Abstract
We previously discovered that a set of 5 microRNAs are concentrated in the nucleolus of rat myoblasts. We now report that several mRNAs are also localized in the nucleoli of these cells as determined by microarray analysis of RNA from purified nucleoli. Among the most abundant of these nucleolus-localized mRNAs is that encoding insulin-like growth factor 2 (IGF2), a regulator of myoblast proliferation and differentiation. The presence of IGF2 mRNA in nucleoli was confirmed by fluorescence in situ hybridization, and RT-PCR experiments demonstrated that these nucleolar transcripts are spliced, thus arriving from the nucleoplasm. Bioinformatics analysis predicted canonically structured, highly thermodynamically stable interactions between IGF2 mRNA and all 5 of the nucleolus-localized microRNAs. These results raise the possibility that the nucleolus is a staging site for setting up particular mRNA-microRNA interactions prior to export to the cytoplasm.
Keywords: microRNA-mRNA interactions, nucleolus, insulin-like growth factor-2
Introduction
mRNA typically exits the nucleus by diffusion from its transcriptional birth and splicing sites culminating in engagement by nuclear pore complexes, without apparent stops at any intranuclear sites along the way.1-6 However, there have been occasional reports of mRNAs being localized in nucleoli. Some of these observations were made during heat shock,7,8 but there also have been reports of mRNAs as well as mRNA splicing factors localized in the nucleoli of healthy cells.9-11 The possibility that some mRNAs might traffic through the nucleolus prior to export was first hinted in a study on the reactivation of the avian erythrocyte nucleus by their fusion with HeLa cells.12 In the resulting heterokaryons, only when a nucleolus had formed in the activated erythrocyte nucleus did mRNAs transcribed from the chicken genome reach the cytoplasm and undergo translation. Although this result is subject to other interpretations, the notion that some mRNAs may circuit through the nucleolus prior to export has remained with us.
During a study of myogenic cell biology we discovered, to our surprise, that a substantial portion of a key microRNA, miR-206, is concentrated in the nucleoli of rat myoblasts,13 and we subsequently found that at least 4 other microRNAs are also concentrated in the nucleoli of these cells.14 More recently, microRNAs have also been found localized in the nucleoli of HeLa and MCF7 cells.15,16 These findings, together with the aforementioned occasional reports of mRNAs in nucleoli of various cell types, led us to investigate the presence of mRNAs in the nucleoli of rat myoblasts. We find that particular mRNAs are concentrated in the nucleoli and have further characterized one of these, the mRNA for insulin-like growth factor-II (IGF2), a potent regulator of myogenic stem cell proliferation. RT-PCR revealed that the nucleolus-associated IGF2 mRNA is spliced, suggesting that it arrives in nucleoli from its nucleoplasmic sites of splicing. Bioinformatics analysis revealed that the 3′ UTR of IGF2 mRNA contains target sites for all 5 of the nucleolus-localized micro-RNAs.
Results and Discussion
To learn whether any mRNAs might be concentrated in the nucleoli of these cells, we isolated nuclei17 and prepared highly purified nucleolar and nucleoplasmic fractions as detailed previously.14 RNA from both nuclear fractions as well as from the cytoplasmic fraction were subjected to denaturing gel electrophoresis and the U3, U2 and U1 RNAs were identified based on their migration. As shown in Figure S1, the nucleolar fraction contained no detectable U2small nuclear RNA and little if any U1, these 2 RNAs being markers for nucleoplasmic splicesomes that are not known to appreciably traffic through nucleoli. In contrast, the nucleoplasmic fraction had no detectable U3 RNA, a RNA species that is involved specifically in pre-rRNA processing in the nucleolus.18 Based on these and our previous quality control results14 we regard these rat myoblast nucleolar and nucleoplasmic fractions to be of very high purity. Nucleolar and nucleoplasmic RNAs were then hybridized to rat genomic microarrays designed to detect the entire transcriptome including multiple splicing variants of both polyadenylated and non-polyadenylated RNAs.
As expected, the microarray analysis revealed small nucleolar RNAs as the most abundant species. Topping the list were 2 familiar, abundant small nucleolar RNAs, 7–2 and U3 and next in abundance were hundreds of less abundant small nucleolar RNAs (Table 1), as expected.19 However, also present were several mRNAs ranging from between 5–10% of the abundance of 7–2 and U3 snoRNAs down to levels much lower (Table 1). The two most abundant mRNAs were α-actin mRNA (which may suggest its precocious transcription in these myoblasts- a point to which we shall return) and insulin-like growth factor-II (IGF2). Because IGF2 has been well-established as autocrine growth factor in myoblasts20,21 we chose it for further study.
Table 1.
