Abstract
AIM: To investigate the biological function of p7 protein and to look for proteins interacting with p7 protein in hepatocytes.
METHODS: We constructed p7 protein bait plasmid by cloning the gene of p7 protein into pGBKT7, then transformed it into yeast AH109 (a type). The transformed yeast was mated with yeast Y187 (α type) containing liver cDNA library plasmid, pACT2 in 2 × YPDA medium. Diploid yeast was plated on synthetic dropout nutrient medium (SD/-Trp-Leu-His-Ade) containing x-α-gal for selection and screening. After extracting and sequencing of plasmids from blue colonies, we performed sequence analysis by bioinformatics.
RESULTS: Fifty colonies were selected and sequenced. Among them, one colony was Homo sapiens signal sequence receptor, seven colonies were Homo sapiens H19, seven colonies were immunoglobulin superfamily containing leucine-rich repeat, three colonies were spermatid peri-nuclear RNA binding proteins, two colonies were membrane-spanning 4-domains, 24 colonies were cancer-associated antigens, four colonies were nucleoporin 214 ku and two colonies were CLL-associated antigens.
CONCLUSION: The successful cloning of gene of protein interacting with p7 protein paves a way for the study of the physiological function of p7 protein and its associated protein.
Keywords: Hepatitis C virus, p7 protein, Interacting proteins, Yeast two-hybrid system
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma throughout the world. Although HCV cannot be incubated efficiently in vitro, several of its key features have been elucidated in the past few years. HCV, cloned successfully via molecule biological technology[1,2], is an enveloped, positive single-stranded RNA (9.6-kb) virus belonging to the Flaviviridae. The HCV genome has only one ORF which is flanked by a 5’ and 3’ noncoding region. The ORF encodes for a large polyprotein precursor of about 3 000 amino acid residues, and this precursor protein is cleaved by the host and viral proteinases to generate at least 10 proteins in the following order: NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. HCV proteins not only function in viral replication but also affect a variety of cellular functions[3-6]. HCV p7, which is located between the E2 and NS2 proteins, is a 63 residue peptide encoded by HCV genome between 2 580-2 768 nt[7]. Although there are studies on the genomic structure, synthesis and function of p7 protein, the role of the HCV p7 protein in the virus life cycle is not known. To gain more information on the HCV p7 protein, and provide some new clues for elucidating the potential biological function of p7, we looked for proteins interacting with p7 protein by screening human liver cDNA library with yeast two-hybrid system.
MATERIALS AND METHODS
Bacteria, yeast strains and plasmids
All yeast strains and plasmids for yeast two-hybrid experiments were obtained from Clontech (Palo Alto, CA, USA) as components of the MATCHMAKER two hybrid system 3. Yeast strain AH109 (MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4△, gal80△, LYS2:GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3: MEL1UAS-MEL1TATA-LacZ) containing pGBKT7-53, coding for DNA-BD/mouse p53 fusing protein and AH109 was used for cloning of bait plasmids, yeast strain Y187 (MATa, ura3-52, his3-200, Ade2-101, trp1-901, leu2-3, 112, gal4△, gal80△, met-, URA3: GAL1UAS-GAL1TATA-lacZ MEL1) containing pTD1-1, in which pACT2 codes for AD/SV40 large T antigen fusing protein and Y187 was used for cloning of library plasmids. Bacterial strain DH5a was used for cloning of every shuttle plasmid. pGBKT7 DNA-BD cloning plasmid, pGADT7 AD cloning plasmid, pGBKT7-53 control plasmid, pGADT7, pGBKT7-Lam control plasmid, pCL1 plasmid were from Clontech Ltd Company (K1612-1). pGEM T vector was from Promega Company, USA.
