Abstract
Poikiloderma with neutropenia (PN) is a rare inherited disorder characterized by poikiloderma, facial dysmorphism, pachyonychia, short stature and neutropenia. The molecular testing of PN patients has identified mutations in the C16orf57 gene, which encodes a protein referred to as USB1 (U Six Biogenesis 1). In this study, we developed a zebrafish model of PN by the microinjection of morpholino antisense oligos to suppress usb1 gene function. Severe morphological defects, including a bent tail, thin yolk extension and reduced body length, were predominant in the Usb1-suppressed embryos (morphants). We also observed significantly decreased number of neutrophils in the morphants by Sudan Black staining. Interestingly, the splicing of genes involved in neutrophil differentiation and development, such as mpx, ncf1, ela3l and npsn, was aberrant in the morphants. However, the splicing of haematopoietic precursors and erythroid-specific genes was unaltered. Importantly, the neutrophil defects were almost completely rescued by co-injection of ela3l mRNA, the most markedly affected gene in the morphants. Our study demonstrated a possible role of USB1 in modulating the tissue-specific gene splicing that eventually leads to the impaired development of neutrophils. This zebrafish model could serve as a valuable tool to investigate the causative role of USB1 in PN pathogenesis.
Keywords: poikiloderma with neutropenia, RNA disease, splicing, U6 biogenesis, zebrafish
Abbreviations
- CHT
caudal haematopoietic tissue
- CRISPR
clustered regularly interspaced short palindromic repeats
- GFP
green fluorescent protein
- hpf
hours post fertilization
- MO
morpholino antisense oligo
- MPN1
mutated in poikiloderma with neutropenia
- PN
poikiloderma with neutropenia
- snRNPs
small nuclear ribonucleoproteins
- sqRT-PCR
semi-quantitative reverse transcription and polymerase chain reaction
- USB1
U Six Biogenesis 1
Introduction
Clericuzio-type poikiloderma with neutropenia (PN) (OMIM# 604173) is a unique genodermatosis that was first observed in the Navajo Indian population.1 It is characterized by poikiloderma together with an erythematous rash on the limbs and face, nail abnormalities, short stature, palmoplantar hyperkeratosis, permanent neutropenia and skeletal defects.1,2 PN patients are eventually susceptible to myelodysplastic syndrome and acute myeloid leukemia.1 A recent report on PN also observed an association with squamous cell carcinoma of the skin.3 The actual number of reported patients is quite limited, mostly because several PN patients have many similar clinical manifestations with dyskeratosis congenita and Rothmund-Thomson syndrome.4,5 To date, 40 patients with PN have been reported,6-16 containing 19 different mutations in the C16orf57 gene that encodes a 265-amino-acid protein, referred to as USB1 (U Six Biogenesis 1).17,18 In some studies, this protein has also been referred to as MPN1 (Mutated in Poikiloderma with Neutropenia).19,20 Several types of mutations, including nonsense, deletion/frame-shift and splice site alterations, have been identified in PN patients (Fig. S1A).
A high-resolution (1.1 Å) crystal structure categorizes human USB1 as a member of the LigT-like superfamily of 2H phosphodiesterases, which is found in the bacteria, archaea and eukarya. In addition, it was demonstrated in vitro that human USB1 is a novel 3′-5′ exoribonuclease that removes terminal oligo(U) and oligo(A) tails at the 3′ end of U6 snRNA and generates the 3′ terminal phosphate modification.21 Deep-sequencing analysis of U6 snRNA from the PN patient lymphoblasts revealed the non-templated addition of more than 2 adenosine nucleotides to the 3′ end of U6, although no significant difference in the steady-state levels of U6 snRNA was observed.21 Interestingly, aberrant 3′ end processing of U6 was observed in the USB1-deficient fission yeasts and the human cells derived from PN patients. In human cells, the steady-state levels of U6 were not affected by the diminished USB1 activity, but in yeasts, the cellular U6 levels were greatly reduced, leading to precursor mRNA (pre-mRNA) splicing defects.18,19 Furthermore, the deep sequencing of the poly(A)+ transcriptome of the PN-patient cells revealed that the pre-mRNA splicing was normal.19,21
These in vitro and in vivo studies suggest that PN manifestations are not derived from common pre-mRNA splicing defects. Instead, they might have resulted from incomplete splicing of the genes expressed in the tissues that are highly affected by the disease, such as neutrophil precursors. To investigate this possibility, we need an animal model that enables us to explore new insight into the molecular pathogenesis of PN. In the last decade, zebrafish (Danio rerio) has been used extensively as a model organism to study vertebrate hematopoiesis owing to its genetic tractability and the embryo transparency that allows simple live-imaging.22,23 In this study, we developed a zebrafish model of PN using a morpholino antisense oligo (MO)-based loss-of-function approach. We observed acute morphological abnormalities and neutrophil reduction in the Usb1-deficient embryos (morphants). In addition, we found that the splicing of neutrophil-specific genes was altered by the usb1 knockdown. Interestingly, the neutrophil defects were almost completely rescued in ela3l mRNA coinjected embryos. Taken together, our results suggest the existence of a regulatory mechanism by which USB1 modulates the splicing of tissue-specific genes and ultimately leads to abnormal tissue development. This study provides a prime model for PN to unravel its pathogenic mechanism and the role of USB1 in disease development.
