Abstract
Background & Aims
Genetic polymorphisms within the interferon lambda (IFN-λ) region are strongly associated with hepatitis C virus (HCV) clearance; the IFNL4-ΔG/TT (rs368234815) polymorphism, which controls generation of the IFN-λ4 protein, is more strongly associated with HCV clearance than rs12979860 (the ‘IL28B variant’). An IFNL3 3′ untranslated region polymorphism (rs4803217) has been proposed as a causal variant that may affect HCV clearance by altering IFNL3 mRNA stability.
Methods
We compared IFNL4-ΔG/TT and rs4803217 for association with response to pegylated-IFN-α/ribavirin in the VIRAHEP-C and HALT-C trials, and spontaneous HCV clearance in the ALIVE, UHS and WIHS studies. Genotyping was performed with TaqMan assays. We compared differences in mean reduction in HCV RNA levels by genotype and haplotype. For HCV clearance, we calculated p-values comparing c-statistics for IFNL4-ΔG/TT and rs4803217 genotypes by a bootstrap approach.
Results
Among European Americans, linkage disequilibrium between IFNL4-ΔG/TT and rs4803217 was strong (r2=0.89–0.99) and there were no significant differences between the variants. In African American (AA) individuals enrolled in VIRAHEP-C, HCV RNA at treatment day 28 was more strongly associated with IFNL4-ΔG/TT than rs4803217 (p=0.003); the IFNL4-ΔG:rs4803217-G haplotype, which includes the putatively favorable IFNL3 allele, was actually associated with the poorest day 28 response (p=0.03, comparison to IFNL4-ΔG:rs4803217-T haplotype). Among AA participants, associations were stronger for IFNL4-ΔG/TT than rs4803217 for undetectable HCV RNA at week 24 in Virahep C (p=0.03) and week 20 in HALT-C (p=0.03), as well as for spontaneous HCV clearance (p=0.048).
Conclusion
IFNL4-ΔG/TT is the primary IFN-λ region polymorphism for impaired HCV clearance.
Keywords: genetics, IL28B, IFNL3, IFNL4, innate immunity, interferon lambda, treatment, viral clearance
Background
Impaired clearance of hepatitis C virus (HCV) is strongly associated with certain genetic polymorphisms in the interferon lambda (IFN-λ) region. The four genes in this chromosomal locus were discovered relatively recently. In 2003, IFNL1, IFNL2 and IFNL3 (previously, IL29, IL28A and IL28B, respectively) were identified by two independent research groups through genome-based computational prediction [1–3]; proteins encoded by these genes are highly similar to each other with 96% amino acid identity between IFN-λ2 and IFN-λ3 [1, 2]. IFNL4 was discovered in 2013 through RNA sequencing of primary human hepatocytes that had been stimulated to mimic HCV infection [4].
Prior to the discovery of IFNL4, a genome-wide association study (GWAS) found single nucleotide polymorphism (SNP) rs12979860 to be the variant most strongly associated with response to treatment with pegylated-IFN-α plus ribavirin for chronic hepatitis C [5]. A number of contemporaneous studies reported consistent findings for both treatment response and spontaneous clearance of HCV [6–9]. Although commonly referred to as an ‘IL28B’ or IFNL3 variant, rs12979860 actually lies in intron 1 of IFNL4 and, therefore, is properly termed IFNL4-rs12979860 [4]. The IFNL4-ΔG/TT frame-shift variant (rs368234815, originally designated ss469415590) lies in exon 1 of IFNL4. The IFNL4-ΔG allele creates an open reading frame for the IFN-λ4 protein, whereas the alternative IFNL4-TT allele eliminates the IFN-λ4 protein [4]. IFNL4-ΔG is in strong linkage disequilibrium (LD) with the unfavorable rs12979860-T allele, however, IFNL4-ΔG is associated with impaired HCV clearance more strongly than rs12979860-T [4, 10, 11], especially in individuals of African ancestry, in whom LD between the two alleles is weakest [4]. Given that IFNL4-ΔG/TT shows the strongest association for genetic differences in viral clearance and that the IFNL4-ΔG allele controls generation of IFN-λ4, IFNL4-ΔG/TT is a very plausible candidate as the causal variant underlying observed genetic associations with HCV clearance.
SNP rs4803217, located in the 3′ untranslated region (UTR) of IFNL3, has also been proposed as a causal variant for HCV clearance [12]. Variants in 3′ UTRs can affect binding of miRNAs and regulation of gene expression [13, 14], and rs4803217-G reportedly decreases degradation of IFNL3 mRNA upon HCV infection through this mechanism [12]. It was proposed, therefore, that rs4803217-G could be associated with enhanced HCV clearance, however, data on rs4803217 genotype and HCV clearance are limited.