Chemical agents and culture media
Taq DNA polymerase was purchased from MBI Company. T4 DNA ligase, EcoRI and BamHI restriction endonuclease were from Takara. c-Myc mAb secreted by 1-9E10.2 hybridoma (ATCC), and goat anti-mouse IgG conjugated with horseradish peroxidase were from Zhongshan Company, China. Lithium acetate, semi-sulfate adenine, acrylamide, and N, N’-bis-acrylamide were from Sigma. TEMED was from Boehringer Mannheim. Tryptone and yeast extracts were from Oxoid. X-α-gal and culture media: YPDA, SD/ -Trp, SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, SD/-Trp/-Leu/-His/-Ade were from Clontech Ltd Company. Protein-G agarose were from Roche. pEGM-T vector was from Promega. RT-PCR kit was from Isotope Company of China. Others were from Sigma Company.
Construction of “bait” plasmid and expression of HCV p7 protein
To make the bait plasmid, HCV-p7 sequences were generated by PCR amplification of HCV plasmid (HCV strain 1b). The plasmid containing coding sequences of all the structural and non-structural proteins was used as the template. The sequence of the primers containing the EcoRI and BamHI restriction enzyme sites are sense primer: 5’-GAA TTC ATG GCT TTG GAG AAC CTC G-3’ and anti-sense primer: 5’-GGA TCC TTA CGC GTA CGC CCG CTG G-3’. The PCR conditions were as follows: at 94°C for 30 s, at 60°C for 30 s, at 72°C for 30 s. Ten nanograms of the 189-bp PCR product were cloned with pGEM-T vector. The primary structure of insert was confirmed by direct sequencing. The fragment of encoding p7 was released from the pEGM-T-p7 by digestion with EcoRI and BamHI, and ligated to pGBKT7. Vector pGBKT7-expressing proteins were fused with amino acids 1-147 of the GAL4 DNA binding domain (DNA-BD), pGADT7-expressing proteins were fused with amino acids 768-881 of the GAL4 activation domain (AD). Plasmid pGBKT7-p7 (Figure 1) containing full-length HCV p7 gene could directly express DNA binding domain, c-Myc and p7 fusion protein. The plasmid was transformed into yeast strain AH109 by using lithium acetate method[8]. Transformed AH109 (bait) was cultured on quadruple dropout media to exclude the auto-activity.
Figure 1.
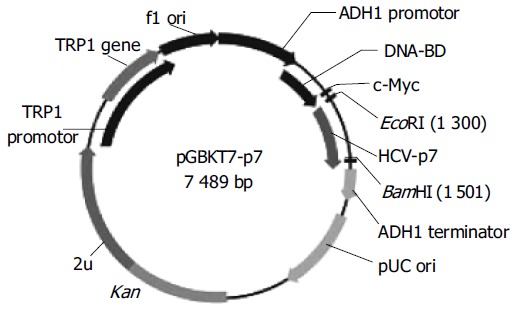
Map of “bait” plasmid pGBKT7-p7.
Western blot analysis
A single isolated colony (1-2 mm in diameter, not older than 5 d) of yeast AH109 transformed with pGBKT7-p7 selected by lithium acetate method was inoculated into 5 mL of SD/-Trp and incubated at 30°C overnight with shaking at 220-250 r/min. The entire overnight culture was inoculated into 50 mL YPDA medium and incubated at 30°C with shaking at 220-250 r/min until the A600 reached 0.4-0.6. The culture was chilled and centrifuged for cell pellets. The yeast protein extracts of p7 were prepared according to urea/SDS method. A part of protein exact was resolved on SDS-polyacrylamide gel and transferred onto nitrocellulose membrane. After being blocked with nonfat dried milk, the membrane was treated with c-Myc mAb for 90 min, then with HRP-goat anti-mouse IgG at the dilution of 1:500 for 60 min. Subsequently the blot was developed by 4-chloro-1-naphthol and H2O2. The yeast AH109 cells with transformed pGBKT7 were used for positive control and the untransformed yeast, AH109 cells were used for negative control.