Results
Zebrafish usb1 gene
USB1, a mutated gene in PN, is highly conserved during evolution. A BLAST search of the zebrafish genome with the human USB1 coding sequence identified a single homologous gene encoding a 276-amino-acid protein (Fig. 1A). The coding region of this gene shares 57% nucleotide and 46% amino acid identity with its human ortholog. Despite the low amino acid identity, the alignment of coding nucleotides revealed that 15 of the 19 mutated nucleotides that have been identified in PN patients are conserved between human USB1 and zebrafish usb1 (Fig. S1A). The comparisons of the exon lengths and the exon-intron structure of zebrafish usb1 with those of human USB1 indicated a high degree of cross-species conservation. The zebrafish usb1 gene possesses 7 exons and is located on chromosome 25 in a region of shared synteny with the human USB1 gene that is located on chromosome 16 (Fig. 1A, S1B).
Figure 1.
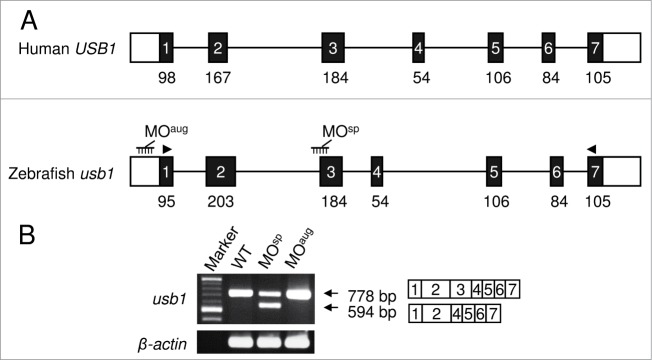
Schematics of human USB1 and zebrafish usb1 gene structures and RT-PCR analysis. (A) A diagrammatic representation of zebrafish usb1 and human USB1 genomic structure. The black and white boxes represent translated and untranslated regions of the exons, respectively. The black lines represent the introns. The numbers below the black bars indicate the exon length (bp). The MOs were designed at the splice site (MOsp) or the translation initiation site (MOaug) to prevent pre-mRNA splicing or to block the translation of usb1. The morpholino binding sites are shown in black combs. The arrowheads indicate the primer binding sites for RT–PCR. The human USB1 and zebrafish usb1 genomic sequences were obtained from the database under the accession numbers NM_024598.3 and NM_001003460.1, respectively. (B) sqRT-PCR analysis of usb1 and β-actin (control) in MO-injected and wild-type embryos. The decreased expression of the full length transcript (778 bp) and the expression of smaller transcript (594 bp) without exon 3 were observed in the MOsp injected embryos. The injection of MOaug had no effect on the splicing. β-actin served as a control.
To determine usb1 expression during zebrafish development, we collected array data from the Gene Expression Omnibus derived from whole embryos, retina and trunk and tail muscle at various stages of the embryonic development (1.5–120 hpf, hours post fertilization). We calculated the fold changes in usb1 expression over β-actin and found a markedly low level of its expression in the early embryonic stage, peaking at 24 hpf followed by a gradual decrease after 52 hpf (Fig. S2A). To verify this expression pattern, we examined the total RNA from wild type embryos at various stages (3–120 hpf) using semi-quantitative reverse transcription and polymerase chain reaction (sqRT-PCR) and confirmed the similar trend of usb1 expression during zebrafish development (Fig. S2B).
Severe developmental defects in Usb1-deficient zebrafish
To investigate the role of Usb1 function in zebrafish development, we knocked down usb1 gene using MOs and analyzed the morphological status of the morphants at various stages of development (Fig. 1A). Injection of the MOsp that targets the 3′-splice site of the second intron altered the splicing and resulted in the exclusion of exon 3 from the mature transcript. In addition, a 50–60% reduction in the expression of usb1 mature transcript was observed in Usb1-suppressed embryos (Fig. 1B). The excision of exon 3 in this aberrant transcript was confirmed by DNA sequencing. These results indicate that the injection of MOsp effectively impeded usb1 expression in zebrafish.
We compared the morphological features of the MOsp and MOaug-injected embryos with wild-type embryos at 25 hpf, a stage when many organs are almost recognizable, and found that the Usb1-deficient embryos displayed abnormal phenotypes, including a thin yolk extension, a bent tail and reduced body length. We also observed the development of other anomalies at 50 hpf in morphants, such as a significantly smaller head and eyes and a defective heart with pericardial edema (Fig. 2). In addition, we found the morphological abnormalities of MOsp-injected embryos were highly similar to those observed in MOaug-injected embryos. The severities of morphological abnormalities were proportional to the MO concentrations (Fig. S3). All the morphants died within a week. Co-injection of MO-resistant usb1 mRNA rescued these developmental defects in the morphants, indicating that the altered phenotypes in the morphants were specifically due to a loss of Usb1 (Fig. 2).
Figure 2.
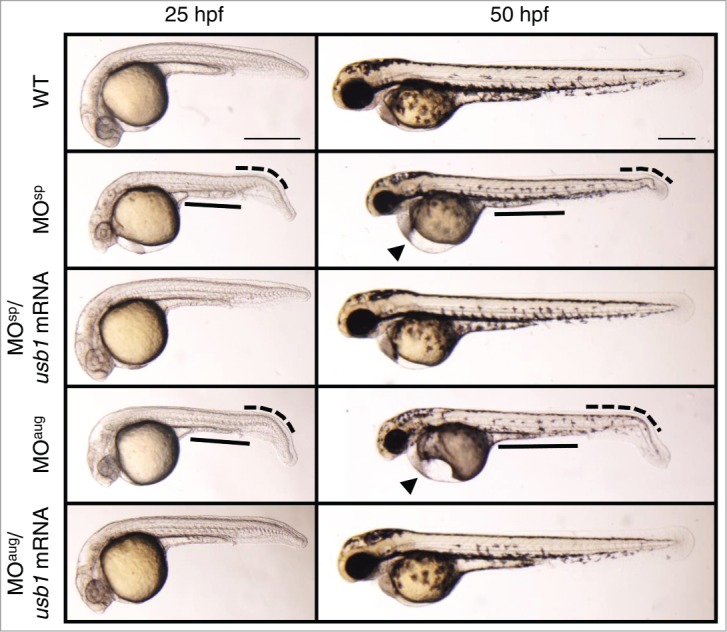
Morphological defects in Usb1-deficient zebrafish. Lateral views of wild-type, MOsp (10 μg/μl) and MOaug (5 μg/μl) injected embryos at 25 and 50 hpf are shown. The morphants displayed a thin yolk extension (black solid line), reduced body length and a bent tail (black dotted curved line) at 25 hpf. The morphants also showed small eyes and pericardial edema (black triangle) at 50 hpf. The co-injection of usb1 mRNA with MOs nearly completely rescued these morphological abnormalities. Scale bars: 200 μm.