Nucleolus-enriched RNAs1
| RNA | Abundance2 | Chromosome | Transcript ID |
|---|---|---|---|
| small nuclear 7-2 (MRP RNase) RNA3 | 13.6 | 5 | J05014 |
| Rattus norvegicus 5.8S ribosomal rRNA3 | 13.3 | – | V01270.1 |
| U3B small nucleolar RNA3 | 12.7 | 10 | K00780 |
| actin, alpha 1, skeletal muscle (Acta1), mRNA | 11.8 | 19 | ENSRNOT00000024084 /// NM_019212 |
| rRNA promoter binding protein (aka ribin), mRNA | 11.6 | 6 | NM_147136.1 ///LOC257642 |
| Ba1-665 mRNA (rat specific-unknown function) | 10.5 | 16 | ENSRNOT00000041882 |
| insulin-like growth factor 2 (Igf2), mRNA | 9.4 | 1 | ENSRNOT00000027602 /// NM_031511 |
| high mobility group AT-hook I, related sequence 1 (hmga-1) mRNA | 9.0 | X | ENSRNOT00000038940 |
| ribosomal protein L36-like mRNA | 8.7 | 7 | ENSRNOG00000033555 |
| histone cluster 1, H2bm (Hist1h2bm) mRNA | 8.3 | 17 | ENSRNOT00000049785 |
| rCG65904-like protein mRNA | 7.3 | 2 | ENSRNOT00000029028 |
| heparan sulfate 6-sulfotransferase 3 mRNA | 7.2 | 15 | GENSCAN00000091390 /// XM_001078545 |
| hydrocephalus-inducing protein (aka hydin) mRNA | 6.5 | 19 | GENSCAN00000040128 /// XM_001077360 |
| olfactory receptor 1320 mRNA3 | 6.4 | 8 | ENSRNOT00000048785 /// NM_001000471 |
| homeo box D13 (Hoxd13) mRNA | 6.3 | 3 | ENSRNOT00000002155 |
| keratin complex 1, acidic, gene 18, mRNA (predicted) | 5.9 | 2 | LOC683212 |
| 14-3-3 protein sigma mRNA (predicted) | 5.7 | 6 | GENSCAN00000032606 /// NM_001013941 |
| peptidylprolyl isomerase (cyclophilin)-like 6 mRNA | 5.7 | 20 | GENSCAN00000029291 /// ENSRNOG00000043186 |
| olfactory receptor 760 mRNA3 | 5.1 | 3 | ENSRNOT00000030909 /// NM_001001069 |
| ceroid-lipofuscinosis, neuronal 5 (Cln5) mRNA | 4.5 | 2 | ENSRNOT00000012928 |
| mast cell protease 8 (Mcpt8) mRNA | 4.4 | 15 | ENSRNOT00000027950 /// NM_021598 |
| vomeronasal 1 receptor, j5 mRNA3 | 4.3 | 1 | ENSRNOT00000040738 /// NM_001008929 |
Nucleolar/nucleoplasmic ratios >1.
Relative nucleolar abundance (log2).
28S and 18S rRNAs, numerous small nucleolar RNAs, several olfactory receptor mRNAs and unannotated RNAs are omitted from this list for clarity.
We sought to corroborate the nucleolar presence of IGF2 mRNA by performing fluorescence in situ hybridization. Figure 1B shows the in situ hybridization patterns obtained using probes to 28 S rRNA, as a positive control. As expected, strong signals were seen both in nucleoli and cytoplasm and, as can be seen, these were RNase-sensitive. No appreciable signal was seen using a scrambled sequence hybridization probe (Fig. 1C). As shown in Figure 1A, IGF2 mRNA signal was observed in the nucleoli in many cells (white arrows), while other cells appeared to have both nucleoplasmic and nucleolar signal. The IGF2 mRNA hybridization was RNase-sensitive (Fig. 1A). An enlargement of a portion of the field is shown in Figure S2. In the course of multiple experiments and average of 70–80% of the cells displayed IGF2 mRNA hybridization signals in the nucleoli. It is possible that the absence of detectable nucleolar signal in the remaining cells reflects cell cycle variation although this was not pursued further. IGF2 mRNA hybridization signal was also present in the cytoplasm of all cells, and immunostaining confirmed the presence of IGF protein (Figure S3)as expected from previous work on the expression of this protein in myoblasts20,21. In additional experiments we confirmed the nucleolar localization of IGF2 mRNA using a second in situ hybridization protocol as described in the Materials and Methods and shown in Figure S4, again revealing strong, RNase-sensitive IGF2 signals in the nucleoli.
Figure 1.