In vitro studies of HCV clearance can be useful for identifying genetic variants that may be functional in humans, however, genetic association studies are needed to determine which variant is more strongly associated with outcome and, therefore, more likely to be causal. Many genetic variants in the IFN-λ region are in strong LD and, for such highly linked variants, it is important to examine the effect in multiple populations, especially populations of African ancestry in which LD tends to be weaker. To address whether IFNL4-ΔG/TT or rs4803217 is the primary functional variant in the IFN-λ region, we compared genetic associations of these polymorphisms for response to treatment of chronic hepatitis C and spontaneous HCV clearance among African American (AA) and European American (EA) individuals.
Methods
Subjects
As previously described, [4] we examined genetic associations in two studies of response to treatment with pegylated IFN-α plus ribavirin that were sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases. The Study of Viral Resistance to Antiviral Therapy of Chronic Hepatitis C (VIRAHEP-C) was designed to compare response to treatment in AA patients with chronic hepatitis C to otherwise similar patients of European ancestry [15]. In VIRAHEP-C, patients with HCV genotype 1 infection who had not undergone previous treatment for chronic hepatitis C received a standard regimen of pegylated IFN-α-2a (180 μg/week) plus ribavirin (1000–1200 mg/day) for up to 48 weeks. Study end points included the following: decrease in HCV RNA levels between baseline and various treatment time points; week 24 response (absence of detectable HCV RNA in serum after 24 weeks of therapy); end-of-treatment response (absence of HCV RNA after 48 weeks of therapy); and sustained virological response (SVR; absence of HCV RNA 24 weeks after treatment was stopped). The Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) Trial was a study of patients with advanced chronic hepatitis C who had failed previous interferon-based treatment. [16, 17] Patients with other liver diseases, human immunodeficiency virus (HIV) infection, active illicit drug use or current alcohol abuse were excluded. During the lead-in phase of HALT-C, patients underwent retreatment with pegylated-IFN-α-2a (180 μg/week) plus ribavirin (1000–1200 mg/day). Subjects with undetectable HCV RNA at week 20 remained on combination treatment through week 48 and were followed until week 72. Subjects with undetectable HCV RNA at weeks 48 and 72 were considered to have an SVR. Investigations of genetic associations in VIRAHEP-C and HALT-C were conducted in those participants who provided written consent for genetic testing. VIRAHEP-C and HALT-C protocols were approved by the Institutional Review Boards (IRBs) of the participating institutions.
Analyses of genetic associations with spontaneous clearance of HCV infection were performed in three studies, as described in previous publications [4, 18]. The AIDS Linked to Intravenous Experience (ALIVE) cohort is an ongoing study that has followed injection drug users (IDUs) in Baltimore since 1988 [19]. HCV infection was established by detection of HCV antibody (anti-HCV) by enzyme immunoassay (EIA) and recombinant immunoblot assay (RIBA [version 3.0]; Novartis). Individuals with cleared HCV infection had anti-HCV (as confirmed by RIBA) and undetectable HCV RNA in serum or plasma without having received any HCV therapy. Written informed consent for genetic testing was obtained from all participants. The study was approved by the IRB at Johns Hopkins Bloomberg School of Public Health.
As previously described, Urban Health Study (UHS) investigators recruited IDUs from street settings in six inner-city San Francisco Bay area neighborhoods from 1986 through 2002, drawing serial cross-sectional samples every six months [20]. Individuals 18 years of age or older were eligible for enrollment if they had injected drugs within the past 30 days or previously enrolled in the UHS study. New participants were screened for visible signs of recent or chronic injection (i.e., venipuncture sites or scars). Interviews were conducted using standardized questionnaires and blood samples were collected from participants who provided written informed consent. The present study included unduplicated IDUs recruited between 1998 and 2000 [21]. Participants who were positive for HCV antibody were divided into two groups based on their HCV RNA result: ‘chronic’ (positive for HCV RNA) or ‘cleared’ (negative for HCV RNA and positive for antibody). All subjects with cleared infection were included in the study and frequency matched to those with chronic infection on the basis of self-reported ethnicity and age. Study procedures were approved by an IRB of the National Cancer Institute and the Committee on Human Subjects Research at the University of California, San Francisco.
The Women’s Interagency HIV Study (WIHS) is a prospective cohort study of HIV-seropositive and at-risk HIV-seronegative women who were enrolled at six clinical sites [22]. Initial enrollment was conducted during 1994–1995, a second recruitment occurred during 2001–02, and a third recruitment period occurred in 2011. Subjects are followed semi-annually with physical exams, specimen collection including blood, and detailed questionnaires regarding health and behavior. The WIHS protocol was approved by each local IRB, and all participants included in this analysis provided written informed consent for genetic testing. The present study focused on the women who were anti-HCV seropositive at the enrollment visit and provided information on race/ethnicity.