Impaired neutrophil development in Usb1-deficient zebrafish
PN is generally associated with characteristic features of poikiloderma and constant neutropenia in all patients.3 To determine the effects of usb1 knockdown on neutrophil development, we injected the MOs into one-cell-stage Tg (mpx:GFP)uwm1 zebrafish embryos in which neutrophils are labeled by a green fluorescent protein (GFP) and analyzed the phenotypes. At 50 hpf, we observed a marked reduction of the green-fluoresce neutrophils in the MO-injected embryos, especially in the caudal haematopoietic tissue (CHT) that is formed around the transient caudal vein plexus where larval neutrophil development takes place (Fig. 3A, illustration). We also performed whole-mount Sudan Black (SB) staining that delineates the neutrophils throughout the embryo and counted the neutrophils in the CHT region (Fig. 3A). At 50 hpf, we found a substantial decrease in the population of neutrophils, especially in the CHT region of Usb1-deficient embryos (Fig. 3). The neutrophil number returned to near normal in the MO-resistant usb1 mRNA-coinjected embryos (Fig. 3B). These results clearly indicate that the development of neutrophils was severely affected by the usb1 knockdown in zebrafish.
Figure 3.
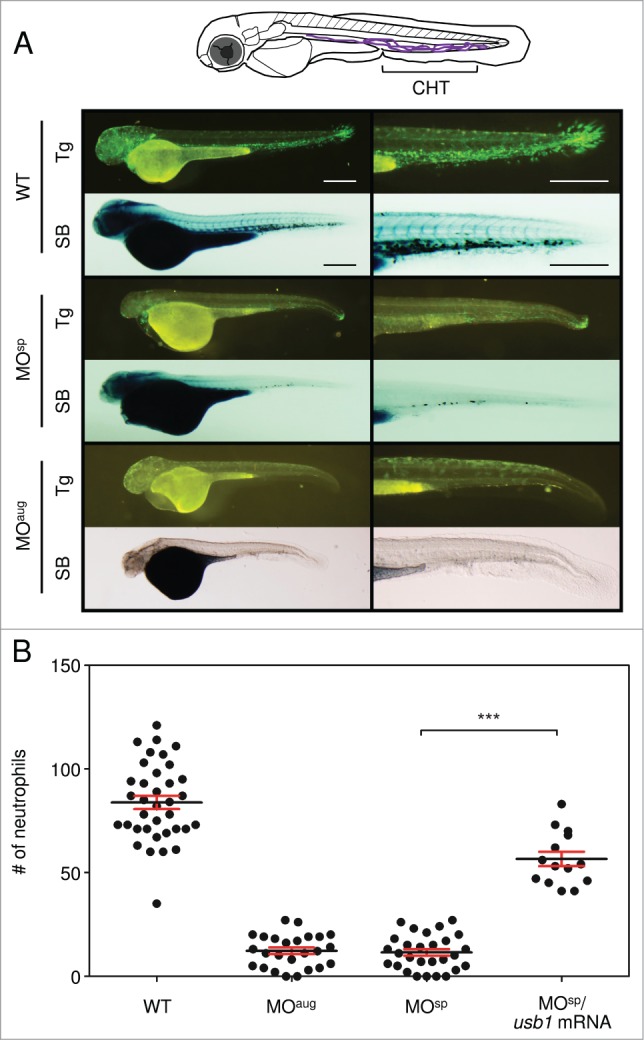
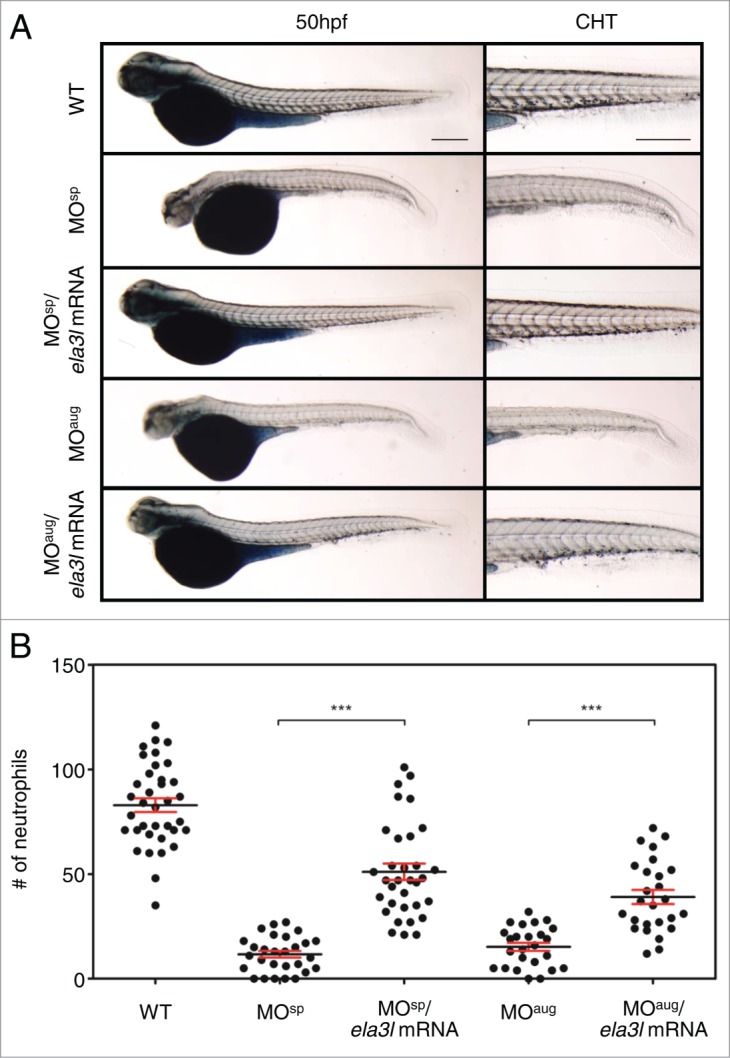
Neutrophil reduction in Usb1-deficient zebrafish. (A) Lateral images of the usb1 MO-injected Tg (mpx:GFP) zebrafish and the whole-mount Sudan Black staining at 50 hpf and close-up images of CHT region. Substantial decreases of green-fluoresce and SB-stained neutrophils, especially in the CHT region, were observed in the Usb1-deficient embryos. Scale bars: 200 μm. (B) Scatter plot showing the mean number of neutrophils at the CHT region in wild type (WT) and morphants at 50 hpf. The number of neutrophils was significantly reduced in the morphants. Co-injection of usb1 mRNA with MOsp returned the neutrophils to near normal. ***P < 0.001 (one way ANOVA with Dunn's multiple comparison test).
Altered splicing of neutrophil-specific genes in Usb1-deficient zebrafish
