs, because coilin depletion results in CBs disintegration in all tested organisms.6,7,17,19 The domain critical for CB formation is the N-terminal 92 amino acids, which is necessary for coilin self-interaction, coilin targeting to CBs, and de novo CB formation.10-12 However, the ability of the N-terminus to form CBs is modulated by the C-terminus and even subtle differences in C-terminal sequence between mouse and human coilin affect CB nucleation.72 The C-terminus interacts with snRNP-specific Sm proteins14-16 and snRNP expression level is a factor important for CB formation.73 We recently showed that increased concentration of unassembled or defective snRNPs triggers formation of CBs in cells that are normally devoid of CBs.37 This suggests that incomplete snRNPs enhance coilin self-interaction properties and induce formation of CBs through an unknown mechanism.
Recent studies have suggested that low-complexity domains, multivalent interactions and RNA are important factors that favor liquid phase separation and the formation of cellular bodies, like nucleoli, that are not delimited by lipid bilayers.74-78 Does CB formation follow a phase separation model? Direct evidence is lacking, though hints exist. CB formation in primary fibroblasts is temperature dependent,79 consistent with the phase separation model.80 Coilin contains short stretches of serines and lysines in the central low complexity region, but their role in CB formation has not been tested. Finally, the role of RNA in CB formation has been documented.33,81,82
Coilin also directly interacts with many CB proteins (Fig. 2) and is therefore widely considered as a molecular hub, bridging otherwise distinct CB components and thereby “gluing” them together via numerous protein-protein interactions.35 Artificial tethering of individual CB components to chromatin induces CB formation, which is consistent with a model that CBs are formed via protein-protein and protein-RNA interactions.83 snRNP-specific proteins are highly active in this assay for CB formation, which is consistent with a role for snRNPs in the context of naturally occurring CBs.37,73
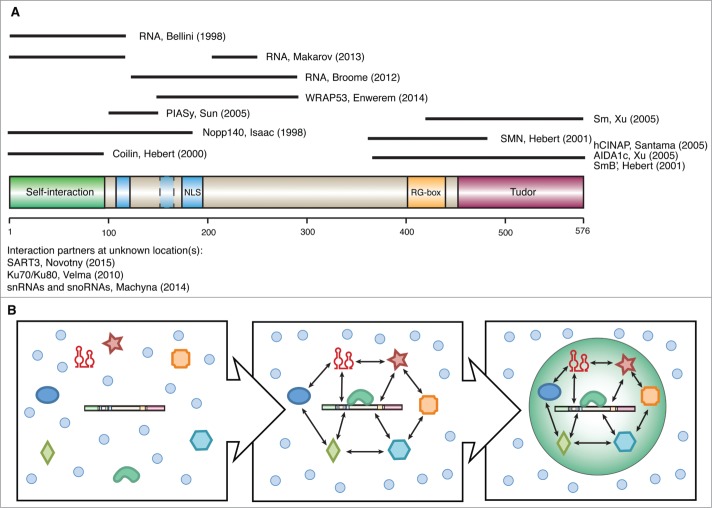
Figure 2.
Coilin interaction partners and the formation of CBs. (A) Schematic representation of human coilin protein showing key structural features: Tudor domain (purple), RG-box (orange), bipartite nuclear localization signal (blue), cryptic nucleolar localization signal (light blue) and self-interaction domain (green). Black bars signify parts of coilin known to interact with indicated proteins/RNA.12,14,15,33,37,48,58,84-86,101-104 (B) The current hypothesis of nuclear body assembly suggests that protein and RNA components (colored shapes) tend to have more interactions (double arrows) among themselves as compared to other molecules (blue circles) in the environment. When the components reach a critical concentration locally, liquid phase separation occurs and creates the nuclear body.