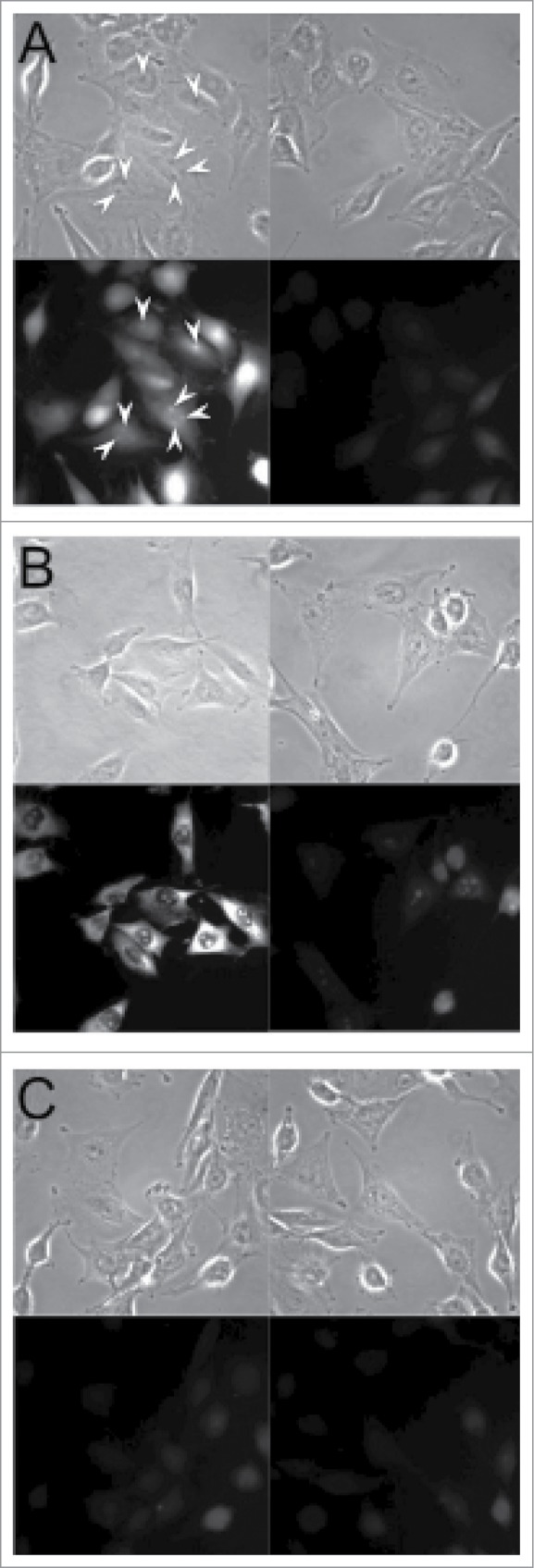
Fluorescence in situ hybridization. - : Cells incubated in buffer prior to hybridization; + : cells pre-incubated with RNase. A: IGF2 mRNA hybridization probes. B: 28 S rRNA probes. C: Scrambled sequence probes (see Materials and Methods). The upper panels in A-C are phase contrast images, the lower panels are the fluorescence microscopy images. Arrows indicate hybridization to IGF2 mRNA in nucleoli. (See also Figure S2).
We next used RT-PCR to interrogate the state of splicing of the nucleolus-localized IGF2 mRNA. As shown in Figure 2, the IGF2 transcripts in the nucleolar fraction generated RT-PCR products consistent with them being spliced mRNAs (compare with the products generated from cytoplasmic RNA). It is to be noted that the nucleoplasmic IGF2 and transcripts also generated RT-PCR products consistent with them being spliced mRNAs, indicating that splicing of IGF2 pre-mRNA is very rapid, whether co- or post-transcriptional. (Our nucleoplasmic fraction contains the chromatin.) Even a method as sensitive as RT-PCR has its limits of detection and thus we cannot conclude that there is no unspliced IGF2 mRNA whatsoever in the nucleolar or nucleoplasmic fractions but it is nonetheless clear that the great majority of IFG2 RNA in both nuclear fractions is the spliced mRNA.
Figure 2.
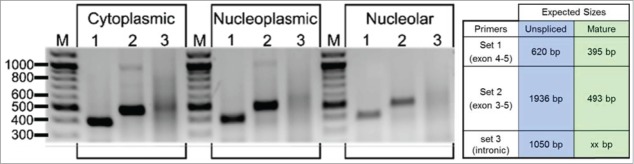
RT-PCR analysis. RNA was isolated from nucleolar, nucleoplasmic and cytoplasmic fractions and subjected to RT-PCR with the primers indicated in the box.
To explore the possibility that the nucleolar localization of IFG2 mRNA might be related to the earlier reported nucleolar abundance of 5 miRNAs in rat myoblasts, we searched the sequence ofIGF2 mRNA for hybridization targets for these miRNAs. As shown in Figure 3, there were predicted strong binding sites for both miR-206-3p and miR-206-5p in the 3′ UTR of IGF2 mRNA. (The terminology “strong binding sites” as used here derives from the rigorous themodynamic parameters incorporated into the Mirza software.36) In addition, strong binding sites in the 3′ UTR of IGF2 mRNA were also predicted for each of the other 4 nucleolus-associated microRNAs: mir-351 (Figure 4), miR-494 and miR-340 (Fig. 5) and miR-664 (Fig. 6). In the latter case, i.e. miR-664, a strong predicted binding site was predicted in both the 3′ and 5′ UTRs. Thus, even though this miRNA is probably generated from a snoRNA,22 it nevertheless has strong target sequences in IGF2 mRNA. The relative strengths of the miRNA target sites are summarized in Table 2.
Figure 3.