It was found recently that USB1 deficiency did not affect either the steady-state levels of U6 snRNA or the general pre-mRNA splicing in PN patients.21 By contrast, USB1 deletion in yeast cells showed lower levels of cellular U6, leading to general pre-mRNA splicing defects.19 To investigate the effects of usb1 knockdown on the pre-mRNA splicing in zebrafish, we examined the total RNA by sqRT-PCR and analyzed the expression of the genes involving the differentiation and development of neutrophils. We observed aberrant transcripts in the neutrophil-specific genes,24,25 including mpx, ncf1, ela3l and npsn, in the morphants (Fig. 4A). The DNA sequencing of these aberrant transcripts confirmed the intron retention in all these genes, suggesting the abnormal regulation of splicing in specific tissues of the morphants.
Figure 4.
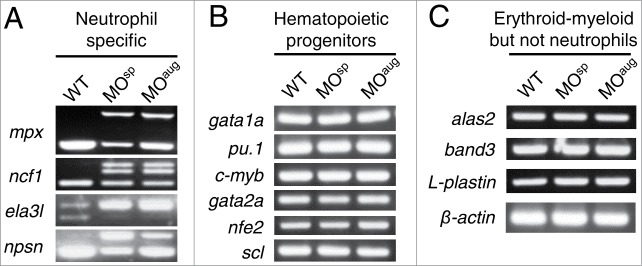
Splicing of haematopoietic genes in Usb1-deficient zebrafish. (A) sqRT-PCR analysis of neutrophil-specific genes, including mpx, ncf1, ela3l and npsn, in the embryos at 25 hpf. The decreased expression of the normal transcripts and the expression of aberrant transcripts retaining the introns were observed in Usb1-deficient embryos. (B–C) sqRT-PCR analysis of haematopoietic - but not neutrophil-specific genes in MO-injected and wild-type embryos. The normal transcripts of haematopoietic progenitors (B) and erythroid and myeloid-but not neutrophil-specific genes (C) were observed in the morphants. β-actin served as a control.
Furthermore, we analyzed the expression of randomly selected genes involving primitive and definitive hematopoiesis (Fig. S4) to establish whether the abnormal splicing activity is restricted to specific tissues in the morphants. The expression of genes involving primitive hematopoiesis,24-26 such as gata1a, pu.1, c-myb, gata2a, nfe2 and scl, were normal in the morphants (Fig. 4B). Similarly, usb1 knockdown had no effect on the expression of erythroid- or myeloid-specific genes24-26 (except neutrophils) including alas2, band-3 and L-plastin (Fig. 4C). In addition, we examined the total RNA by northern blot analysis and found no difference in the U6 snRNA levels between wild-type and Usb1-deficient embryos (Fig. S5). These results suggest that the aberrant pre-mRNA splicing of neutrophil-specific genes induces neutropenia during zebrafish development.