Laboratory
Genotyping of IFNL4-ΔG/TT (rs368234815) was performed previously in the Laboratory of Translational Genomics, NCI with a custom TaqMan allelic discrimination genotyping assay (Life Technologies, Foster City, California) [4, 18]. For the present study, genotyping of IFNL3-rs4803217 was performed in the same laboratory with a custom TaqMan assay (Life Technologies, Foster City, California) as previously described [23]. Genotyping was performed by staff who had no knowledge of the clinical phenotype or racial ancestry of the study subjects. For WIHS and UHS, duplicate samples were genotyped for rs4803217. The panel of blinded specimens from WIHS included 43 samples that were tested in duplicate for rs4803217 with 95.3% concordance. For UHS, 130 specimens were tested in duplicate for rs4803217 and 99.2% of the pairs produced concordant results.
In VIRAHEP-C, serum HCV RNA concentrations on day 0, 1, 2, 7, 14, and 28 had been measured with the COBAS Amplicor Hepatitis C Virus Monitor Test, 2.0™ assay (sensitivity 600 international units (IU)/ml; Roche Molecular Diagnostics, Alameda, CA) [24]. In HALT-C, serum samples that had undetectable HCV RNA by that assay were also tested using the qualitative Roche COBAS Amplicor HCV Test, v. 2.0 assay (lower limit of detection, 100 IU/mL; Roche Molecular Diagnostics, Alameda, CA).
Statistical Analysis
We examined LD of IFNL3-rs4803217 with IFNL4-ΔG/TT and with rs12979860 by calculating the correlation coefficient (r2) using Haploview software (version 4.2) [25].
Statistical comparisons of IFNL4-ΔG/TT and rs4803217 were limited to subjects whose DNA specimen was successfully genotyped for both variants. Analyses were stratified by race (self-reported) to control potential confounding. We used the Kruskal-Wallis test to compare median HCV RNA levels between genotypes for each variant (e.g., IFNL4-TT/TT versus IFNL4-ΔG/ΔG). As previously described [4], we compared mean HCV RNA levels in each of the three IFNL4-ΔG/TT genotype groups (i.e., ΔG/ΔG, ΔG/TT, TT/TT) with the respective rs4803217 genotype groups (TT, GT, GG). To determine global statistical significance of these three mean differences, we computed the covariance matrix of the mean differences using a bootstrap procedure. We re-sampled individuals in the study with replacement, and then computed the three differences of the mean RNA levels in the three genotype groups in this bootstrap dataset. We repeated this calculation 10,000 times and used the bootstrap replicates to compute the covariance matrix of the mean differences of the original sample. This covariance matrix was used to compute a three degrees-of-freedom Wald statistic to test the null hypothesis that there was no difference in mean HCV RNA decreases for IFNL4-ΔG/TT and rs4803217.
For dichotomous outcomes (e.g., SVR, spontaneous clearance) we calculated the odds ratio (OR) and accompanying Wald chi-square p-value (Proc Logistic, SAS, Cary, NC) in logistic models that were adjusted for pre-treatment HCV RNA level and fibrosis stage. We examined both general and additive genetic models. We determined the area under the receiver operating characteristic curve (AUROC or c-statistic) [26] for genotypes of each variant for each outcome and computed a p-value for differences in AUROC values (i.e., c-statistic) based on a chi-square test (1 df) that used a bootstrap variance estimate computed by resampling subjects with replacement and repeating the AUROC computations for each bootstrap sample. Haplotype analyses were conducted using the function haplo.glm from the package haplo.stats in R.
Results
Linkage Disequilibrium
We examined LD between the rs4803217, IFNL4-ΔG/TT and rs12979860 variants in the AA and EA participants in this study (Supplementary Figure 1). LD between the IFNL4-ΔG and rs4803217-T alleles was weaker in the AA participants (r2=0.77) than in the EA subjects (r2=0.90), whereas, LD between the rs4803217 and rs12979860 SNPs was the same (r2=0.92) in the AA and EA subjects.
African American Participants
Treatment Response
AA participants in VIRAHEP-C (n=170) and HALT-C (n=142) are described in Table 1. Patients in the two studies were similar with regard to age (medians, 49 and 51 years for VIRAHEP-C and HALT-C, respectively), HCV genotype (all or virtually all genotype 1) and serum HCV RNA levels (medians, 6.4 log10 IU/mL); most participants in both studies were male. By design, HALT-C enrolled patients with more advanced hepatic fibrosis such that 89.4% had an Ishak fibrosis score >3, compared to 33.5% in VIRAHEP-C. Frequencies of the unfavorable IFNL4-ΔG allele and of rs4803217-T were higher in HALT-C than VIRAHEP-C, reflecting that, by design, HALT-C participants had failed previous treatment with IFN-α, whereas VIRAHEP-C participants were treatment naive. LD between IFNL4-ΔG and rs4803217-T was similar in VIRAHEP-C (r2=0.79) and HALT-C (r2=0.74).