Screening of liver cell cDNA library by yeast two-hybrid system
One large (2-3 mm), fresh (< 2 mo old) colony of AH109 (bait) was inoculated into 50 mL of SD/-Trp and incubated at 30°C overnight (16-24 h) with shaking at 250-270 r/min. Then the cells were spun by centrifuging the entire 50 mL culture at 1 000 r/min for 5 min. After supernatant was decanted, the cell pellet was resuspended in the residual liquid by vortexing. A human liver cDNA library was cloned into pACT2 and yeast reporter strain Y187 (Clontech Co.). The entire AH109 (bait) culture and 1 mL human liver cDNA library (1 × 106 CFU/mL) were combined and cultured in a 2-L sterile flask and 45 mL of 2´YPDA/Kan was added and swirled gently. After 20 h mating, the cells were spun re-suspended and spread on 50 large (150 mm) plates containing 100 mL of SD/-Ade/-His/-Leu/-Trp (QDO). After growth for 6–15 d, the yeast colonies were transferred onto the plates containing X-α-gal to check for expression of the MEL1 reporter gene (blue colonies).
Plasmid isolation from yeast and transformation of E. coli with yeast plasmid
Approximately 1 × 106 colonies were screened and 50 positive clones were identified. Yeast plasmid was isolated from positive yeast colonies with lyticase method (Clontech Co.), and transformed into super-competence E. coli DH5α by a chemical method. Transformants were plated on ampicillin SOB selection media and grown under selection. Subsequently, pACT2-cDNA constructs were re-isolated following the standard protocol, analyzed by restriction digestion and sequencing in.
Bioinformatic analysis
After the positive colonies were sequenced, the sequences were blasted with GenBank to analyze the function of the genes (http://www.ncbi.nlm.gov.blast).
Confirmation of the true interaction in yeast
To confirm the true protein–protein interaction and exclude false positives, the plasmids of positive colonies were transformed into yeast strain Y187, and then mating experiments were carried out by mating with yeast strain AH109 containing pGBKT7-p7 or pGBKT7-Lam. After mating, the diploids of yeast were plated on SD/-Ade-His-Leu-Trp (QDO) covered with X-α-gal to test the specificity of interactions.
RESULTS
Identification of recombinant plasmid
The full length sequences of HCV p7 were generated by PCR amplification of HCV plasmid (HCV strain 1b), sequenced and analyzed by comparing to vector NTI 6 and BLAST database homology search (http://www.ncbi. nlm.nih.gov/blast). After being cut by EcoRI/BamHI, the fragment was in-frame ligated into pGBKT7 EcoRI/BamHI site. Restriction enzyme analysis of pGBKT7-p7 plasmid with EcoRI/BamHI yielded two bands: 7 300 bp empty pGBKT7 and 189 bp HCV p7. The product of plasmid was amplified by PCR. Analysis of the PCR reaction products by agarose gel electrophoresis showed the clear bands with the expected size (189 bp of p7). Sequences of the PCR products were correct (Figures 2A and B).
Figure 2.
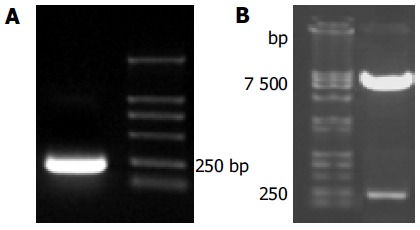
Fragment-p7 amplified by PCR (A) and pGBKT7-p7 cut by EcoRI/ BamHI (B).
Expression of “bait” fusion protein
Yeast strain AH109 transformed with pGBKT7-p7 could stably express the fusion protein at high level (Figure 3) and could only grow on SD/-Trp medium but not on QDO medium. Thus, the transformed yeast could be used for yeast hybrid analysis.
Figure 3.
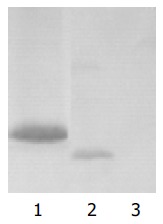
Expression of HCV p7 in yeast shown by Western blotting. Lane 1: HCV p7 protein; lane 2: positive control; lane 3: negative control.
