Abstract
AIM: To observe the variation of DNA polymerase β (polβ) in esophageal carcinoma.
METHODS: Thirty specimens containing adjacent normal epithelial tissues were collected from patients in Linzhou region (a high risk area for esophageal squamous carcinoma) and 25 specimens were from a non-high risk area. Total RNA was extracted from the samples and reverse transcription polymerase chain reaction (RT-PCR) was performed. PCR products were cloned and sequenced to investigate the polβ gene with DNASIS and OMIGA. Statistical significance was evaluated using the χ2 test.
RESULTS: High-incidence area group: polβ gene variation was detected in 13 of 30 esophageal carcinoma tissue specimens, and only one variation was found in 30 corresponding adjacent normal tissue specimens. Non high-incidence area group: polβ gene variation was detected in 5 of 25 esophageal carcinoma tissue specimens, and no variation was found in 25 corresponding adjacent normal tissue specimens. The incidence of polβ gene variation observed in the high-incidence area group was significantly higher than in the non-high incidence area group. Two mutation hot spots (454-466 and 648-670 nt) and a 58 bp deletion (177-234 nt) were found.
CONCLUSION: Variations of polβ perform different functions between the high-incidence areas and the other areas, and may play a more important role in the high-incidence areas.
Keywords: DNA polymerase β, Esophageal carcinoma, Gene mutation
INTRODUCTION
Esophageal carcinoma occurs frequently in China, especially in the mountainous Taihang area. Epidemiology and laboratory studies suggest that the carcinogenesis and progression of esophageal carcinoma are probably associated with some gene mutations[1,2]. Some researches indicate that the ability of polβ to repair DNA damage reduces in peripheral blood of esophageal carcinoma patients and that obvious chromosome changes occur in tumor cells[3-5]. Therefore, there must be DNA damage repair in the development of esophageal carcinoma. However, variations of DNA replication and repair enzymes in esophageal carcinoma, especially the mutation of polβ is rare. Thus, we made a preliminary analysis on the mutation of polβ in esophageal carcinoma.
MATERIALS AND METHODS
Specimens
High-incidence area group: Specimens of 30 esophageal squamous carcinomas (serial numbers H1-H30) and matched adjacent normal tissues were obtained from patients in Linzhou region of northern China, a well-recognized high-risk area for esophageal carcinoma.
Non-high incidence area group: Specimens of 25 esophageal squamous carcinomas (serial numbers N1-N25) and corresponding adjacent normal tissues were obtained from patients who underwent surgery at Cancer Hospital of Henan Province and the First Affiliated Hospital of Zhengzhou University.
All patients were histopathologically diagnosed to be infiltrative squamous carcinoma cases. The tissues were frozen in liquid nitrogen immediately after surgery.
RT-PCR
A pair of primers for PCR was designed to amplify the total polβ gene according to the sequence of M13140 in GenBank. Primer P1 (sense): 5’ ATGAGCAAACG GAG-GGCGCCG 3’; Primer P2 (antisense): 5’ TCATTCGCT-CCGGTCCTTGG 3’. The primer was synthesized by Shanghai Sangon Co., Ltd.
Total RNA was extracted with the QIAGEN RNA extraction kit. Five microliters of total RNA was transcribed into cDNA using 0.2 μmol/L primer P2 , 0.2 μmol/L dNTP, RNasin 40 U, 1 ×buffer (Promega), and 2 U AMV in a final volume of 30 μL. In PCR assay, the PCR reaction mixture consisted of 1 ×PCR buffer (PE), 200 μmol/L dNTP, 20 pmoL of each primer, 2 U of Golden Taq DNA polymerase (PE). The mixture was pre-incubated for 5 min at 94°C, followed by amplification at 94°C for 50s, 56°C for 50s, and at 72°C for 60s, for 30 cycles. A final extension was performed at 72°C for 7 min.
DNA cloning and sequencing
The PCR products from all specimens were excised from 0.8% agarose gels, and the desired fragments were purified using a DNA gel extraction kit (Promega). The purified fragments were cloned into a pGEM-T plasmid vector, and then transformed into E. coli JM109 competent cells. Plasmid DNA was extracted from the positive clones and sequenced using a PE 377 sequencer. The sequences were analyzed by DNASIS and OMEGA.
Statistical analysis
Statistical significance was evaluated using the χ2 test. P < 0.05 was considered statistically significant. Statistical analysis was performed with SPSS 11.0.
RESULTS
PT-PCR analysis of whole polβ gene
In the high-incidence group, PCR products were obviously smaller than 1 008 bp in six carcinoma specimens. The correct length of polβ gene was obtained by amplifying the other specimens (Figure 1).