Table 1.
Characteristics of African American participants in the VIRAHEP-C and HALT-C trials of response to treatment with pegylated-interferon-α plus ribavirin
| Characteristic | VIRAHEP-C (n = 170) | HALT-C (n = 142) | ||
|---|---|---|---|---|
| Age (Median, IQR) | 49.1 | 45.6 – 52.8 | 51 | 46.0 – 56.0 |
| Male (n, %) | 111 | 65.3 | 81 | 57.0 |
| HCV Genotype 1 (n, %) | 170 | 100.0 | 137 | 96.5 |
| HCV RNA level (log10 IU) (median, IQR) | 6.4 | 5.6 – 6.7 | 6.4 | 6.1 – 6.8 |
| Ishak Fibrosis Score | ||||
| Missing (n, %) | 1 | 0.6 | ||
| 0–2 (n, %) | 112 | 65.9 | 15 | 10.6 |
| 3–4 (n, %) | 49 | 28.8 | 81 | 57.0 |
| 5–6 (n, %) | 8 | 4.7 | 46 | 32.4 |
| IFNL4-ΔG allele frequency (%) | 66.2 | 69.7 | ||
| IFNL3 rs4803217-T allele frequency (%) | 60.6 | 64.8 | ||
| Linkage disequilibrium (r2) | 0.79 | 0.74 | ||
IQR, Interquartile range
IU, International units
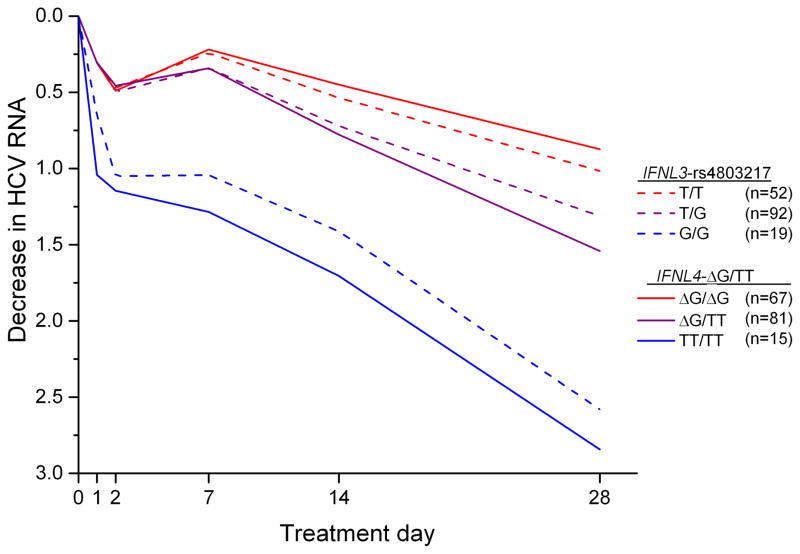
In VIRAHEP-C, the decrease in HCV RNA during the first 28 days of treatment was associated more strongly with IFNL4-ΔG/TT than rs4803217 (p=0.003, difference in mean values; Figure 1). For example, at treatment day 28, the viral decline for the most favorable IFNL4 genotype (TT/TT) was 2.84 log10 IU/ml compared to 2.58 log10 IU/ml for the corresponding rs4803217-G/G genotype and, for the unfavorable genotypes, the decline was 0.87 log10 IU/ml for IFNL4-ΔG/ΔG and 1.02 log10 IU/ml for rs4803217-T/T (Supplementary Table 1).
Figure 1.
Median decrease in HCV RNA levels (log10 international units (IU)/ml) in African American participants in the VIRAHEP-C study during the first 28 days of treatment with pegylated-interferon-α plus ribavirin. P = 0.003 for the mean differences in HCV RNA levels at day 28 for each of the three IFNL4-ΔG/TT (rs368234815) genotype groups relative to the respective IFNL3 rs4803217 genotype groups.