Screening of liver cell cDNA library
We isolated the plasmids from the blue colonies containing only pGBKT7-p7 and one library plasmid other than other plasmids. Because plasmid pACT2-cDNA contains two restriction endonuclease sites of BglII on both sides of multiple cloning sites, the gene fragments of the liver cell cDNA library (pACT2-cDNA) screened were released by BglII digestion (Figure 4). The gene fragments of different lengths in Figure 4 proved, that these screened clones were positive colonies growing on SD/-Trp/-Leu/-His/-Ade culture medium after mating.
Figure 4.
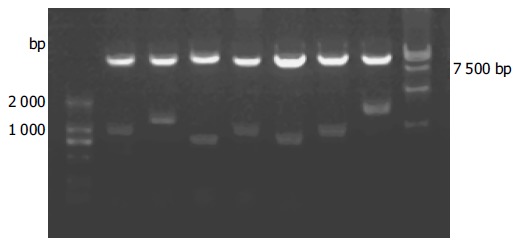
Identification of different colonies with BglII digestion.
Analysis of cDNA sequence and homology
We obtained a total of 50 positive colonies growing on the selective SD/-trp-leu-his-ade/X-α-gal medium. These colonies were prescreened by BglII digestion to make sure that only colonies with different inserts were subjected to sequencing. Fifty colonies from cDNA library were sequenced. Using the BLAST program at the National Center for Biotechnology Information, the 50 sequences had a high similarity to known genes. The data are presented in Table 1.
Table 1.
Comparison between positive clones and similar sequences in GenBank
| High similarity to known genes | Number of similar | Homolog (%) |
| Homo sapiens signal sequence receptor, delta | 1 | 100 |
| Homo sapiens H19, imprinted maternally expressed untranslated mRNA | 7 | 96-99 |
| Homo sapiens immunoglobulin superfamily containing leucine-rich repeat | 7 | 98 |
| Homo sapiens spermatid perinuclear RNA binding protein | 3 | 100 |
| Homo sapiens membrane-spanning 4-domains, subfamily A, member 6A (MS4A6A) | 2 | 97-98 |
| Homo sapiens colon cancer-associated antigen | 24 | 94-100 |
| Homo sapiens nucleoporin 214 ku | 4 | 86 |
| Homo sapiens CLL-associated antigen KW-13 | 2 | 99 |
DISCUSSION
The precise mechanism of early HCV infection remains largely unknown. We tried to obtain a sufficient amount of free virions from the plasma of infected individuals and to establish a robust in vitro system for virus propagation. In HCV polyprotein, HCV p7 is located downstream of the envelope glycoprotein, E2 and upstream of the NS2 protein and begins at the position 747 of HCV H strain polyprotein. The generation of HCV p7 is supposed to be catalyzed by a host signal peptidase localized in the endoplasmic reticulum[9]. The p7 polypeptide of HCV is a small hydrophobic polyprotein of unknown function and contains 63 amino acids. Its encoding gene is present between structural and non-structural proteins, but it is still not clear whether p7 is a structural protein or a non-structural protein. Studies on the subcellular localization of HCV proteins indicate that most of them associate with ER membranes[10-12]. HCV p7 comprising two transmembrane alpha helices linked by a small charged cytoplasmic loop, is indeed membrane-associated and mostly located in the endoplasmic reticulum[7]. HCV p7 is an integral membrane polypeptide. Pulse-chase analysis showed that a large proportion of p7 stays in an early compartment of the secretory pathway. Carrere-Kremer et al.,’s studies[7] showed that p7 has a double membrane-spanning topology, the amino- and carboxyl-terminal tails are oriented toward the ER and should therefore also be accessible from the extracellular environment. The C-terminal transmembrane domain of p7 is a signal sequence and the amino- and/or carboxyl-terminal intraluminal tails of p7 contain sequences with genotype-specific function[13]. The data indicate that p7 is a polytopic membrane protein that could play a functional role in several compartments of the secretory pathway.