Rescuing neutrophil defects by co-injection of ela3l mRNA in Usb1-deficient zebrafish
We observed that the Usb1 deficiency in zebrafish lead to defective neutrophil development and altered splicing of neutrophil-specific genes. To determine whether co-injection of mRNA of neutrophil-specific genes could rescue the neutrophil reduction in Usb1-deficient embryos, we injected ela3l mRNA, the most markedly affected neutrophil-specific gene in the morphants, with usb1 MO into the embryos. We observed a substantial recovery of the neutrophil defects in the ela3l mRNA-coinjected embryos, as evidenced by the significant increase in the population of neutrophils in the CHT region (Fig. 5A). The recovery was observed in >50% of Usb1-deficient embryos (Fig. 5B, S6). These results suggest that the decreased neutrophils in the Usb1-deficient embryos were derived from the aberrant splicing of neutrophil-specific genes in zebrafish.\raster="rgFigKRNB_A_1017240_F0005_B"
Discussion
In this study, we reported the development of an in vivo model of PN by morpholino-mediated usb1 knockdown. We showed that the loss of usb1 gene function in zebrafish resulted in a marked reduction in neutrophils and abnormal phenotypes that partially recapitulate the PN defects.1 Furthermore, we established that the neutrophil defects were derived from the incomplete splicing of neutrophil-specific genes in the morphants. Consistently, the splicing was unaltered in haematopoietic precursors and erythroid-or-myeloid-specific genes (except neutrophils). Moreover, co-injection of ela3l mRNA, one of the most aberrantly spliced transcript in Usb1-deficient zebrafish, rescued neutrophil defects. Although we also observed partial rescue of morphological abnormalities, this might be due to the different roles of elastase in other tissues such as exocrine pancreas.27-29 Overall, our results demonstrated that Usb1 deficiency in zebrafish recapitulates the PN phenotype with constant neutropenia that might have been derived from the similar mechanism of defective splicing of tissue-specific genes, as seen in Usb1-suppressed embryos. This presumption is due to the fact that haematopoietic differentiation and development in zebrafish closely resembles hematopoiesis in humans.23,30 In addition, zebrafish is a suitable and powerful animal model system for studying various haematopoietic disorders,31,32 primarily because the embryos can survive for several days, even in the absence of blood cells.33 Although we focused on neutrophil defects in this study, the usb1 gene is universally expressed in zebrafish at different stages of development, and when suppressed, embryos show severe morphological abnormalities. Owing to the several symptoms associated with PN, this zebrafish model is appropriate for studying the molecular mechanisms underlying its pathogenesis.
The regulation of pre-mRNA splicing events is crucial for correct haematopoietic lineage specification.34,35 Nuclear pre-mRNA splicing is catalyzed by a multi-protein/RNA complex called the major spliceosome that consists of 5 small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U5 and U6.36,37 The spliceosome recognizes intron-exon boundaries and removes intervening introns via 2 transesterification reactions that result in the ligation of 2 adjacent exons.38,39 It was recently disclosed that USB1 acts as 3′–5′ RNA exonuclease that trims the oligouridine tail of U6 snRNA and generates terminal 2′,3′-cyclic phosphate groups. The USB1 dysfunction resulted in the non-templated addition of 2 or more adenosine nucleotides to the 3′ end of U6 was established by the deep-sequencing analysis of U6 snRNA from PN patients. Despite the loss of USB1 activity, the steady state levels of U6 snRNA were unaffected in PN patients.21 This might be attributed to the presence of multiple and dispersed U6 snRNA genes that have varied transcriptional efficiencies in humans.40 Although we have not determined the 3′ end sequence of U6 snRNA in this study, instead we observed that the expression of U6 snRNA was not altered by the usb1 knockdown in zebrafish. As zebrafish has multiple copies of U6 snRNA genes, we presume that zebrafish might also possess a similar mechanism of differential transcriptional efficiency that is present in humans. Although USB1 functions as a U6 biogenesis factor, how it controls target-specific pre-mRNA splicing in Usb1-deficient zebrafish remains to be fully explained.
Previous studies in zebrafish have shown that dysfunction of the splicing apparatus or the deficiency of splicing factors lead to tissue-specific alternative splicing that affects gene expression in specific organs during early embryogenesis. Loss of the usp39 gene, a component of the RNA splicing machinery, leads to rb1 mRNA splicing defects and pituitary lineage expansion in zebrafish.41 Deficiency of the zebrafish rbfox genes disrupts splicing regulatory proteins that regulate muscle-specific alternative splicing, which is essential for proper differentiation and function of vertebrate muscle.42,43 In addition, tri-snRNP dysfunction, by silencing the systemic splicing factors prpf31 and prpf4, led to specific defects in retinal gene expression in a zebrafish model of retinitis pigmentosa, a hereditary eye disease that causes blindness due to a progressive loss of photoreceptors in the retina.44 In vivo mutation of pre-mRNA processing factor 8 (Prpf8) in an ENU-induced zebrafish mutant, Cephalophonus, led to accumulation of aberrantly spliced transcripts retaining both U2- and U12-type introns that cause impaired myeloid differentiation in zebrafish.45
In summary, our results suggest that Usb1 deficiency in zebrafish affects the splicing of specific transcripts that elicit defects in specific tissues, such as neutrophils. However, understanding how the USB1 protein recognizes the tissue-specific transcripts during pre-mRNA splicing, is of special importance in the context of several symptoms associated with PN disease. A genome-wide RNA-Seq analysis in mutant zebrafish revealed a large set of specific target genes that changed their alternative splicing patterns in the absence of the U1C protein.46 Similarly, we expect that the full transcriptome analysis of usb1-mutant zebrafish, which we are now generating using the CRISPR (clustered regularly interspaced short palindromic repeats) RNA-guided Cas9 nuclease system,47 will allow us to resolve this question.
Materials and Methods
Zebrafish maintenance
Zebrafish (wild-type AB line) were raised and maintained according to standard laboratory conditions48 in the Bio-resource Division at the Frontier Science Research Center, University of Miyazaki, Japan. Current Japanese rules do not require approval for research on zebrafish embryos. The embryos were raised in E3 embryo medium at 28.5°C. The transgenic zebrafish line Tg(mpx:GFP) was purchased from the Zebrafish International Resource Center (ZIRC; http://zebrafish.org/zirc/fish/lineAll.php).