Coilin: A non-canonical RNA binding protein
Despite the fact that coilin does not contain a canonical RNA binding motif, its X. laevis coilin was shown to interact with poly-U and poly-G in vitro. It was noted by the same authors that the N-terminal self-interaction domain may contain a degenerate RNA recognition motif.84 Later, Makarov et al. showed that plant coilin binds U1 snRNA in vitro but the RNA binding domain was not unambiguously determined. The authors also showed that the U1 snRNA induces multimerization of Arabidopsis coilin in vitro, suggesting that RNA binding modulates coilin self-interaction.85 Finally, human coilin was shown to bind RNA in vitro and co-precipitated several RNAs from cell extracts, though it was unclear whether this interaction was direct86,87; these studies suggest that the RNA binding domain is localized in the central, least conserved part of coilin.87
Direct evidence that coilin binds RNA in vivo was recently provided by iCLIP, in which 0 Å crosslinking with UV light followed by immunopurification of coilin-RNA adducts permitted sequencing of bound RNAs.33 This study identified hundreds of short ncRNA including snRNA, snoRNA and telomerase RNA, as coilin binding partners in vivo. Interestingly, coilin prefers RNA stem-loop regions for binding.33 This may explain why previous work investigating coilin binding to homopolymer RNAs revealed low affinity interactions. Interestingly, extensive coilin-RNA interactions revealed by iCLIP agrees with original findings: CBs were first observed in neuronal sections by silver staining, which deposits on RNA-rich structures.88 In addition, coilin was first discovered as a target of autoantibodies, which often recognize ribonucleoprotein complexes or nucleic acid binding proteins (snRNPs, hnRNP proteins, histones, Ro, La, SR proteins etc.).
Post-transcriptional modifications
Many aspects of coilin function are modulated by posttranscriptional modifications. Arginines in the RG box can be symmetrically methylated. In the dimethylated state, arginines are bound by the SMN Tudor domain and this interaction mediates association of nuclear gems and CBs.14,89,90 Cells lacking 5′-methylthioadenosine phosphorylase, a key enzyme of the methionine salvage pathway, have reduced methylation activity and coilin is re-localized to nucleoli.91 However, this result has to be cautiously interpreted because 2 methyltransferases, PRMT5 and PRMT7, that symmetrically methylate arginines in Sm proteins, are important for snRNP biogenesis92 and ongoing snRNP biogenesis may be critical for CB formation.93,94
Coilin is also heavily phosphorylated and phosphorylation controls CB disassembly at mitosis, proper CB assembly after mitosis and coilin localization.12,79,95,96 Perhaps contributing to cell cycle variation in CB number, cdk2/cyclin E and the nuclear phosphatase PPM1G phosphorylate and dephosphorylate coilin in vitro.97,98 It was suggested that the interplay between coilin phosphorylation and dephosphorylation affects coilin self-association and interaction with snRNPs, SMN and RNAs.16,98-100
Future perspective
During the first 25 years, we learned that coilin is a tricky protein to deal with. Coilin often proves insoluble when expressed or otherwise purified, and little structural information exists. Coilin depletion always impacts CB size and number, yet experiments in model organisms have yielded mixed phenotypes ranging from embryonic lethality in zebrafish to no apparent phenotype in flies and plants. Future establishment of additional direct approaches to analyzing coilin structure and function will be instructive. It is encouraging that global mapping of protein and RNA interaction partners did not identify surprisingly novel proteins or RNAs,33 raising optimism that coilin's interactors are known and molecular function(s) will be revealed soon. A screen for synthetic interactions in model organisms might provide great insight, alongside bottom-up approaches in vitro. We also need to devise experiments that would identify the physical principles that hold CBs together.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
David Stanek was supported by a Fulbright scholar fellowship, the Academy of Sciences of the Czech Republic (RVO68378050) and the Czech Science Foundation (P305/12/G034).