At present, studies are carried on the biological functions of HCV p7 protein in the virus life cycle. HCV p7 protein forms hexamer cation ion channels in black lipid membranes. The function of p7 is supposed to be similar to viroporin, which mediates cation permeability across membranes and are important for virion release or maturation[14,15]. Mutants with deletions of all or part of p7 and a mutant with substitutions of two conserved residues in the cytoplasmic loop are not viable. Thus, p7 is essential for infectivity of HCV[13]. HCV p7 regulates the internalization properties of HCV structural proteins[16]. It has been hypothesized that lack of p7 may lead to conformational changes of envelope proteins, as reported for closely related virus, that are perhaps important for cell binding and entry of the HCV virion. Studies of HCV subgenomic replicons showed that p7 is not critical for RNA replication[17,18].
Transfection with RNA transcripts from an infectious BVDV cDNA clone with in-frame deletions or point mutations of p7 could not produce infectious virus, but the infectivity could be restored by providing p7 in transmembrane. HCV p7 has characteristics similar to those of a group of proteins called viroporins[13,15]. These proteins form ion channels that might be of importance for virus assembly and/or release or maturation. Mutation of a conserved basic loop located between the two predicted transmembrane alpha helices rendered HCV p7 non-functional as an ion channel. The intra-cellular localization of p7 was unaffected by this mutation[19,20]. The HCV-like particles (HCV-LPs) without p7 (p7-HCV-LP) have been shown to be more potent in inducing cellular immune responses with a Th1 bias than HCV-LP with p7[21]. The study showed that p7-HCV-LP can also induce both humoral and cellular immunities in AAD mice[22].
Protein-protein interactions occur in a wide variety of biological processes and essentially control the cell fate from division to death. Yeast two-hybrid assays represent a versatile tool to study protein interactions in vivo. Yeast two-hybrid system 3 based on the system originally designed by Fields and Song, takes advantage of the properties of the GAL4 protein of the yeast Saccharomyces cerevisiae. GAL4-based assay uses yeast transcription factor, GAL4 for detection of protein interactions by transcriptional activation. GAL4 possesses a characteristic phenomenon that the transactivation function can be restored when the factor’s DNA-binding domain (DBD) and its transcription-AD are brought together by two interacting heterologous proteins. GAL4-yeast two-hybrid assay uses two expression vectors, one uses DBD and the other uses AD. The GAL4-DBD fuses to a protein ‘X’ and a GAL4-AD fuses to a protein ‘Y’ to form the bait and the target of the interaction trap, respectively. A selection of host cells with different reporter genes and different growth selection markers provides a means to detect and confirm protein-protein interactions and has significantly fewer false positives[23-26].
To investigate the role of HCV p7 in pathogenesis of HCV and physiologic function in hepatocytes, yeast two hybrid system 3 is used to screen the proteins interacting with p7 protein. In this study, the “bait” plasmid pGBKT7-p7 was transformed into yeast strain AH109. In order to further confirm the expression of HCV p7 protein in AH109 yeast strain, we performed the experiment of Western blot and a strong expression of the HCV p7 protein was observed. After the “bait” plasmid pGBKT7-p7 yeast strain AH109 was mated with liver cDNA library yeast strain Y187, the diploid yeast cells were plated on QDO media containing X-α-gal, 50 true positives were obtained. By sequencing analysis of isolated library plasmids, we got the sequences of 50 genes with known functions. Eight of them were respectively associated with genesis of tumor and immunity, regulation of cell life cycle, and the way of signal transmission.