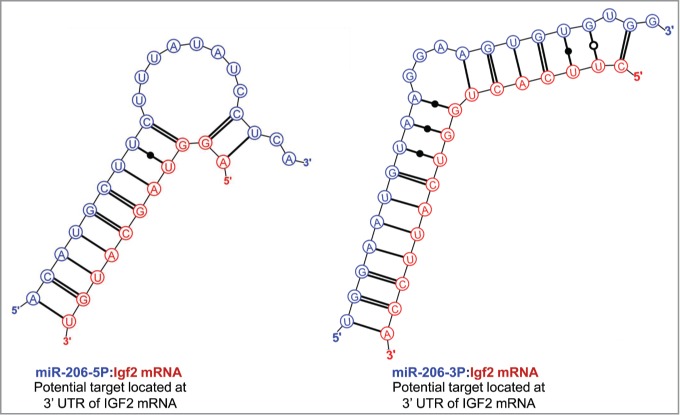
Predicted binding sites of miR-206 in the 3′-UTR of IGF2 mRNA.
Figure 4.
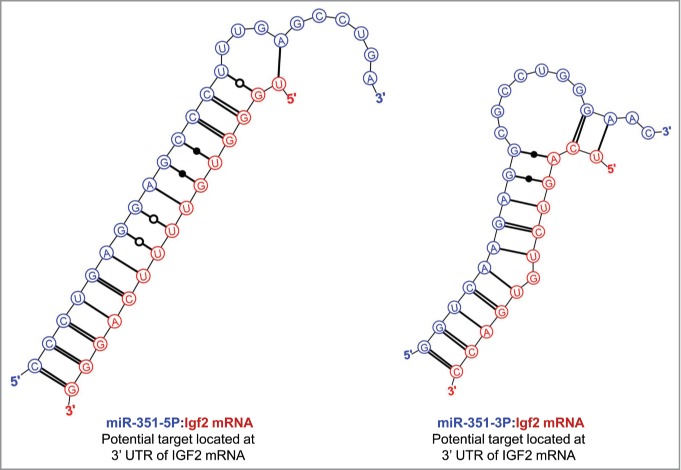
Predicted binding sites of miR-351 in the 3′-UTR of IGF2 mRNA.
Figure 5.
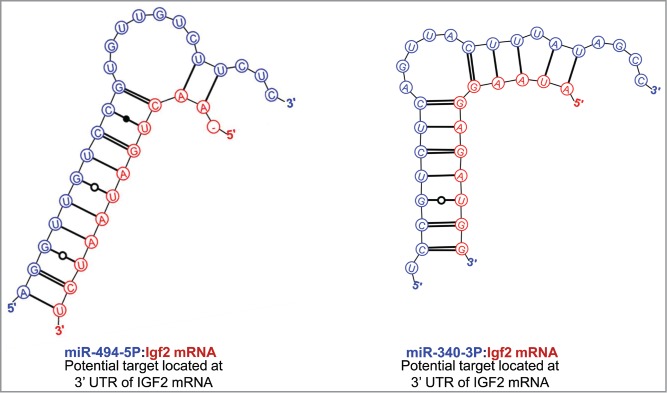
Predicted binding sites of miR-494 and miR-340 in the 3′-UTR of IGF2 mRNA.
Figure 6.
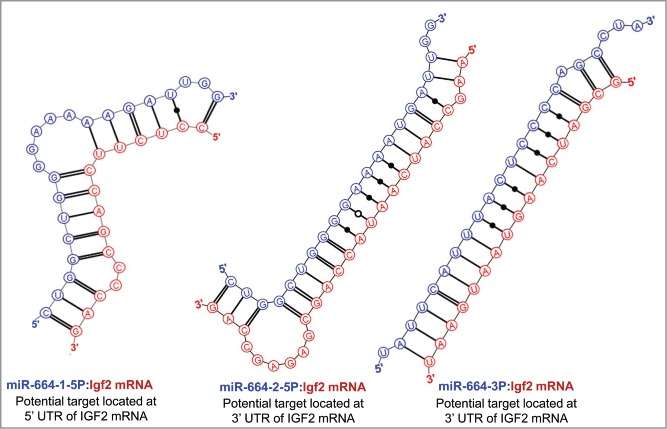
Predicted binding sites of miR-664 in the 5′- and 3'-UTRs of IGF2 mRNA.
Table 2.