To examine whether rs4803217 might modify the effect of IFNL4-ΔG/TT, we examined compound genotypes and haplotypes based on both variants. In contrast to the hypothesis that the rs4803217-G allele might affect viral clearance favorably, the 15 VIRAHEP-C patients with the IFNL4-ΔG/ΔG: rs4803217-T/G compound genotype had the least decline in HCV RNA at day 28 of treatment (0.62 log10 IU/ml; p=0.03 compared to patients with the IFNL4-ΔG/ΔG: rs4803217-T/T genotype [Table 2]). HCV RNA decline was also lower in individuals with the IFNL4-ΔG/TT: rs4803217-G/G genotype compared to those with the IFNL4-ΔG/TT: rs4803217-T/G genotype, although that difference was not statistically significant (p=0.08). A haplotype analysis of the decrease in HCV RNA levels at day 28 of treatment yielded similar results (Supplementary Table 2). As expected, the IFNL4-TT: rs4803217-G haplotype was associated with a much greater decrease in HCV RNA than the IFNL4-ΔG: rs4803217-T haplotype (coefficient, 0.73; p=0.0001). The haplotype that paired the unfavorable IFNL4-ΔG allele with the putatively favorable rs4803217-G allele was associated with a smaller decline in HCV RNA than IFNL4-ΔG: rs4803217-T (coefficient, −0.50; p=0.03).
Table 2.
Median decrease in HCV RNA levels (log10 international units (IU)/ml) in African American participants in the VIRAHEP-C study at day 28 of treatment with pegylated-interferon-α plus ribavirin, by compound genotype for IFNL4-ΔG/TT (rs368234815) and IFNL3 rs4803217.
| IFNL4-ΔG | rs4803217 | N | HCV RNA | P-value |
|---|---|---|---|---|
| ΔG/ΔG | T/T | 52 | 1.02 | |
| ΔG/ΔG | T/G | 15 | 0.62 | 0.03* |
| ΔG/TT | T/G | 77 | 1.58 | |
| ΔG/TT | G/G | 4 | 0.82 | 0.08** |
| TT/TT | G/G | 15 | 2.84 |
Compares patients with the IFNL4-ΔG/ΔG: rs4803217-T/G genotype to those with the IFNL4-ΔG/ΔG: rs4803217-T/T genotype
Compares patients with the IFNL4-ΔG/TT: rs4803217-G/G genotype to those with the IFNL4-ΔG/TT: rs4803217-T/G genotype
We also compared IFNL4-ΔG/TT and rs4803217 for associations with undetectable HCV RNA at several treatment time points in VIRAHEP-C and in HALT-C (Table 3). In VIRAHEP-C, the association was significantly stronger for IFNL4-ΔG/TT than rs4803217 at treatment week 24 (p=0.03, difference in AUROC; Supplementary Table 3), at end of treatment (p=0.02) and for SVR (p=0.03). A similar pattern was seen for treatment response in the HALT-C trial (Table 3), although only the finding at week 20, reached statistical significance (p=0.03, difference in AUROC; Supplementary Table 3). In haplotype analyses for the three treatment time points in VIRAHEP-C and in HALT-C (Supplementary Table 4), associations with response were weaker for IFNL4-ΔG: rs4803217-G than IFNL4-ΔG: rs4803217-T, but none of the associations were statistically significant. The corresponding compound genotypes analyses lacked statistical power (data not shown).
Table 3.
Comparison of IFNL4-ΔG/TT (rs368234815) and IFNL3 rs4803217 for genotype associations among African-American participants for association with response to treatment with pegylated interferon-alfa plus ribavirin, VIRAHEP-C and HALT-C Trial.
| Treatment Time Point
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Week 20/24* | End-of-Treatment | SVR | |||||||||
|
|
|||||||||||
| Variant | Genotype | N | Response Rate (%) | aOR | p-value | Response Rate (%) | aOR | p-value | Response Rate (%) | aOR | p-value |
| VIRAHEP-C (n = 170)** | |||||||||||
|
| |||||||||||