Morpholino injections
To knock down usb1, we used 2 types of MOs that were obtained from Gene Tools, LLC (USA). The MOs were designed to target either the splice site (MOsp, 5′-AGGATCATCTGAAATTTAGGCAGGA-3′) in the intron 2/exon 3 boundary region to interrupt usb1 splicing or the complementary sequence between -60 and -35 nucleotides from the translation start site (MOaug, 5′-TAGAAGAATGTCATCTCAGACACGT-3′) to inhibit Usb1 protein expression. MOs were injected into one-cell-stage embryos at varied concentrations (MOsp at 5, 10 and 20 μg/μl; MOaug at 2.5, 5 and 10 μg/μl), using an IM-30 Electric Microinjector (Narishige, Japan).
Sudan Black staining
Sudan Black staining was performed as previously described.49 Two-day-old embryos were fixed with 4% paraformaldehyde (Polysciences, Warrington, PA) in phosphate-buffered saline (PBS) for overnight at 4°C and incubated in Sudan Black solution (Sigma-Aldrich, France) for 20 minutes after rinsing thoroughly in PBS. Then, the embryos were washed extensively in 70% ethanol and rehydrated in PBS containing 0.1% Tween 20 (PBS-T). The stained neutrophils in the CHT region were counted under a stereomicroscope (Olympus, SZX12). Statistical analyses were performed using GraphPad Prism (Ver. 5) software.
In vitro mRNA synthesis and rescue experiments
Zebrafish usb1 and ela3l mRNAs were synthesized from their full-length cDNA sequences (GenBank accession numbers NM_001003460 and NM_001024408, respectively) following a previously described protocol.22 The synthesized mRNAs were injected into the embryos at a concentration of 500 ng/μl.
Semi-quantitative RT-PCR
The total RNA was extracted from 25 hpf morphants and control embryos using TRIZOL reagent (Sigma-Aldrich, USA) according to the manufacturer's instructions. One microgram of total RNA was reverse-transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) with random primers. The PCR analysis was performed according to the FastStart Taq DNA polymerase (Roche, Germany) using specific primers (Table S1).
Northern blot analysis
The total RNA (10 μg/lane) was separated on a 1.5% denaturing agarose gel and blotted according to standard procedures.50 Then, the blots were hybridized overnight at 65°C in modified Church-Gilbert hybridization buffer (0.5 M NaHPO4, 1 mM EDTA, 0.5% BSA and 7% SDS) containing LNA (locked nucleic acid) probes labeled with digoxigenin (DIG) using the DIG oligonucleotide Tailing Kit (Roche, Germany). The sequence of the U6 LNA probe is 5′-aAagAtgGaaCgcTtcAcgAatTtgCgtGt-3′ (uppercase letters indicate the LNAs).
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interests.
Acknowledgments
We thank Dr. Maki Yoshihama and Ms. Yukari Nakajima for their advice and useful discussions.
Funding
This work was supported by JSPS KAKENHI Grants 2591003 (N.K.) and 24591556 (T.U.) and a grant from the Takeda Science Foundation (T.U.).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Clericuzio C, Hoyme HE, Aase JM. Immune deficient poikiloderma: a new genodermatosis. Am J Hum Genet 1991; 49:A661 [Google Scholar]
- 2. Larizza L, Negri G, Colombo EA, Volpi L, Sznajer Y. Clinical utility gene card for: poikiloderma with neutropenia. Eur J Hum Genet 2013; 21; PMID:22669413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodgers W, Ancliff P, Ponting CP, Sanchez-Pulido L, Burns S, Hayman M, Kimonis V, Sebire N, Bulstrode N, Harper JI. Squamous cell carcinoma in a child with clericuzio-type poikiloderma with neutropenia. Br J Dermatol 2013; 168:665-7; PMID:22924337; http://dx.doi.org/ 10.1111/bjd.12016 [DOI] [PubMed] [Google Scholar]
- 4. Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and rothmund-thomson syndrome. Hum Mol Genet 2010; 19:4453-61; PMID:20817924; http://dx.doi.org/ 10.1093/hmg/ddq371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Hove JLK, Jaeken J, Proesmans M, Boeck KD, Minner K, Matthijs G, Verbeken E, Demunter A, Boogaerts M. Clericuzio type poikiloderma with neutropenia is distinct from rothmund-thomson syndrome. Am J Med Genet 2005; 132A:152-8; PMID:15558713; http://dx.doi.org/ 10.1002/ajmg.a.30430 [DOI] [PubMed] [Google Scholar]
- 6. Mostefai R, Morice-Picard F, Boralevi F, Sautarel M, Lacombe D, Stasia MJ, McGrath J, Taïeb A. Poikiloderma with neutropenia, clericuzio-type, in a family from Morocco. Am J Med Genet 2008; 146A:2762-9; PMID:18925663; http://dx.doi.org/ 10.1002/ajmg.a.32524 [DOI] [PubMed] [Google Scholar]
- 7. Concolino D, Roversi G, Muzzi GL, Sestito S, Colombo EA, Volpi L, Larizza L, Strisciuglio P. Clericuzio type poikiloderma with neutropenia syndrome in three sibs with mutations in the C16orf57 gene: delineation of the phenotype. Am J Med Genet 2010; 152A:2588-94; PMID:20734427; http://dx.doi.org/ 10.1002/ajmg.a.33600 [DOI] [PubMed] [Google Scholar]
- 8. Arnold AW, Itin PH, Pigors M, Kohlhase J, Bruckner-Tuderman L, Has C. Poikiloderma with neutropenia: a novel C16orf57 mutation and clinical diagnostic criteria. Br J Dermatol 2010; 163:866-9; PMID:20618321; http://dx.doi.org/ 10.1111/j.1365-2133.2010.09929.x [DOI] [PubMed] [Google Scholar]