Summary of predicted miRNA Targets in IGF2 mRNA1
| Target ID | miRNA | miRNA Sequence 5’-3’ | Target Quality | Target Sequence 5’-3’ |
|---|---|---|---|---|
| ENSRNO_IGF2_mRNA_51 | rno-miR-664-1-5p | CUGGCUGGGGAAAAAGAUUGG | 23.071 | CUUCAUCCUCUUCCAGCCCCAGCGGCCUCCUUAUCCAACUUCAGGUACCA |
| ENSRNO_IGF2_mRNA_1076 | rno-miR-664-3p | UAUUCAUUUACUCCCCAGCCUA | 123.88 | UCAUAAUCCCAUUCCCUAUGUAACGGGGGCAGCGAUCAAGUAAUGAAUGC |
| ENSRNO_IGF2_mRNA_1601 | rno-miR-351-5p | UCCCUGAGGAGCCCUUUGAGCCUGA | 19.102 | CGUGGGAUGGGUGUUUUCAGGGGCAUUUGCUGACCAUCCUCUGUGUCCCC |
| ENSRNO_IGF2_mRNA_1101 | rno-miR-664-2-5p | CUGGCUGGGGAAAAUGAUUGG | 24.847 | AUCAAGCCAUCAAUACCAGCGAGAGCCAGUAACACCGGCUAGAGCCAUCA' |
| ENSRNO_IGF2_mRNA_651 | rno-miR-206-3p | UGGAAUGUAAGGAAGUGUGUGG | 76.522 | AACAAUAGCCGCCCAAACUCUUUCUUCACUGGUCAUUCCAUCACAAAUGU |
| ENSRNO_IGF2_mRNA_351 | rno-miR-351-3p | GGUCAAGAGGCGCCUGGGAAC | 10.592 | UAUGUGGUAAUUCUGCAAUGUAGUACCAUCAGUCUGUGACCUCCUCUUGA |
| ENSRNO_IGF2_mRNA_501 | rno-miR-494-5p | AGGUUGUCCGUGUUGUCUUCUC | 11.874 | CAAAUUGGCUUAAGAAACUCCAUAACUGAUAAUCUAAAAAUUAAAUAACC |
| ENSRNO_IGF2_mRNA_601 | rno-miR-206-5p | ACAUGCUUCUUUAUAUCCUCAU | 15.066 | AGAAGAAAGGAAGGGGACCCAAAAUUUUGCAGGUAGCAUGUCAUUGCUUC |
| ENSRNO_IGF2_mRNA_851 | rno-miR-340-3p | UCCGUCUCAGUUACUUUAUAGCC | 13.73 | AGUCAGUUUGGCCAGAUAAGGAGAUGGCACUGCCAAGUGAUACAUGCUAC |
miRNA targets in IGF2 mRNA were located using MIRZA.
Translational regulation by microRNAs is typically negative. However, since IGF2 protein is abundantly present in these myoblasts (Figure S3) and continues to be present after myotube differentiation (Figure S5), the possibility of positive regulation of IGF2 mRNA by miR-206 arises. In this respect we have been struck by the fact that a well-established case of microRNA positive regulation, that of mRNA encoding the KLF-4 transcription factor in human mammary epithelial cells, has been demonstrated to involve none other than miR-206.23 Even more striking is the fact that the 3′ UTR of human KLF-4 mRNA contains a miR-206 target site that is highly homologous to one we have detected in the rat IGF2 mRNA (Fig. 7). A second finding that is highly relevant here is that in murine myogenesis, IGF2 mRNA translation is indeed known to be under positive control.24 The factor involved in establishing in this positive control is the murine homolog of the C. elegans RNA-binding protein Lin-28, which in that organism acts in collaboration with the microRNA lin-4.25 This again points to positive regulation of IGF2 by a microRNA route. A third finding of relevance is that miR-206 has been implicated in the positive translational regulation of liver LXRα receptor mRNA in human macrophages.26 While these findings together with the results presented here are not sufficient to conclude that
Figure 7.

Comparison of the predicted rat myoblast miR-206:IGF2 mRNA interaction and that of miR-206 and KLF4 mRNA in human cells.23
IFG2 mRNA translation in rat myoblasts is under positive control by miR-206, they are entirely consistent with this possibility and seem to constitute a provocative case.
It is to be emphasized that the 5 nucleolus-associated microRNAs have targets in mRNAs other than IGF2 mRNA. Thus, miR-206 was found to have targets in the 3′ UTR of α-tubulin mRNA, and the other 4 nucleolus-associated microRNAs were found to have targets in the 3′ UTRs of both histone 2A mRNA and ß-actin mRNA (Mirza program analyses, data not shown). This is consistent with the well-established fact that most microRNAs have targets in many different mRNAs.27 We observed another very abundant mRNA in the myoblast nucleoli, viz. one encoding a hallmark of differentiated muscle, α-actin (Table 1). Like IGF2 mRNA, it contains strong miR-206 binding sites in its 3′-UTR (data not shown) and yet, in contrast to IGF2 mRNA, is not appreciably translated in growing myoblasts.28 This suggests that its precocious transcription is accompanied by a temporary suppression of its translation, mediated perhaps by miR-206. Such negative regulation would be relieved later in the myogenic differentiation timetable. Thus, it is conceivable that both positive and negative translation-regulatory miRNA:mRNA interactions may be staged in the nucleolus, an intriguing hypothesis that warrants further investigation. Although in the studies on microRNA positive control of translation cited above,23,24,26 the intracellular localization of the envisioned miRNA:mRNA complexes was not investigated, our results intriguingly point to the nucleolus as an intracellular site where the assembly of positive (and perhaps also negative) regulatory microRNA:mRNA complexes might be staged. Whether this might occur before the miRNA and/or the mRNA had already once visited the cytoplasm is a matter for speculation, which nothing inhibits us from posing here. The return of microRNAs to the nucleus is well documented.29
Since the plurifunctional nucleolus hypothesis was first articulated30 (and where “pluri’” was used at that time to simply mean “more than one”), at least 2 roles of the nucleolus beyond ribosome synthesis have been established: assembly of the nascent signal recognition particle31 and a ribosome synthesis-independent role in cell cycle progression.32 On the basis of the present study it appears that there may be a third function of the nucleolus, the staging of certain mRNA-microRNA interactions, perhaps prior to any translational opportunity in the cytoplasm. We may therefore now consider prefixing “multi” (for at least 3) to the plurifunctional nucleolus concept.