We screened Homo sapiens membrane-spanning 4-domains, subfamily A (MS4A6A). Tetraspanins are a large superfamily of cell surface membrane proteins characterized by their four transmembrane domains, and expressed in a wide variety of cell types and formed two different circularity structures outside cellular membrane. Tetraspanins interact with diverse important proteins, such as integrins, immunoreceptors, and signaling molecules and have functional roles in processes. They promote cellular growth and signal transduction and are possibly related to viral adhesion and entry, cellular cancerization, tumor cell invasion, and metastasis[27,28]. The tetraspanin web refers to a network of molecular interactions involving tetraspanins and other molecules. The functions of net are complex and multiple. The CD81 is one of the tetraspanins and widely expressed in a variety of cell types. Todres et al.[29], and Bartosch et al.[30], showed that CD81 is likely to be the receptor protein of HCV infecting target cells, and the structures of CD81 and other subfamilies of tetraspanins are partly homologous. Because CD37 can regulate directly B cell function and interact between T and B cells, and regulate humoral immunity[31,32]. CD53 relates to apoptosis and tumor formation. CD63 relates to maturation of human dendritic cells and modulates differential distribution of associated MHC class II molecules during maturation of dendritic cells[33]. Migration of activated hepatic stellate cells (HSC) is a key event in the progression of liver fibrosis. The tetraspanin CD151 molecule plays a key role as a regulator of α6β1 integrin adhesion strengthening and involves in HSC migration, adhesion, and proliferation[34,35]. Human CD81 directly enhances Th1 and Th2 cell activation, but preferentially induces proliferation of Th2 cells upon long-term stimulation. CD81 cross-linking can also induce adhesion in B and T cells and decrease the threshold for B cell activation via the immunoglobulin (Ig) receptor, which produces autoantibodies and form corresponding immunopathologic damage[36]. In this study, it was shown that HCV p7 could bind to tetraspanins and the functions of p7 are complex.
Another important protein interacting with p7 protein from liver cDNA library is Homo sapiens nucleoporin 214 ku (NUP214). Nuclear pore complexes (NPCs) are large protein structures spanning the double membrane of the eukaryotic nucleus that serves as a site for translocation of macromolecules between the nucleus and the cytoplasm. NPCs include a family of 50-100 proteins termed as nucleoporins (Nups). Studies in the past several years have demonstrated that individual Nups play a unique role in regulating NPC function and nucleo-cytoplasmic transport of proteins and RNAs. The functions of individual Nups are associated with specific human diseases. NUP214, also named as CAN protein, is an oncogene and one of the components of NPCs in eukaryotic cells. It contains FG repeat sequences of special proteins of NPCs, which bind directly to receptors that transport substrates through the NPC. NUP214 is located at cytoplasmic side of the NPC and plays a role in numerous pathways, including cell cycle progression, control of gene expression, oncogenesis, and transport of nucleoplasmic. 3’ extremity of the gene forms an integral gene with DEK gene on chromosome 6, which is correlative to acute myelocytic leukemia and myelodysplastic syndrome[37-39].
We also screened Homo sapiens Ig superfamily (IgSF) containing leucine-rich repeat. The IgSF is one of the largest families of protein domains in this genome and one of the major families in other multicellular eukaryotes. The members of the superfamily are involved in a variety of functions including cell-cell recognition, cell-surface receptors, muscle structure and the immune system[40]. The IgSF containing a leucine-rich repeat (LRR) is named as ISLR. The ISLR gene is mapped on human chromosome 15q23-q24 by fluorescence in situ hybridization. It is a protein with a molecular mass of 46 ku and contains a LRR, with conserved flanking sequences and a C2-type Ig-like domain. These domains are important for protein-protein interaction or cell adhesion, therefore it is possible that the ISLR may also interact with other proteins or cells[41].
HCV p7 protein also interacts with Homo sapiens signal sequence receptor δ, Homo sapiens H19, imprinted maternally-expressed untranslated mRNA, Homo sapiens spermatid perinuclear RNA binding protein, Homo sapiens colon cancer-associated antigen and Homo sapiens CLL-associated antigen KW-13. The significance of interactions of the proteins with HCV p7 should be further studied in vivo and in vitro.