- 9. Tanaka A, Morice-Picard F, Lacombe D, Nagy N, Hide M, Taïeb A, McGrath J. Identification of a homozygous deletion mutation in C16orf57 in a family with clericuzio-type poikiloderma with neutropenia. Am J Med Genet 2010; 152A:1347-8; PMID:20503306 [DOI] [PubMed] [Google Scholar]
- 10. Clericuzio C, Harutyunyan K, Jin W, Erickson RP, Irvine AD, McLean WH, Wen Y, Bagatell R, Griffin TA, Shwayder TA, et al. Identification of a novel C16orf57 mutation in athabaskan patients with poikiloderma with neutropenia. Am J Med Genet 2011; 155A:337-42; PMID:21271650; http://dx.doi.org/ 10.1002/ajmg.a.33807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dereure O. Mutations of C16orf57 gene have been identified in the poikiloderma-neutropenia syndrome and in a specific subset of congenital dyskeratosis with normal-length telomeres. Ann Dermatol Venereol 2011; 138:362-3; PMID:21497268 [DOI] [PubMed] [Google Scholar]
- 12. Chantorn R, Shwayder T. Poikiloderma with neutropenia: report of three cases including one with calcinosis cutis. Pediatr Dermatol 2012; 29:463-72; PMID:21967010; http://dx.doi.org/ 10.1111/j.1525-1470.2011.01513.x [DOI] [PubMed] [Google Scholar]
- 13. Colombo EA, Bazan JF, Negri G, Gervasini C, Elcioglu NH, Yucelten D, Altunay I, Cetincelik U, Teti A, Del Fattore A, et al. Novel C16orf57 mutations in patients with poikiloderma with neutropenia: bioinformatic analysis of the protein and predicted effects of all reported mutations. Orphanet J Rare Dis 2012; 23:7-7; PMID:22269211; http://dx.doi.org/ 10.1186/1750-1172-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piard J, Holder-Espinasse M, Aral B, Gigot N, Rio M, Tardieu M, Puzenat E, Goldenberg A, Toutain A, Franques J, et al. Systematic search for neutropenia should be part of the first screening in patients with poikiloderma. Eur J Med Genet 2012; 55:8-11; PMID:21872685; http://dx.doi.org/ 10.1016/j.ejmg.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 15. Farruggia P, Indaco S, Dufour C, Lanza T, Mosa C, Macaluso A, Milioto M, D'Angelo P, Lanciotti M. Poikiloderma with neutropenia: A case report and review of the literature. J. Pediatr Hematol Oncol 2014; 36:297-300; PMID:23823120; http://dx.doi.org/ 10.1097/MPH.0b013e31829f35e7 [DOI] [PubMed] [Google Scholar]
- 16. Koparir A, Gezdirici A, Koparir E, Ulucan H, Yilmaz M, Erdemir A, Yuksel A, Ozen M. Poikiloderma with neutropenia: genotype-ethnic origin correlation, expanding phenotype and literature review. Am J Med Genet 2014; 165A:2535-40; PMID:25044170; http://dx.doi.org/ 10.1002/ajmg.a.36683 [DOI] [PubMed] [Google Scholar]
- 17. Volpi L, Roversi G, Colombo EA, Leijsten N, Concolino D, Calabria A, Mencarelli MA, Fimiani M, Macciardi F, Pfundt R, et al. Targeted next-generation sequencing appoints C16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am J Hum Genet 2010; 86:72-6; PMID:20004881; http://dx.doi.org/ 10.1016/j.ajhg.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kuciński I, Ginalski K, Dziembowski A. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev 2012; 26:1911-25; PMID:22899009; http://dx.doi.org/ 10.1101/gad.193169.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. Mpn1, Mutated in poikiloderma with neutropenia, is a conserved 3′-5′ RNA exonuclease processing U6 snRNA. Cell Rep 2012; 2:855-65; PMID:23022480; http://dx.doi.org/ 10.1016/j.celrep.2012.08.031 [DOI] [PubMed] [Google Scholar]
- 20. Shchepachev V, Azzalin CM. The Mpn1 RNA exonuclease: cellular functions and implication in disease. FEBS Letters 2013; 587:1858-62; PMID:23684637; http://dx.doi.org/ 10.1016/j.febslet.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 21. Hilcenko C, Simpson PJ, Finch AJ, Bowler FR, Churcher MJ, Jin L, Packman LC, Shlien A, Campbell P, Kirwan M, et al. Aberrant 3′ oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood 2013; 121:1028-38; PMID:23190533; http://dx.doi.org/ 10.1182/blood-2012-10-461491 [DOI] [PubMed] [Google Scholar]
- 22. Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of diamond-blackfan anemia. Human Mol Genet 2008; 17:3204-11; PMID:1865374; http://dx.doi.org/ 10.1093/hmg/ddn216 [DOI] [PubMed] [Google Scholar]
- 23. Jing L, Zon LI. Zebrafish as a model for normal and malignant hematopoiesis. Dis Model Mech 2011; 4:433-8; PMID:21708900; http://dx.doi.org/ 10.1242/dmm.006791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, Sheng Y, Chen Y, Ruan Z, Jiang CL, et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci USA 2004; 101:16240-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Brown AD, Zon LI, Fleming MD, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood 2001; 98:643-51; PMID:11468162; http://dx.doi.org/ 10.1182/blood.V98.3.643 [DOI] [PubMed] [Google Scholar]
- 26. de Jong JLO, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet 2005; 39:481-501; PMID:16285869; http://dx.doi.org/ 10.1146/annurev.genet.39.073003.095931 [DOI] [PubMed] [Google Scholar]