One might ask why the formation of a positive (or negative) translational regulatory complex between a particular mRNA and a microRNA would need to be staged in the nucleolus, instead of just having the 2 molecules find each other in the cytoplasm (or in the nucleoplasm). We speculate that a nucleolus-localization factor or factors are bound to the miRNAs concentrated there (and perhaps these factor(s) brought them there) and that these special miRNPs then complex with nucleolar IFG2 mRNA (and likely other nucleolus-visiting mRNAs) to commit this transiently nucleolus-nested mRNA to translational up-regulation once in the cytoplasm. This idea has formal analogy with the exon-junction complex, which by its deposition on spliced mRNAs tags them for translational competency.32 But in addition, this hypothesis supposes that a visit to the nucleolus would allow the assembly of positive ribonucleoprotein regulatory complexes in a non-competitive environment, whereas formation of these complexes in the cytoplasm would be subject to competing binding reactions, for example ones dominated by high concentrations of other miRNPs that would negatively regulate IGF2 mRNA translation. The result would be a miRNA:mRNA complex that reaches the cytoplasm tagged and ready for rapid translation. Further studies will be needed to assess these provocative ideas.
Materials and Methods
Cell culture and fractionation
Rat L6 myoblasts were cultured as previously described13,14 and nucleolar, nucleoplasmic and cytoplasmic fractions were isolated as detailed previously.14 RNA from the nucleolar and nucleoplasmic fractions was electrophoresed in denaturing 10% polyacrylamide gels followed by staining with ethidium bromide to assess the quality of both nucleolar (U3 RNA) and nucleoplasmic (U1 and U2 RNAs) fractions. For myogenic differentiation of myoblasts (Figure S5), cells were transferred to medium containing 2% horse serum and cultured for 5 days.
RNA microarray analysis
RNA from the nucleolar and nucleoplasmic fractions was analyzed on Affymetrix rat genomic microarrays (GeneChip® Rat Exon ST Array) at the UMass Medical School Core RNA.
Sequencing Facility. Normalizations were performed using the Sketch-Quantile system within the Affymetrix Expression Console Program.
RT-PCR
RNA was purified from nucleolar, nucleoplasmic and cytoplasmic fractions of L6 cells with DNase I treatment using a Qiagen RNeasy kit. First strand cDNAs were synthesized from RNA using the reverse primer IGF2-Rv1 (5′-TAATTTGGTTCACTGATGGTTGCTGGAC-3′) and SuperScript III reverse transcriptase (Invitrogen, Grand Island, NY). The resulting cDNAs were used as templates for PCR reactions. IGF2 amplification from exon 4 to 5 was done with primer set 1: IGF2-Fw2 (5′-GCAGGCCTTCAAGCCGTG-3′) and IGF2-Rv1. Amplification of exons 3 to 5 was done with primer set 2: IGF2-Fw5 (5′-GTGCTGCATCGCTGCTTACC-3′) and IGF-Rv1. Amplification of unspliced IGF2 transcripts employed the primer set 3: IGF2-Fw3-Intr3 (5′-CAGGCTCAAGGTGACACCAGG-3′) and IGF2-Rv1. All reactions were run in presence of Herculase II Fusion DNA Polymerase (Agilent Technologies, Santa Clara, CA). Standard PCR conditions consisted of initial denaturation at 95°C for 2 min, followed by 35 cycles at 95°C (30 s), 52°C (30 s) and 72°C (60 s) and final elongation at 72°C (5 min). Products were visualized by 1% agarose gel electrophoresis followed by ethidium bromide staining.
Fluorescence in situ hybridization
In situ hybridization to IGF2 mRNA was carried out using the DNA probes listed below. The underlined bases denote 5-aminohexylthymines which were labeled with Cy3 dye as previously described.13
IGF2-probe-1: 5′-GAGTCTCGCTGGGGCGGTAAGCAGCGATGCAGC-3′
IGF2-probe-2: 5′-GTAGACACGTCCCTCTCGGACTTGGCGGGGGTG-3′
Oligos with the reverse polarity sequences of the IGF2 probes 1 and 2 were used as negative controls:
Scrambled-1: 5′-CGAGCTAGCGACGAATGGCGGGGTCGCTCTGAG-3′
Scrambled-2: 5′-GTGGGGGCGGTTCAGGCTCTCCCTGCACAGATG-3′
Positive control probes were for 28 S rRNA and SRP RNA as detailed in our previous studies.33,34
28 S-1: 5′-GTTACCGGCACTGGACGCCTCGCGGCGCCCATC-3′
28 S-2: 5′-ATGGCTCTCCGCACCGGACCCCGGTCCCGACTC-3′
SRP-1: 5′-TTCCCACTACTGATCAGCACGGGAGTTTTGACCTT-3′
SRP-2: 5′-GCTCCCGGGAGGTCACCATATTGATGCCGAACTT-3′
Cells on coverslips were fixed with 4% (v/v) formaldehyde and permeabilized with cold (4°C) acetone. In situ hybridization and RNase pre-treatment, when desired, were performed as detailed previously13 except that after hybridization, cells were washed with 40% formamide/2× SSC (1× SSC = 0.15 M NaCl/0.015 mM sodium citrate, pH 7.0) and then 20% formamide/1× SSC for 30 min. each at 37°C followed by 3 15-min washes in 1× SSC at room temperature. An alternative in situ hybridization protocol was performed (Figure S4) in which cells were permeabilized with CSK buffer (10 mM PIPES, pH 6.8, 300 mM sucrose, 25 mM NaCl, 1 mM EGTA, 5 mM MgCl2,) containing 0.5% Triton X-100 and 2 mM vanadyl ribonucleoside complex, and then fixed in 4% formaldehyde.