Footnotes
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
Supported by the National Natural Scientific Foundation, No. C03011402, No. C30070690; the Research and Technique Foundation of PLA during the 9th-five year plan period, No. 98D063; the Launching Foundation for Student Studying Abroad of PLA, No. 98H038; and the Youth Research and Technique Foundation of PLA during the 10th-five year plan period, No. 01Q138; and the Research and Technique Foundation of PLA during the 10th-five year plan period, No. 01MB135
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Kato N. Molecular virology of hepatitis C virus. Acta Med Okayama. 2001;55:133–159. doi: 10.18926/AMO/32025. [DOI] [PubMed] [Google Scholar]
- 3.De Francesco R, Neddermann P, Tomei L, Steinkühler C, Gallinari P, Folgori A. Biochemical and immunologic properties of the nonstructural proteins of the hepatitis C virus: implications for development of antiviral agents and vaccines. Semin Liver Dis. 2000;20:69–83. doi: 10.1055/s-2000-9504. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52–59. doi: 10.1002/jmv.10545. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann H. [Virology of hepatitis C virus] Wien Med Wochenschr. 2000;150:463–466. [PubMed] [Google Scholar]
- 6.Kato N. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microb Comp Genomics. 2000;5:129–151. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- 7.Carrère-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J Virol. 2002;76:3720–3730. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol Cell Biochem. 1997;172:67–79. doi: 10.1023/A:1006859319926. [DOI] [PubMed] [Google Scholar]
- 9.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 10.Hügle T, Fehrmann F, Bieck E, Kohara M, Kräusslich HG, Rice CM, Blum HE, Moradpour D. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 11.Itakura J, Sakamoto N, Enomoto N. [Structure and functions of HCV non-structural proteins] Nihon Rinsho. 2004;62 Suppl 7:64–68. [PubMed] [Google Scholar]
- 12.Op De Beeck A, Voisset C, Bartosch B, Ciczora Y, Cocquerel L, Keck Z, Foung S, Cosset FL, Dubuisson J. Characterization of functional hepatitis C virus envelope glycoproteins. J Virol. 2004;78:2994–3002. doi: 10.1128/JVI.78.6.2994-3002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai A, Claire MS, Faulk K, Govindarajan S, Emerson SU, Purcell RH, Bukh J. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc Natl Acad Sci USA. 2003;100:11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlović D, Neville DC, Argaud O, Blumberg B, Dwek RA, Fischer WB, Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc Natl Acad Sci USA. 2003;100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/S0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 16.Saunier B, Triyatni M, Ulianich L, Maruvada P, Yen P, Kohn LD. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J Virol. 2003;77:546–559. doi: 10.1128/JVI.77.1.546-559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada T, Tautz N, Thiel HJ. E2-p7 region of the bovine viral diarrhea virus polyprotein: processing and functional studies. J Virol. 2000;74:9498–9506. doi: 10.1128/JVI.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol. 2002;76:4008–4021. doi: 10.1128/JVI.76.8.4008-4021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol. 2004;85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 20.Fischer WB, Sansom MS. Viral ion channels: structure and function. Biochim Biophys Acta. 2002;1561:27–45. doi: 10.1016/S0304-4157(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 21.Lechmann M, Murata K, Satoi J, Vergalla J, Baumert TF, Liang TJ. Hepatitis C virus-like particles induce virus-specific humoral and cellular immune responses in mice. Hepatology. 2001;34:417–423. doi: 10.1053/jhep.2001.26523. [DOI] [PubMed] [Google Scholar]