- 27. Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp and elastaseB mRNA as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn 2004; 230:165-73; PMID:15108321; http://dx.doi.org/ 10.1002/dvdy.20032 [DOI] [PubMed] [Google Scholar]
- 28. Wan H, Korzh S, Li Z, Mudumana SP, Korzh V, Jiang YJ, Lin S, Gong Z. Analyses of pancreas development by generation of gfp transgenic zebrafish using an exocrine pancreas-specific elastaseA gene promoter. Exp Cell Res 2006; 312:1526-39; PMID:16490192; http://dx.doi.org/ 10.1016/j.yexcr.2006.01.016 [DOI] [PubMed] [Google Scholar]
- 29. Yee NS, Lorent K, Pack M. Exocrine pancreas development in zebrafish. Dev Biol 2005; 284:84-101; PMID:15963491; http://dx.doi.org/ 10.1016/j.ydbio.2005.04.035 [DOI] [PubMed] [Google Scholar]
- 30. Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood 2008; 111:3331-42; PMID:18182572; http://dx.doi.org/ 10.1182/blood-2007-10-052761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ellett F, Lieschke GJ. Zebrafish as a model for vertebrate hematopoiesis. Curr Opin Pharmacol 2010; 10:563-70; PMID:20538521; http://dx.doi.org/ 10.1016/j.coph.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 32. Martin CS, Moriyama A, Zon LI. Hematopoietic stem cells, hematopoiesis and disease: lessons from the zebrafish model. Genome Med 2011; 3:83; PMID:22206610; http://dx.doi.org/ 10.1186/gm299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. North TE, Zon LI. Modeling human hematopoietic and cardiovascular diseases in zebrafish. Dev Dyn 2003; 228:568-83; PMID:14579393; http://dx.doi.org/ 10.1002/dvdy.10393 [DOI] [PubMed] [Google Scholar]
- 34. Yamamoto ML, Clark TA, Gee SL, Kang JA, Schweitzer AC, Wickrema A, Conboy JG. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood 2009; 113:3363-70; PMID:19196664; http://dx.doi.org/ 10.1182/blood-2008-05-160325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. English MA, Lei L, Blake T, Wincovitch SM, Sr, Sood R, Azuma M, Hickstein D, Liu PP. Incomplete splicing, cell division defects and hematopoietic blockage in dhx8 mutant zebrafish. Dev Dyn 2012; 241:879-89; PMID:22411201; http://dx.doi.org/ 10.1002/dvdy.23774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 2003; 12:5-14; PMID:12887888; http://dx.doi.org/ 10.1016/S1097-2765(03)00270-3 [DOI] [PubMed] [Google Scholar]
- 37. Blencowe BJ. Alternative splicing: new insights from global analyses. Cell 2006; 126:37-47; PMID:16839875; http://dx.doi.org/ 10.1016/j.cell.2006.06.023 [DOI] [PubMed] [Google Scholar]
- 38. Will MC, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13:290-301; PMID:11343899; http://dx.doi.org/ 10.1016/S0955-0674(00)00211-8 [DOI] [PubMed] [Google Scholar]
- 39. Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/ 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 40. Domitrovich AM, Kunkel GR. Multiple, dispersed human U6 small nuclear RNA genes with varied transcriptional efficiencies. Nucleic Acids Res 2003; 31:2344-52; PMID:12711679; http://dx.doi.org/ 10.1093/nar/gkg331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rios Y, Melmed S, Lin S, Liu NA. Zebrafish usp39 mutation leads to rb1 mRNA splicing defect and pituitary lineage expansion. PLOS Genet 2011; 7:e1001271; PMID:21249182; http://dx.doi.org/ 10.1371/journal.pgen.1001271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 2003; 22:905-12; PMID:12574126; http://dx.doi.org/ 10.1093/emboj/cdg089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol 2011; 359:251-61; PMID:21925157; http://dx.doi.org/ 10.1016/j.ydbio.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linder B, Dill H, Hirmer A, Brocher J, Lee GP, Mathavan S, Bolz HJ, Winkler C, Laggerbauer B, Fischer U. Systemic splicing factor deficiency causes tissue-specific defects: a zebrafish model for retinitis pigmentosa. Hum Mol Genet 2011; 20:368-77; PMID:21051334; http://dx.doi.org/ 10.1093/hmg/ddq473 [DOI] [PubMed] [Google Scholar]
- 45. Keightley MC, Crowhurst MO, Layton JE, Beilharz T, Markmiller S, Varma S, Hogan BM, de Jong-Curtain TA, Heath JK, Lieschke GJ. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Letters 2013; 587:2150-7; PMID:23714367; http://dx.doi.org/ 10.1016/j.febslet.2013.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosel TD, Hung LH, Medenbach J, Donde K, Starke S, Benes V, Rätsch G, Bindereif A. RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J 2011; 30:1965-76; PMID:21468032; http://dx.doi.org/ 10.1038/emboj.2011.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JJ-R, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; http://dx.doi.org/ 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). Univ. of Oregon Press, Eugene: 2000 [Google Scholar]
- 49. Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 2008; 111:132-41; PMID:17875807; http://dx.doi.org/ 10.1182/blood-2007-06-095398 [DOI] [PubMed] [Google Scholar]
- 50. Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 2010; 38:e98; PMID:20081203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