Figure 1.
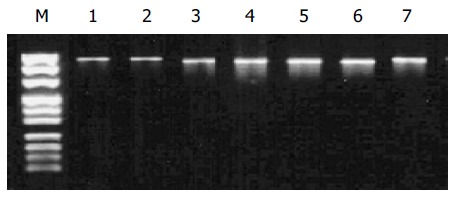
RT-PCR amplification of polβ gene. Lanes 1, 2 and 7: Normal size of PCR products of H1, H2 and H3; lanes 3-6: shorter size of PCR product of H5, H8, H10, and H16; M: DNA marker (from top to bottom: 1 000, 900, 800, 700, 600, 500, 400, 300, 200, 100 bp).
Result of sequencing
Point mutations and deletion of polβ gene were detected in the high-incidence area group (Table 1 and Figure 2). Overall, polβ gene variations were found in 13 of 30 esophageal carcinoma tissue specimens. A 58 bp(177-234 nt) deletion was detected in six tumor tissue specimens (Figure 3). Only one variation was found in the corresponding adjacent normal tissue (Table 1).
Table 1.
Mutation analysis of polβ in high-incidence areas
| Specimen |
Gene mutation |
|
| Carcinoma | Corresponding adjacent normal tissue | |
| H1 | - | - |
| H2 | - | - |
| H3 | - | - |
| H4 | 660 nt A→G | - |
| H5 | 462 nt G→T, | - |
| 177-234 nt deletion | ||
| H6 | - | - |
| H7 | 64 nt G→C, 665 nt T→C | - |
| H8 | 462 nt G→T, 660 nt A→G, | 660 nt A→G |
| 177-234 nt deletion | ||
| H9 | - | - |
| H10 | 177-234 nt deletion, | - |
| 454 nt T→C 466 nt G→A | ||
| H11 | - | - |
| H12 | 375 nt A→G | - |
| 177-234 nt deletion | ||
| H13 | - | - |
| H14 | - | |
| H15 | 454 nt T→C, 466 nt G→A | - |
| H16 | 177-234 nt deletion | - |
| H17 | 737 nt A→T, 740 nt A→G | - |
| H18 | - | - |
| H19 | - | - |
| H20 | - | - |
| H21 | 462 nt G→T | - |
| H22 | - | - |
| H23 | - | - |
| H24 | - | - |
| H25 | 177-234 nt deletion, | - |
| 660 nt A→G | ||
| H26 | 177-234 nt deletion | - |
| H27 | - | - |
| H28 | 613 nt A→T | - |
| H29 | - | - |
| H30 | - | - |
Figure 2.
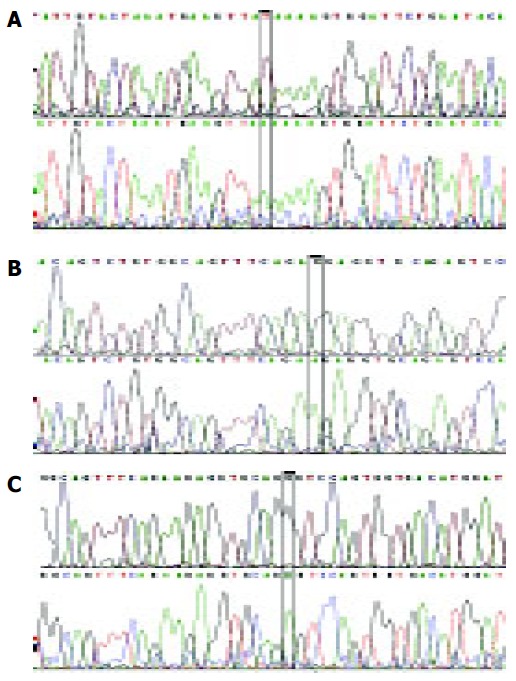
Mutations in the polβ gene. A: 660 nt A→G mutation in H4 carcinoma; B: 670 nt A→G mutation in N1 carcinoma; C: 613 nt A→T mutation in H28 carcinoma.
Figure 3.