| IFNL4-ΔG/TT | ΔG/ΔG | 71 | 35.2 | Ref. | 32.4 | Ref. | 21.1 | Ref. | |||
| ΔG/TT | 83 | 55.4 | 2.81 | 0.004 | 44.6 | 1.88 | 0.07 | 31.3 | 2.01 | 0.08 | |
| TT/TT | 16 | 68.8 | 5.10 | 0.01 | 56.3 | 2.93 | 0.07 | 43.8 | 3.28 | 0.05 | |
| Additive | 2.48 | 0.001 | 1.77 | 0.03 | 1.87 | 0.03 | |||||
|
| |||||||||||
| rs4803217 | T/T | 56 | 39.3 | Ref. | 35.7 | Ref. | 25.0 | Ref. | |||
| T/G | 94 | 51.1 | 2.01 | 0.06 | 41.5 | 1.53 | 0.25 | 28.7 | 1.53 | 0.3 | |
| G/G | 20 | 60.0 | 3.02 | 0.05 | 50.0 | 2.12 | 0.17 | 35.0 | 2.01 | 0.24 | |
| Additive | 1.81 | 0.03 | 1.47 | 0.14 | 1.44 | 0.20 | |||||
|
| |||||||||||
| HALT-C Trial (n = 142) | |||||||||||
|
| |||||||||||
| IFNL4-ΔG/TT | ΔG/ΔG | 64 | 9.4 | Ref. | 7.8 | Ref. | 3.1 | Ref. | |||
| ΔG/TT | 70 | 18.6 | 3.38 | 0.047 | 14.3 | 2.70 | 0.13 | 8.6 | 4.41 | 0.13 | |
| TT/TT
|
8 | 50.0 | 22.74 | 0.002 | 37.5 | 13.58 | 0.01 | 25.0 | 24.24 | 0.02 | |
| Additive | 4.29 | 0.002 | 3.38 | 0.01 | 4.86 | 0.02 | |||||
|
| |||||||||||
| rs4803217 | T/T | 59 | 10.2 | Ref. | 8.5 | Ref. | 3.4 | ||||
| T/G | 66 | 18.2 | 3.12 | 0.07 | 13.6 | 2.56 | 0.16 | 7.6 | 4.32 | 0.14 | |
| G/G | 17 | 29.4 | 6.43 | 0.03 | 23.5 | 5.04 | 0.05 | 17.6 | 10.66 | 0.04 | |
| Additive | 2.59 | 0.01 | 2.27 | 0.04 | 3.21 | 0.03 | |||||
In VIRAHEP-C response is at 24 weeks after initiation of treatment; in HALT-C response is at 20 weeks after initiation of treatment
One subject is lost from the adjusted analyses for VIRAHEP-C due to missing data for fibrosis
SVR, sustained virological response; aOR, odds ratio adjusted for pre-treatment HCV RNA level and fibrosis stage
Spontaneous Clearance
Characteristics of participants in ALIVE, UHS and WIHS are presented in Table 4. Those enrolled in UHS were somewhat older (median, 46 years) compared to those enrolled in the other two studies (medians, ~40 years). By design, all WIHS participants were female (most participants in ALIVE and UHS were male) and all participants in ALIVE and UHS had a history of injection drug use (WIHS, 81.5%). The prevalence of HIV-1 infection ranged from 85.9% in WIHS, which selectively enrolled HIV-1-infected women, to 12.4% in UHS. The r2 values for LD between IFNL4-ΔG and rs4803217-T were: ALIVE, 0.75; UHS, 0.77; WIHS, 0.80.
Table 4.
Characteristics of African American participants in the studies of spontaneous HCV clearance: ALIVE, UHS, WIHS.
| ALIVE (n = 593) | UHS (n = 363) | WIHS (n = 519) | ||||
|---|---|---|---|---|---|---|
| Age, median (IQR) | 40.3 | 35.9 – 44.5 | 46 | 42.0 – 50.0 | 40.7 | 37.2 – 44.3 |
| Male (%) | 443 | 74.7 | 227 | 62.5 | 0 | 0.0 |
| Injection drug use [ever] (%) | 593 | 100.0 | 363 | 100.0 | 423 | 81.5 |
| HIV-1-infected (%) | 318 | 53.6 | 45 | 12.4 | 446 | 85.9 |
| IFNL4-ΔG allele frequency (%) | 63.9 | 58.3 | 61.8 | |||
| IFNL3 rs4803217-T allele frequency (%) | 60.6 | 56.2 | 57.2 | |||
| Linkage disequilibrium(r2) | 0.75 | 0.77 | 0.80 | |||
IQR, Interquartile range
Table 5 shows associations for IFNL4-ΔG/TT and rs4803217 with spontaneous clearance in the three individual studies and pooled results. In the combined analysis of 1,475 AA participants, comparison of IFNL4-TT/TT to IFNL4-ΔG/ΔG yielded an OR of 4.14, whereas the OR for the corresponding comparison of rs4803217 genotypes was 3.22. Overall, the AUROC was 0.63 for the three IFNL4-ΔG/TT genotypes and 0.61 for the rs4803217 genotypes (p=0.048, difference in AUROC). Results from each individual study were generally consistent with the pooled analysis (Table 5).
Table 5.