References
- 1.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 2000; 16:273–300; PMID:11031238; http://dx.doi.org/ 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- 2.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 1991; 195:27–37; PMID:2055273; http://dx.doi.org/ 10.1016/0014-4827(91)90496-H [DOI] [PubMed] [Google Scholar]
- 3.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med 1991; 173:1407–19; PMID:2033369; http://dx.doi.org/ 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol 1990; 104:120–7; PMID:2088441; http://dx.doi.org/ 10.1016/1047-8477(90)90066-L [DOI] [PubMed] [Google Scholar]
- 5.Tuma RS, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol 1993; 122:767–73; PMID:8349728; http://dx.doi.org/ 10.1083/jcb.122.4.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell 2006; 17:2942–51; PMID:16624863; http://dx.doi.org/ 10.1091/mbc.E05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell 2009; 20:1661–70; PMID:19158395; http://dx.doi.org/ 10.1091/mbc.E08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beven AF, Simpson GG, Brown JW, Shaw PJ. The organization of spliceosomal components in the nuclei of higher plants. J Cell Sci 1995; 108 (Pt 2):509–18; PMID:7768997 [DOI] [PubMed] [Google Scholar]
- 9.Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol 2006; 172:875–84; PMID:16533947; http://dx.doi.org/ 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell 1994; 5:1119–27; PMID: 7532471; http://dx.doi.org/ 10.1091/mbc.5.10.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol 1995; 131:817–31; PMID:7490287; http://dx.doi.org/ 10.1083/jcb.131.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 2000; 11:4159–71; PMID:11102515; http://dx.doi.org/ 10.1091/mbc.11.12.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shanbhag R, Kurabi A, Kwan JJ, Donaldson LW. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett 2010; 584:4351–6; PMID:20875822; http://dx.doi.org/ 10.1016/j.febslet.2010.09.034 [DOI] [PubMed] [Google Scholar]
- 14.Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 2001; 15:2720–9; PMID:11641277; http://dx.doi.org/ 10.1101/gad.908401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schümperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005; 114:155–66; PMID:16003501; http://dx.doi.org/ 10.1007/s00412-005-0003-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB'. Chromosoma 2010; 119:205–15; PMID:19997741; http://dx.doi.org/ 10.1007/s00412-009-0249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293–307; PMID:11470819; http://dx.doi.org/ 10.1083/jcb.200104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One 2009; 4:e6171; PMID:19587784; http://dx.doi.org/ 10.1371/journal.pone.0006171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 2010; 17:403–9; PMID: 20357773; http://dx.doi.org/ 10.1038/nsmb.1783 [DOI] [PubMed] [Google Scholar]
- 20.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 1992; 117:1–14; PMID:1532583; http://dx.doi.org/ 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol 1993; 121:715–27; PMID:8491767; http://dx.doi.org/ 10.1083/jcb.121.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. Embo J 2004; 23:3000–9; PMID:15257298; http://dx.doi.org/ 10.1038/sj.emboj.7600296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 2004; 166:1015–25; PMID:15452143; http://dx.doi.org/ 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 2008; 19:2534–43; PMID:18367544; http://dx.doi.org/ 10.1091/mbc.E07-12-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 2004; 117:4423–33; PMID:15316075; http://dx.doi.org/ 10.1242/jcs.01308 [DOI] [PubMed] [Google Scholar]
- 26.Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 2003; 160:505–16; PMID:12578909; http://dx.doi.org/ 10.1083/jcb.200210087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2'-O- methylation and pseudouridylation guide RNAs. Embo J 2002; 21:2746–56; PMID:12032087; http://dx.doi.org/ 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J 2003; 22:1878–88; PMID:12682020; http://dx.doi.org/ 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 2006; 17:4972–81; PMID:16987958; http://dx.doi.org/ 10.1091/mbc.E06-06-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novotny I, Blazikova M, Stanek D, Herman P, Malinsky J. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 2011; 22:513–23; PMID:21177826; http://dx.doi.org/ 10.1091/mbc.E10-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 2009; 20:5250–9; PMID: 19846657; http://dx.doi.org/ 10.1091/mbc.E09-09-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deryusheva S, Gall JG. Novel small Cajal-body-specific RNAs identified in Drosophila: probing guide RNA function. RNA 2013; 19:1802–14; PMID: 24149844; http://dx.doi.org/ 10.1261/rna.042028.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler P, Neugebauer KM. Global identification of coilin binding partners reveals hundreds of small non-coding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389–99; PMID:25514182; http://dx.doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 34.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonné R, et al.. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777–87; PMID:15574332; http://dx.doi.org/ 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 35.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 2013; 4:17–34; PMID:23042601; http://dx.doi.org/ 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- 36.Konig H, Matter N, Bader R, Thiele W, Muller F. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 2007; 131:718–29; PMID:18022366; http://dx.doi.org/ 10.1016/j.cell.2007.09.043 [DOI] [PubMed] [Google Scholar]
- 37.Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Švéda M, Knejzlík Z, Staněk D. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Rep 2015; pii: S2211-1247(14)01059–6; PMID:25600876 [DOI] [PubMed] [Google Scholar]
- 38.Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 2006; 17:944–54; PMID:16319170; http://dx.doi.org/ 10.1091/mbc.E05-09-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647–52; PMID:14981093; http://dx.doi.org/ 10.1083/jcb.200310138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukowiak AA, Narayanan A, Li ZH, Terns RM, Terns MP. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA 2001; 7:1833–44; PMID:11780638 [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 2004; 15:81–90; PMID:14528011; http://dx.doi.org/ 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 2006; 17:955–65; PMID:16339074; http://dx.doi.org/ 10.1091/mbc.E05-09-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson RL, Li J, Culp BR, Terns RM, Terns MP. A Cajal body-independent pathway for telomerase trafficking in mice. Exp Cell Res 2010; 316:2797–809; PMID:20633556; http://dx.doi.org/ 10.1016/j.yexcr.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47–57; PMID:19285445; http://dx.doi.org/ 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009; 323:644–8; PMID:19179534; http://dx.doi.org/ 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. Embo J 2003; 22:4283–93; PMID: 12912925; http://dx.doi.org/ 10.1093/emboj/cdg394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 2011; 25:11–6; PMID:21205863; http://dx.doi.org/ 10.1101/gad.2006411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enwerem II, Velma V, Broome HJ, Kuna M, Begum RA, Hebert MD. Coilin association with Box C/D scaRNA suggests a direct role for the Cajal body marker protein in scaRNP biogenesis. Biol Open 2014; 3:240–9; PMID:24659245; http://dx.doi.org/ 10.1242/bio.20147443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; http://dx.doi.org/ 10.1371/journal.pbio.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 2007; 27:882–9; PMID:17889662; http://dx.doi.org/ 10.1016/j.molcel.2007.07.020 [DOI] [PubMed] [Google Scholar]
- 51.Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, Artandi SE. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 2014; 159:1389–403; PMID:25467444; http://dx.doi.org/ 10.1016/j.cell.2014.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 2012; 32:2384–95; PMID:22547674; http://dx.doi.org/ 10.1128/MCB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 2012; 150:481–94; PMID:22863003; http://dx.doi.org/ 10.1016/j.cell.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Deng Z, Jiang S, Hu Q, Liu H, Songyang Z, Ma W, Chen S, Zhao Y. Human cells lacking coilin and Cajal bodies are proficient in telomerase assembly, trafficking and telomere maintenance. Nucleic Acids Res 2015; 43:385–95; PMID:25477378; http://dx.doi.org/ 10.1093/nar/gku1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartova E, Foltankova V, Legartova S, Sehnalova P, Sorokin DV, Suchankova J, Kozubek S. Coilin is rapidly recruited to UVA-induced DNA lesions and gamma-radiation affects localized movement of Cajal bodies. Nucleus 2014; 5:460–8; PMID:24859326; http://dx.doi.org/ 10.4161/nucl.29229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol 2006; 175:401–13; PMID:17088425; http://dx.doi.org/ 10.1083/jcb.200604099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henriksson S, Rassoolzadeh H, Hedstrom E, Coucoravas C, Julner A, Goldstein M, Imreh G, Zhivotovsky B, Kastan MB, Helleday T, et al.. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev 2014; 28:2726–38; PMID:25512560; http://dx.doi.org/ 10.1101/gad.246546.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velma V, Carrero ZI, Cosman AM, Hebert MD. Coilin interacts with Ku proteins and inhibits in vitro non-homologous DNA end joining. FEBS Lett 2010; 584:4735–9; PMID:21070772; http://dx.doi.org/ 10.1016/j.febslet.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilder AS, Do PM, Carrero ZI, Cosman AM, Broome HJ, Velma V, Martinez LA, Hebert MD. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol Biol Cell 2011; 22:1070–9; PMID:21289084; http://dx.doi.org/ 10.1091/mbc.E10-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velma V, Carrero ZI, Allen CB, Hebert MD. Coilin levels modulate cell cycle progression and gammaH2AX levels in etoposide treated U2OS cells. FEBS Lett 2012; 586:3404–9; PMID:22986342; http://dx.doi.org/ 10.1016/j.febslet.2012.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 1999; 9:126–35; PMID:10021385; http://dx.doi.org/ 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- 62.Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 1995; 59:473–85; PMID:8749717; http://dx.doi.org/ 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- 63.Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 1997; 25:4740–7; PMID:9365252; http://dx.doi.org/ 10.1093/nar/25.23.4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schul W, Adelaar B, van Driel R, de Jong L. Coiled bodies are predisposed to a spatial association with genes that contain snoRNA sequences in their introns. J Cell Biochem 1999; 75:393–403; PMID:10536363; http://dx.doi.org/ 10.1002/(SICI)1097-4644(19991201)75:3%3c393::AID-JCB5%3e3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 65.Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell 1998; 9:1025–36; PMID:9571237; http://dx.doi.org/ 10.1091/mbc.9.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell 1999; 10:1653–63; PMID:10233169; http://dx.doi.org/ 10.1091/mbc.10.5.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell 2001; 12:565–76; PMID:11251071; http://dx.doi.org/ 10.1091/mbc.12.3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell 2000; 11:2987–98; PMID:10982395; http://dx.doi.org/ 10.1091/mbc.11.9.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499–509; PMID:11489914; http://dx.doi.org/ 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095–103; PMID: 18070915; http://dx.doi.org/ 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915–9; PMID:7597053; http://dx.doi.org/ 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci 2003; 116:303–12; PMID:12482916; http://dx.doi.org/ 10.1242/jcs.00211 [DOI] [PubMed] [Google Scholar]
- 73.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 2001; 114:4407–19; PMID:11792806 [DOI] [PubMed] [Google Scholar]
- 74.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753–67; PMID:22579281; http://dx.doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013; 155:1049–60; PMID:24267890; http://dx.doi.org/ 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al.. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012; 483:336–40; PMID: 22398450; http://dx.doi.org/ 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep 2013; 5:918–25; PMID:24268778; http://dx.doi.org/ 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al.. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012; 149:768–79; PMID:22579282; http://dx.doi.org/ 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 79.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841–52; PMID:7679389; http://dx.doi.org/ 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol 2015; 25(5):641–6; PMID:25702583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tuma RS, Roth MB. Induction of coiled body-like structures in Xenopus oocytes by U7 snRNA. Chromosoma 1999; 108:337–44; PMID:10591993; http://dx.doi.org/ 10.1007/s004120050385 [DOI] [PubMed] [Google Scholar]