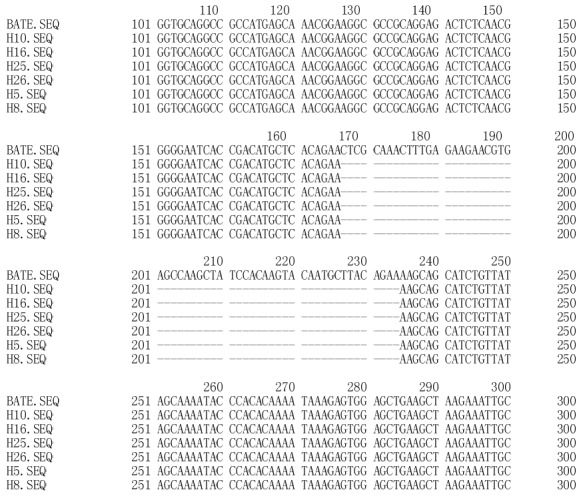
Comparison between wild type polβ gene fragment and six gene fragments with deletion (177→234 nt).
Polβ gene point mutations were detected in 5 of 25 esophageal carcinoma tissue specimens, and no variation was found in corresponding adjacent normal tissues in the non-high incidence area group (Table 2).
Table 2.
Mutation analysis of polβ in low-incidence areas
| Specimen |
Gene mutation |
|
| Carcinoma | Corresponding adjacent normal tissue | |
| N1 | 670 nt A→G | – |
| N2 | 660 nt A→G | – |
| N8 | 660 nt A→G | – |
| N16 | 613 nt A→T | – |
| N22 | 670 nt A→G | – |
The incidence of polβ gene variation observed in the high-incidence area group was significantly higher than that in the non-high incidence area group (P = 0.007, χ2 test).
Variation of amino acid caused by gene mutations
Twelve kinds of variation in the polβ gene were found in the present study, including 11 point mutations and a 58 bp deletion (Table 3). The translation of polβ was interrupted due to the emergence of a termination codon at 117 nt caused by 462 nt G→T mutation. Mutations at 665, 737, and 740 nt were synonymous mutations, which would not change the amino acids. The other seven point mutations caused replacement of amino acids.
Table 3.
Gene and amino acid variations in polβ
| Mutation code | Gene variation | Amino acid variation | Mutation |
| 1 | 375 nt A→G | 88 nt Ile→Val | |
| 2 | 454 nt T→C | 114 nt Phe→Ser | |
| 3 | 462 nt G→T | 117 nt Glu→termination | Termination |
| codon | mutation | ||
| 4 | 466 nt G→A | 118 nt Gly→Glu | |
| 5 | 613 nt A→T | 167 nt Lys→Ile | |
| 6 | 648 nt G→C | 179 nt Gly→Arg | |
| 7 | 660 nt A→G | 183 nt Arg→Gly | |
| 8 | 665 nt T→C | 184 nt Gly→Gly | Synonymous |
| mutation | |||
| 9 | 670 nt A→G | 186 nt Glu→Gly | |
| 10 | 737 nt A→T | 208 nt Pro→Pro | Synonymous |
| mutation | |||
| 11 | 740 nt A→G | 209 nt Lys→Lys | Synonymous |
| mutation | |||
| 12 | 177–234 nt deletion | Frameshift | |
| mutation | |||
| Termination | |||
| mutation |
DISCUSSION
DNA polβ is one of the four recognized, vertebrate, cellular, DNA polymerizing enzymes. Two features of polβ from various species may play a key role. First, the structure of polβ is highly conserved from the standpoint of both polypeptide size and amino acid sequence. Second, in cultured mammalian cells, the level of polβ enzymatic activity is low and independent of cell-cycle stage[6,7]. Hence, polβ is considered as a constitutively expressed “housekeeping” enzyme required for DNA metabolic events other than replicative synthesis of genomic DNA. DNA synthesis during DNA repair and recombination are examples of such events, and the idea that polβ is involved in some types of DNA repair is supported by various studies with DNA polymerase inhibitors[8-11]. Recent observations have shown that the variation of polβ occurs in some tumors such as colorectal carcinoma, bladder carcinoma, breast carcinoma, prostate carcinoma and non-small cell lung cancer. The variation rate is particularly high in colon carcinoma, being more than 80% (5/6)[12-17,20]. Some studies indicate that the accumulation of proto-oncogene and tumor suppressor gene variations perhaps leads to tumor. However, the initial molecular defects causing accumulated mutations and inducing cancer are not well understood. Interestingly, there is a higher mutation rate of polβ, p53 and ras in colorectal carcinoma. A genetic disease, xeroderma pigmentosum, is associated with ERCC, a kind of DNA repair gene[18-21], and these patients are more susceptible to skin carcinoma[22,23]. The above findings suggest that there is a correlation between human tumors and the damage, maladjustment and defects in the DNA repair system.