- 22.Murata K, Lechmann M, Qiao M, Gunji T, Alter HJ, Liang TJ. Immunization with hepatitis C virus-like particles protects mice from recombinant hepatitis C virus-vaccinia infection. Proc Natl Acad Sci USA. 2003;100:6753–6758. doi: 10.1073/pnas.1131929100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng JJ, Rojas M, Bacon W, Stickney JT, Ip W. Methods to study protein-protein interactions. Methods Mol Biol. 2005;289:341–358. doi: 10.1385/1-59259-830-7:341. [DOI] [PubMed] [Google Scholar]
- 24.Osman A. Yeast two-hybrid assay for studying protein-protein interactions. Methods Mol Biol. 2004;270:403–422. doi: 10.1385/1-59259-793-9:403. [DOI] [PubMed] [Google Scholar]
- 25.Gietz RD, Woods RA. Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol Biol. 2002;185:471–486. doi: 10.1385/1-59259-241-4:471. [DOI] [PubMed] [Google Scholar]
- 26.Zhen Z. [Progress in proteomics] Sheng Wu Gong Cheng Xue Bao. 2001;17:491–493. [PubMed] [Google Scholar]
- 27.Tarrant JM, Robb L, van Spriel AB, Wright MD. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 2003;24:610–617. doi: 10.1016/j.it.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Carloni V, Mazzocca A, Ravichandran KS. Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene. 2004;23:1566–1574. doi: 10.1038/sj.onc.1207287. [DOI] [PubMed] [Google Scholar]
- 29.Todres E, Nardi JB, Robertson HM. The tetraspanin superfamily in insects. Insect Mol Biol. 2000;9:581–590. doi: 10.1046/j.1365-2583.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 30.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 31.van Spriel AB, Puls KL, Sofi M, Pouniotis D, Hochrein H, Orinska Z, Knobeloch KP, Plebanski M, Wright MD. A regulatory role for CD37 in T cell proliferation. J Immunol. 2004;172:2953–2961. doi: 10.4049/jimmunol.172.5.2953. [DOI] [PubMed] [Google Scholar]
- 32.Knobeloch KP, Wright MD, Ochsenbein AF, Liesenfeld O, Löhler J, Zinkernagel RM, Horak I, Orinska Z. Targeted inactivation of the tetraspanin CD37 impairs T-cell-dependent B-cell response under suboptimal costimulatory conditions. Mol Cell Biol. 2000;20:5363–5369. doi: 10.1128/MCB.20.15.5363-5369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engering A, Kuhn L, Fluitsma D, Hoefsmit E, Pieters J. Differential post-translational modification of CD63 molecules during maturation of human dendritic cells. Eur J Biochem. 2003;270:2412–2420. doi: 10.1046/j.1432-1033.2003.03609.x. [DOI] [PubMed] [Google Scholar]
- 34.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME. Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc Natl Acad Sci USA. 2003;100:7616–7621. doi: 10.1073/pnas.1337546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzocca A, Carloni V, Sciammetta S, Cordella C, Pantaleo P, Caldini A, Gentilini P, Pinzani M. Expression of transmembrane 4 superfamily (TM4SF) proteins and their role in hepatic stellate cell motility and wound healing migration. J Hepatol. 2002;37:322–330. doi: 10.1016/S0168-8278(02)00175-7. [DOI] [PubMed] [Google Scholar]
- 36.Maecker HT. Human CD81 directly enhances Th1 and Th2 cell activation, but preferentially induces proliferation of Th2 cells upon long-term stimulation. BMC Immunol. 2003;4:1. doi: 10.1186/1471-2172-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cronshaw JM, Matunis MJ. The nuclear pore complex: disease associations and functional correlations. Trends Endocrinol Metab. 2004;15:34–39. doi: 10.1016/j.tem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004;165:823–833. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teichmann SA, Chothia C. Immunoglobulin superfamily proteins in Caenorhabditis elegans. J Mol Biol. 2000;296:1367–1383. doi: 10.1006/jmbi.1999.3497. [DOI] [PubMed] [Google Scholar]
- 41.Yoon IK, Kim HK, Kim YK, Song IH, Kim W, Kim S, Baek SH, Kim JH, Kim JR. Exploration of replicative senescence-associated genes in human dermal fibroblasts by cDNA microarray technology. Exp Gerontol. 2004;39:1369–1378. doi: 10.1016/j.exger.2004.07.002. [DOI] [PubMed] [Google Scholar]