African American participants from ALIVE, UHS and WIHS with chronic or cleared HCV infection, genotype proportions for IFNL4-ΔG/TT (rs368234815) and IFNL3 rs4803217 variants, and corresponding Mantel-Haenszel (M-H) odds ratios, 95% confidence intervals (95% CI) and p-values for each association.
| Variant | IFNL4-ΔG/TT | rs4803217 | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Genotype | ΔG/ΔG | ΔG/TT | TT/TT | T/T | T/G | G/G | |
| ALIVE | Chronic (%) [N= 534] | 43.3 [231] | 45.9 [245] | 10.9 [58] | 40.6 [217] | 44.8 [239] | 14.6 [78] |
| Cleared (%) [N= 59] | 23.7 [14] | 39.0 [23] | 37.3 [22] | 23.7 [14] | 30.5 [18] | 45.8 [27] | |
| Odds ratio | Ref. | 1.55 | 6.26 | Ref. | 1.17 | 5.37 | |
| 95% CI | - | 0.78–3.08 | 3.02–12.98 | - | 0.57–2.45 | 2.68–10.76 | |
| p-value | 0.21 | 8.3×10−7 | 0.67 | 2.2×10−6 | |||
| UHS | Chronic (%) [N=276] | 36.6 [101] | 50.0 [138] | 13.4 [37] | 32.2 [89] | 52.5 [145] | 15.2 [42] |
| Cleared (%) [N=87] | 24.1 [21] | 47.1 [41] | 28.7 [25] | 25.3 [22] | 47.1 [41] | 27.6 [24] | |
| Odds ratio | Ref. | 1.43 | 3.25 | Ref. | 1.14 | 2.31 | |
| 95% CI | 0.80–2.57 | 1.63–6.49 | 0.64–2.05 | 1.17–4.59 | |||
| p-value | 0.23 | 8.4×10−4 | 0.65 | 0.02 | |||
| WIHS | Chronic (%) [N=437] | 41.2 [180] | 46.5 [203] | 12.4 [54] | 36.2 [158] | 46.5 [203] | 17.4 [76] |
| Cleared (%) [N=82] | 31.7 [26] | 32.9 [27] | 35.4 [29] | 26.8 [22] | 37.8 [31] | 35.4 [29] | |
| Odds ratio | Ref. | 0.92 | 3.72 | Ref. | 1.10 | 2.74 | |
| 95% CI | - | 0.52–1.65 | 2.02–6.85 | - | 0.61–1.97 | 1.48–5.08 | |
| p-value | 0.78 | 2.0×10−5 | 0.76 | 0.001 | |||
| TOTAL | Chronic (%) [N=1,247] | ||||||
| Cleared (%) [N=228] | |||||||
| M-H Odds ratio | Ref. | 1.25 | 4.14 | Ref. | 1.15 | 3.22 | |
| 95% CI | 0.88–1.77 | 2.81–6.11 | 0.80–1.64 | 2.20–4.72 | |||
| p-value | 0.21 | 7.2×10−13 | 0.45 | 2.0×10−9 | |||
In contrast to the haplotype analysis of treatment response, in which IFNL4-ΔG: rs4803217-G was less favorable than IFNL4-ΔG: rs4803217-T, we saw no difference between these haplotypes for spontaneous HCV clearance (OR, 0.97; 95% CI, 0.56–1.67; p=0.90; Supplementary Table 5). Similarly, the pooled compound genotype analysis for spontaneous viral clearance showed no difference between the IFNL4-ΔG/ΔG: rs4803217-G/G genotype and the IFNL4-ΔG/ΔG: rs4803217-T/G genotype (OR, 0.96; p=0.93; Supplementary Table 6). Consistent with our finding that rs4803217-G actually appeared to be unfavorable for treatment response in the presence of IFNL4-ΔG, individuals with the IFNL4-ΔG/TT: rs4803217-T/T genotype were more likely to have cleared HCV than those with IFNL4-ΔG/TT: rs4803217-T/G genotype (OR, 2.87), however, that finding was of borderline statistical significance (p=0.06) and there was no difference in clearance between those with the IFNL4-ΔG/TT: rs4803217-G/G and IFNL4-ΔG/TT: rs4803217-T/G genotypes (OR, 0.92; p=0.85).
European American Participants
We also examined associations for IFNL4-ΔG/TT and rs4803217 genotypes among European American participants, however, strong LD between the variants limited the statistical power for those comparisons. Among the 183 European American patients enrolled in VIRAHEP-C, LD was nearly complete (r2 = 0.99) and, therefore, genotype comparisons were not informative (data not shown). For HALT-C (n=743), r2 between IFNL4-ΔG and rs4803217-T was 0.89; IFNL4-ΔG/TT was somewhat more strongly associated with treatment response than rs4803217 (Supplementary Tables 7 and 8). Similarly, with regard to spontaneous HCV clearance among the European American participants, r2 values were 0.90 and 0.89 in UHS and WIHS, respectively, and the association with spontaneous clearance was somewhat stronger for IFNL4-ΔG/TT than rs4803217 (Supplementary Tables 9 and 10).