- 82.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13:167–73; PMID: 21240286; http://dx.doi.org/ 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- 83.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713–7; PMID:18948503; http://dx.doi.org/ 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- 84.Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell 1998; 9:2987–3001; PMID:9763457; http://dx.doi.org/ 10.1091/mbc.9.10.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Makarov V, Rakitina D, Protopopova A, Yaminsky I, Arutiunian A, Love AJ, Taliansky M, Kalinina N. Plant coilin: structural characteristics and RNA-binding properties. PLoS One 2013; 8:e53571; PMID:23320094; http://dx.doi.org/ 10.1371/journal.pone.0053571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broome HJ, Hebert MD. In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One 2012; 7:e36300; PMID:22558428; http://dx.doi.org/ 10.1371/journal.pone.0036300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Broome HJ, Hebert MD. Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J Mol Biol 2013; 425:713–24; PMID:23274112; http://dx.doi.org/ 10.1016/j.jmb.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cajal RY. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol (Madrid) 1903; 2:129–221 [Google Scholar]
- 89.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell 2002; 3:329–37; PMID:12361597; http://dx.doi.org/ 10.1016/S1534-5807(02)00222-8 [DOI] [PubMed] [Google Scholar]
- 90.Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol 2002; 159:957–69; PMID:12486110; http://dx.doi.org/ 10.1083/jcb.200207028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 2010; 119:527–40; PMID:20449600; http://dx.doi.org/ 10.1007/s00412-010-0276-7 [DOI] [PubMed] [Google Scholar]
- 92.Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 2007; 178:733–40; PMID:17709427; http://dx.doi.org/ 10.1083/jcb.200702147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Girard C, Neel H, Bertrand E, Bordonne R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res 2006; 34:2925–32; PMID:16738131; http://dx.doi.org/ 10.1093/nar/gkl374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Lührmann R. Ongoing U snRNP Biogenesis Is Required for the Integrity of Cajal Bodies. Mol Biol Cell 2006; 17:3221–31; PMID: 16687569; http://dx.doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res 1997; 230:84–93; PMID:9013710; http://dx.doi.org/ 10.1006/excr.1996.3380 [DOI] [PubMed] [Google Scholar]
- 96.Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res 1998; 243:290–304; PMID:9743589; http://dx.doi.org/ 10.1006/excr.1998.4135 [DOI] [PubMed] [Google Scholar]
- 97.Liu J, Hebert MD, Ye Y, Templeton DJ, Kung H, Matera AG. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci 2000; 113 (Pt 9):1543–52; PMID:10751146 [DOI] [PubMed] [Google Scholar]
- 98.Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 2009; 122:1872–81; PMID:19435804; http://dx.doi.org/ 10.1242/jcs.044040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Broome HJ, Carrero ZI, Douglas HE, Hebert MD. Phosphorylation regulates coilin activity and RNA association. Biol Open 2013; 2:407–15; PMID: 23616925; http://dx.doi.org/ 10.1242/bio.20133863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carrero ZI, Velma V, Douglas HE, Hebert MD. Coilin phosphomutants disrupt Cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS One 2011; 6:e25743; PMID:21991343; http://dx.doi.org/ 10.1371/journal.pone.0025743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun J, Xu H, Subramony SH, Hebert MD. Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci 2005; 118:4995–5003; PMID:16219678; http://dx.doi.org/ 10.1242/jcs.02613 [DOI] [PubMed] [Google Scholar]
- 102.Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol 1998; 142:319–29; PMID:9679133; http://dx.doi.org/ 10.1083/jcb.142.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santama N, Ogg SC, Malekkou A, Zographos SE, Weis K, Lamond AI. Characterization of hCINAP, a novel coilin-interacting protein encoded by a transcript from the transcription factor TAFIID32 locus. J Biol Chem 2005; 280:36429–41; PMID:16079131; http://dx.doi.org/ 10.1074/jbc.M501982200 [DOI] [PubMed] [Google Scholar]
- 104.Xu H, Hebert MD. A novel EB-1/AIDA-1 isoform, AIDA-1c, interacts with the Cajal body protein coilin. BMC Cell Biol 2005; 6:23; PMID:15862129; http://dx.doi.org/ 10.1186/1471-2121-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]