Coverslips were dehydrated through a 70%, 90%, 100% ethanol series and air-dried followed by incubation with ∼20 ng of each oligo in 40% formamide containing tRNA and salmon sperm DNA (5 μg/μl each). Cells were washed 3 times with 40% formamide/2× SSC, then 2× SSC for 5 min each and then once with 4× SSC/0.1% Tween-20. Coverslips were mounted in Prolong Gold Antifade (Invitrogen, Carlsbad, CA) and image acquisition and analysis was as previously described.13
Immunostaining
This was performed as previously detailed13 using a polyclonal rabbit IgG for IGF2 (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:25 dilution followed by visualization using a Cy3-conjugated donkey anti-rabbit IgG.
Bioinformatics
The genomic rat IGF2 sequence as well as that of rat IGF2 mRNA and its component exons were obtained from the UCSC genome database (http://genome.ucsc.edu), NCBI database and/or Rat Genome Database-RGD (http://rgd.mcw.edu). The desired rat micro-RNA sequences were obtained from miRBase (Kozomara and Griffiths-Jones, NAR, 2011) (http://mirbase.org). Searches for targets of the 5 nucleolus-associated microRNAs and IGF2 mRNA and other mRNAs were performed with the Mirza program34, as refined in the University of Massachusetts Medical School Bioinformatics Core Facility and RNA Therapeutics Institute.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Hanhui Ma in the Pederson laboratory for very constructive advice and encouragement, and Alper Kucukural and Manuel Garber in the UMass Medical School Bioinformatics Core Facility for their expert assistance with Mirza. T.P. also thanks Victor Ambros of this institution for supportive input during the course of this project. We dedicate this paper to the memory of Eric Hogan.14
Funding
This research was supported by NSF grant MCB-00445841 to TP.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website: http://www.tandfonline.com/kncl
References
- 1. Politz JC, Browne ES, Wolf DE, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci USA 1998; 95:6043-6048; PMID:9600914; http://dx.doi.org/ 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Politz JC, Tuft RA, Pederson T, Singer RH. Movement of poly(A) RNA throughout the interchromatin space in living cells. Curr Biol 1999; 9:285-291; PMID:10209094; http://dx.doi.org/ 10.1016/S0960-9822(99)80136-5 [DOI] [PubMed] [Google Scholar]
- 3. Daneholt B. Pre-mRNP particles:from gene to nuclear pore. Curr Biol 1999; 9:R412-R415; PMID:10359690; http://dx.doi.org/ 10.1016/S0960-9822(99)80256-5 [DOI] [PubMed] [Google Scholar]
- 4. Politz JC, Pederson T. Movement of mRNA from transcription sites to nuclear pores. J Struc Biol 2000; 129:252-257; http://dx.doi.org/ 10.1006/jsbi.2000.4227 [DOI] [PubMed] [Google Scholar]
- 5. Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science 2004; 304:1797-1800; PMID:15205532; http://dx.doi.org/ 10.1126/science.1099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mor A, Ben-Yishay R, Shav-Tal Y. On the right track:following the nucleo-cytoplasmic path of an mRNA. Nucleus 2010; 1:492-498; PMID:21327092; http://dx.doi.org/ 10.4161/nucl.1.6.13515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tani T, Derby RJ, Hiraoka Y, Spector DL. Nucleolar accumulation of poly(A)+ RNA in heat-shocked yeast cells:implication of nucleolar involvement in mRNA transport. Mol Biol Cell 1995; 6:1515-1534; PMID:8589453; http://dx.doi.org/ 10.1091/mbc.6.11.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Názer E, Verdύn RE, Sánchez DO. Severe heat shock induces nucleolar accumulation of mRNAs in Trypanosoma cruzi. PLoS One 2012; 7:e43715; http://dx.doi.org/ 10.1371/journal.pone.0043715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. John HA, Patrinou-Georgoulas M, Jones KW. Detection of myosin heavy chain mRNA during myogenesis in tissue culture by in vitro and in situ hybridization. Cell 1977; 12:501-508; PMID:912755; http://dx.doi.org/ 10.1016/0092-8674(77)90126-X [DOI] [PubMed] [Google Scholar]
- 10. Bond VC, Wold BJ. Nucleolar localization of myc transcripts. Mol Cell Biol 1993; 13:3221-3230; PMID:7684491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. The Plant Cell 2009; 21:2045-2057; PMID:19602621; http://dx.doi.org/ 10.1105/tpc.109.067736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sidebottom E, Harris H. The role of the nucleolus in the transfer of RNA from nucleus to cytoplasm. J Cell Sci 1969; 5:351-364. [Google Scholar]