The present results showed that some mutations, such as 462 nt G→T and deleting 177-234 nt, could lead to the abnormal amino acid of polβ followed by abnormal protein structure and lack of DNA repair activity. From the histopathological diagnosis, we found that cancer cells involving the two mutations were more malignant. In addition, there were three synonymous mutations at 665 nt (T→G), 737 nt (A→T) and 740 nt (A→G), which may be the single nucleotide polymorphism (SNP)[24-29].
Polβ gene variation in the high-incidence area group was detected in 13 of 30 esophageal carcinoma tissue specimens. The mutation rate of this group was 43.3%. Polβ gene variation in the non-high incidence area group was found in 5 of 25 esophageal carcinoma tissue specimens, and the mutation rate was 20%. Thus, there was a significant difference in mutation rates between these two groups (P < 0.05). Twelve types of polβ gene variation were found in high-incidence area, but only three types were found in the non-high incidence area, suggesting that damage of the DNA repair system via alteration of this gene may contribute to the development of esophageal carcinoma, and that polβ may play a more important role in the high-incidence area.
By analyzing the variation site of polβ, we found that three point mutations were located in one region (454-466 nt), and four point mutations were located in another region (648-670 nt). The two regions are probably the mutation hot spots of the polβ gene of esophageal carcinoma. Moreover, the deletion of 58 bp was found in 6 of 20 specimens from the high-incidence area (31.6% frequency), but was not found in the non-high incidence area. Hence, this deletion may be one of the main variations of polβ gene in esophageal carcinoma. Our results prove that polβ mutations do exist in esophageal carcinoma, and are probably correlated to the development of esophageal carcinoma.
It is suggested that during such genetic evolution, the DNA repair system plays an important role in cancer development[30-32]. Although the relationship between mutations of polβ and alterations in proto-oncogenes or tumor suppressor genes is currently not clear, polβ gene mutation is involved in a subset of human esophageal carcinoma, especially in the high-incidence area.
Footnotes
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
Supported by the National Natural Science Foundation of China, No. 39870287
References
- 1.Okuda E, Osugi H, Morimura K, Takada N, Takemura M, Fukushima S, Higashino M, Kinoshita H. Detection of p53 gene mutations in human esophageal squamous cell carcinomas using a p53 yeast functional assay: possible difference in esophageal carcinogenesis between the young and the elderly group. Clin Cancer Res. 2001;7:600–606. [PubMed] [Google Scholar]
- 2.Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, Wang LD, Yang CS. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 3.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J Biol Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Shimada Y, Hashimoto Y, Kaganoi J, Kan T, Watanabe G, Murakami Y, Imamura M. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer Res. 2003;63:6320–6326. [PubMed] [Google Scholar]
- 5.Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Mori M, Croce CM. Allele loss and promoter hypermethylation of VHL, RAR-beta, RASSF1A, and FHIT tumor suppressor genes on chromosome 3p in esophageal squamous cell carcinoma. Cancer Res. 2003;63:3724–3728. [PubMed] [Google Scholar]
- 6.Kim SJ, Beard WA, Harvey J, Shock DD, Knutson JR, Wilson SH. Rapid segmental and subdomain motions of DNA polymerase beta. J Biol Chem. 2003;278:5072–5081. doi: 10.1074/jbc.M208472200. [DOI] [PubMed] [Google Scholar]
- 7.Widen SG, Kedar P, Wilson SH. Human beta-polymerase gene. Structure of the 5'-flanking region and active promoter. J Biol Chem. 1988;263:16992–16998. [PubMed] [Google Scholar]
- 8.Cabelof DC, Raffoul JJ, Yanamadala S, Guo Z, Heydari AR. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002;23:1419–1425. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- 9.Lavrik OI, Kolpashchikov DM, Prasad R, Sobol RW, Wilson SH. Binary system for selective photoaffinity labeling of base excision repair DNA polymerases. Nucleic Acids Res. 2002;30:e73. doi: 10.1093/nar/gnf073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Servant L, Cazaux C, Bieth A, Iwai S, Hanaoka F, Hoffmann JS. A role for DNA polymerase beta in mutagenic UV lesion bypass. J Biol Chem. 2002;277:50046–50053. doi: 10.1074/jbc.M207101200. [DOI] [PubMed] [Google Scholar]