- 13. Politz JCR, Zhang F, Pederson T. MicroRNA-206 co-localizes with ribosome-rich regions in the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA 2006; 103:18957-18962; http://dx.doi.org/ 10.1073/pnas.0609466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Politz JCR, Hogan E, Pederson T. MicroRNAs with a nucleolar location. RNA 2009; 15:1705-1715; PMID:19628621; http://dx.doi.org/ 10.1261/rna.1470409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, Xue CJ, Poon LM, Lam YW. Dynamic localization of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One 2013; 8:e70869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bai B, Liu H, Laiho M. Small RNA expression and deep sequencing analysis of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio 2014; 4:441-449; PMID:24918059; http://dx.doi.org/ 10.1016/j.fob.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mellon I, Bhorjee JS. Isolation and characterization of nuclei and purification of chromatin from differentiating cultures of rat skeletal muscle. Exp Cell Res 1982; 137:141-154; PMID:7035194; http://dx.doi.org/ 10.1016/0014-4827(82)90016-7 [DOI] [PubMed] [Google Scholar]
- 18. Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 1990; 60:697-908; PMID:2107022; http://dx.doi.org/ 10.1016/0092-8674(90)90338-F [DOI] [PubMed] [Google Scholar]
- 19. Ellis JC, Brown DD, Brown JW. The small nucleolar ribonucleoprotein (snoRNP) database. RNA 2010:16:664-666; http://dx.doi.org/ 10.1261/rna.1871310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florini JR, Ewton DZ, Magri KA. Hormones, growth factors and myogenic differentiation. Ann Rev Physiol 1991; 53:201-216; http://dx.doi.org/ 10.1146/annurev.ph.53.030191.001221 [DOI] [PubMed] [Google Scholar]
- 21. Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev 1996; 17:481-517; PMID:8897022 [DOI] [PubMed] [Google Scholar]
- 22. Scott MS, Avollo F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comp Biol 2009; 5:e1000507; PMID:19763159; http://dx.doi.org/ 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin C-C, Liu L-Z, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A KLF4-miRNA autoregulatory feedback loop can promote or inhibit protein translation depending on cell context. Mol Cell Biol 2011; 31:2513-2527; PMID:21518959; http://dx.doi.org/ 10.1128/MCB.01189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 2009; 21:461-469; PMID:19278845; http://dx.doi.org/ 10.1016/j.ceb.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan . Lon-28 binds IGF2 mRNA and participates in skeletal myogenesis by increasing translational efficiency. Genes Dev 2007; 1125-1138; PMID:17473174; http://dx.doi.org/ 10.1101/gad.415007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vinod M, Chennamsetty I, Colin S, Belloy L, De Paoli F, Schaider H, Graier WF, Frank S, Kratky D, Staels B, et al. MiR-206 controls LXRα expression and promotes LXR-mediated cholesterol efflux in macrophages. Biochim Biophys Acta 2014; 1841:827-835; PMID:24603323; http://dx.doi.org/ 10.1016/j.bbalip.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farh KK, Grimson AA, Jan c, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 2005; 310:1817-1821; PMID:16308420; http://dx.doi.org/ 10.1126/science.1121158 [DOI] [PubMed] [Google Scholar]
- 28. Bagchi T, Larson DE, Sells BH. Cytoskeletal association of muscle-specific mRNAs in differentiating L6 rat myoblasts. Exp Cell Res 1987; 168:160-172; PMID:3780870; http://dx.doi.org/ 10.1016/0014-4827(87)90425-3 [DOI] [PubMed] [Google Scholar]
- 29. Schraivogel D, Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem Sci 2014; 39:420-431; PMID:25131816; http://dx.doi.org/ 10.1016/j.tibs.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 30. Pederson T. The plurifunctional nucleolus. Nucleic Acids Res 1998; 26:3871-3876; PMID:9705492; http://dx.doi.org/ 10.1093/nar/26.17.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pederson T, Politz JC. The nucleolus and the four ribonucleoproteins of translation. J Cell Biol 2000; 148:1091-1095; PMID:10725320; http://dx.doi.org/ 10.1083/jcb.148.6.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsai RY, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J 2014; 28:3290-3296; PMID:24790035; http://dx.doi.org/ 10.1096/fj.14-254680 [DOI] [PubMed] [Google Scholar]
- 33. Tange TØ, Nott A. Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol 2004; 16:279-284; PMID:15145352; http://dx.doi.org/ 10.1016/j.ceb.2004.03.012 [DOI] [PubMed] [Google Scholar]
- 34. Politz JCR, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell 2003; 14:4805-4812; PMID:12960421; http://dx.doi.org/ 10.1091/mbc.E03-06-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zweib C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA 2000:97, 55-80; PMID:10618370; http://dx.doi.org/ 10.1073/pnas.97.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat. Meth 2013; 10:253-25. [DOI] [PubMed] [Google Scholar]
- 37. Reddy R, Busch H. 1998. In:“Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles,” Birnstiel M.L., ed. Springer, Berlin, pp. 1-39. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