- 11.Idriss HT, Al-Assar O, Wilson SH. DNA polymerase beta. Int J Biochem Cell Biol. 2002;34:321–324. doi: 10.1016/s1357-2725(01)00131-5. [DOI] [PubMed] [Google Scholar]
- 12.SenGupta DN, Zmudzka BZ, Kumar P, Cobianchi F, Skowronski J, Wilson SH. Sequence of human DNA polymerase beta mRNA obtained through cDNA cloning. Biochem Biophys Res Commun. 1986;136:341–347. doi: 10.1016/0006-291x(86)90916-2. [DOI] [PubMed] [Google Scholar]
- 13.Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Canitrot Y, Bieth A, Garès M, Wright M, Delsol G, Loeb LA, et al. Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis. Cancer Res. 2002;62:3511–3514. [PubMed] [Google Scholar]
- 14.Muniappan BP, Thilly WG. The DNA polymerase beta replication error spectrum in the adenomatous polyposis coli gene contains human colon tumor mutational hotspots. Cancer Res. 2002;62:3271–3275. [PubMed] [Google Scholar]
- 15.Thompson TE, Rogan PK, Risinger JI, Taylor JA. Splice variants but not mutations of DNA polymerase beta are common in bladder cancer. Cancer Res. 2002;62:3251–3256. [PubMed] [Google Scholar]
- 16.Tompkins JD, Nelson JL, Hazel JC, Leugers SL, Stumpf JD, Foster PL. Error-prone polymerase, DNA polymerase IV, is responsible for transient hypermutation during adaptive mutation in Escherichia coli. J Bacteriol. 2003;185:3469–3472. doi: 10.1128/JB.185.11.3469-3472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamoto H, Miyagi Y, Ishikawa T, Ichikawa Y, Hosaka M, Kubota Y. DNA polymerase beta gene mutation in human breast cancer. Int J Cancer. 1999;83:708–709. doi: 10.1002/(sici)1097-0215(19991126)83:5<708::aid-ijc24>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 18.Selfridge J, Hsia KT, Redhead NJ, Melton DW. Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res. 2001;29:4541–4550. doi: 10.1093/nar/29.22.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 20.Dobashi Y, Shuin T, Tsuruga H, Uemura H, Torigoe S, Kubota Y. DNA polymerase beta gene mutation in human prostate cancer. Cancer Res. 1994;54:2827–2829. [PubMed] [Google Scholar]
- 21.Lindstrom UM, Chandrasekaran RA, Orbai L, Helquist SA, Miller GP, Oroudjev E, Hansma HG, Kool ET. Artificial human telomeres from DNA nanocircle templates. Proc Natl Acad Sci USA. 2002;99:15953–15958. doi: 10.1073/pnas.252396199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada NA, Farber RA. Induction of a low level of microsatellite instability by overexpression of DNA polymerase Beta. Cancer Res. 2002;62:6061–6064. [PubMed] [Google Scholar]
- 23.Saxowsky TT, Matsumoto Y, Englund PT. The mitochondrial DNA polymerase beta from Crithidia fasciculata has 5'-deoxyribose phosphate (dRP) lyase activity but is deficient in the release of dRP. J Biol Chem. 2002;277:37201–37206. doi: 10.1074/jbc.M206654200. [DOI] [PubMed] [Google Scholar]
- 24.Hartenstine MJ, Goodman MF, Petruska J. Weak strand displacement activity enables human DNA polymerase beta to expand CAG/CTG triplet repeats at strand breaks. J Biol Chem. 2002;277:41379–41389. doi: 10.1074/jbc.M207013200. [DOI] [PubMed] [Google Scholar]
- 25.Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J Biol Chem. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 26.Jezewska MJ, Galletto R, Bujalowski W. Dynamics of gapped DNA recognition by human polymerase beta. J Biol Chem. 2002;277:20316–20327. doi: 10.1074/jbc.M200918200. [DOI] [PubMed] [Google Scholar]
- 27.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase iota by controlled proteolysis. J Biol Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda T, Vande Berg BJ, Bebenek K, Osheroff WP, Wilson SH, Kunkel TA. The base substitution fidelity of DNA polymerase beta-dependent single nucleotide base excision repair. J Biol Chem. 2003;278:25947–25951. doi: 10.1074/jbc.C300170200. [DOI] [PubMed] [Google Scholar]
- 29.Le Page F, Schreiber V, Dherin C, De Murcia G, Boiteux S. Poly(ADP-ribose) polymerase-1 (PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in the absence of DNA polymerase beta. J Biol Chem. 2003;278:18471–18477. doi: 10.1074/jbc.M212905200. [DOI] [PubMed] [Google Scholar]
- 30.Harrigan JA, Opresko PL, von Kobbe C, Kedar PS, Prasad R, Wilson SH, Bohr VA. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J Biol Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 31.Heidenfelder BL, Topal MD. Effects of sequence on repeat expansion during DNA replication. Nucleic Acids Res. 2003;31:7159–7164. doi: 10.1093/nar/gkg920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]