Discussion
In vitro studies demonstrated that the rs4803217-G allele within the 3′UTR of IFNL3 decreased HCV-induced degradation of IFNL3 mRNA and, on that basis, rs4803217 was proposed as a causal variant that improves HCV clearance [12]. In the present study, we compared rs4803217 to IFNL4-ΔG/TT, a functional variant that controls generation of the IFN-λ4 protein. Among participants of African ancestry, associations with HCV clearance (response to treatment of chronic hepatitis C with pegylated IFN-α plus ribavirin and spontaneous HCV clearance) were consistently stronger for IFNL4-ΔG/TT than rs4803217. LD between these variants was high in subjects of European ancestry and we observed no statistically significant differences between IFNL4-ΔG/TT and rs4803217 in that population. Based on information from the 1000 Genomes project, LD between IFNL4-ΔG/TT and rs4803217 was complete in Asians (r2=1.0), which indicates these variants will provide equal predictive value in patients of Asian ancestry. These results provide additional evidence that the IFNL4-ΔG/TT variant is the best genetic predictor of HCV clearance.
This study provides no indication that the rs4803217-G allele improves HCV clearance. In fact, it offers some evidence that rs4803217-G may have a negative effect on HCV clearance, since the haplotype consisting of IFNL4-ΔG and rs4803217-G was associated with even poorer HCV RNA decline at day 28 of treatment than the IFNL4-ΔG:rs4803217-T haplotype. We could not confirm an unfavorable association for the IFNL4-ΔG:rs4803217-G haplotype in the analysis of spontaneous clearance, however, our study provided no evidence that rs4803217-G increases HCV clearance.
To our knowledge, this is the first study comparing IFNL4-ΔG/TT and rs4803217 for HCV clearance. Two smaller studies compared rs4803217 to rs12979860, a variant that is in strong LD with rs4803217 and which is associated with HCV clearance less strongly than IFNL4-ΔG/TT [4, 10]. Among 197 HCV/HIV-1 co-infected patients from Barcelona, associations for response to treatment with pegylated IFN-α plus ribavirin were comparable for rs4803217 and rs12979860 genotypes [27]. Those findings are consistent with the data from EA individuals in our study. An analysis of spontaneous HCV clearance in an Egyptian population (n=261) suggested a slightly weaker genotype association for rs4803217 than rs12979860 [28].
The available evidence strongly supports the IFNL4-ΔG/TT polymorphism and generation of the IFN-λ4 protein as the explanation for observed associations of IFN-λ region variants with HCV clearance. As demonstrated here and previously [4, 10, 11, 18], IFNL4-ΔG/TT is the best variant for predicting HCV clearance across a range of populations. Furthermore, this polymorphism is highly functional – the IFNL4-ΔG allele generates the IFN-λ4 protein, while the frameshift caused by the IFNL4-TT allele abrogates that protein [4]. Furthermore, structural variation in IFN-λ4 appears to affect HCV clearance. A non-synonymous IFNL4 polymorphism (rs117648444-T/C, Pro70Ser) that exists only on a haplotype with the IFNL4-ΔG allele generates an alternative form of IFN-λ4 protein [4]. Individuals with the IFN-λ4-S70 variant have rates of treatment response and spontaneous HCV clearance that are intermediate between those for patients with IFN-λ4-P70 and those who generate neither form of IFN-λ4 (i.e., those with the IFNL4-TT/TT genotype) [29, 30], which is consistent with the observation that the IFN-λ4-S70 protein is less biologically active than IFN-λ4-P70 [29]. In addition, the IFNL4-TT allele explains the signature of positive selection that has been observed in the IFN-λ chromosomal region (and evidence of selection is stronger for IFNL4-ΔG/TT than rs4803217)[31, 32]. Selection at this locus resulted in IFNL4-TT being the major form in Asia, while IFNL4-ΔG is the major allele in Africa [4]; IFNL4-ΔG/TT is among the top 0.5% of polymorphisms with regard to genome-wide differentiation between African and East Asian populations [31]. Selection for IFNL4-TT may not have been driven by HCV infection [33], however, the data are consistent with a favorable role for IFNL4-TT in viral infection.
This study was not designed to elucidate how the IFN-λ4 protein might impair HCV clearance and that functional mechanism remains unclear [33]. Interferons are generally considered antiviral cytokines and there is evidence that IFN-λ4 has anti-viral activity [4, 34], yet genetic association studies link generation of IFN-λ4 to impaired HCV clearance. Although IFN-λ4 most closely resembles IFN-λ3, these proteins share only 29% amino acid identity [4]. Like other members of the IFN-λ family, IFN-λ4 signals through the IFN-λ receptor complex, induces expression of interferon-stimulated genes via the JAK-STAT signaling pathway and demonstrates anti-viral activity [4, 34], yet, in contrast to IFN-λ1-3, IFN-λ4 is only weakly secreted [4, 34]. Additional work is needed to resolve the apparent paradox of IFN-λ4 and HCV clearance.
Regimens based on direct-acting antiviral agents (DAA) are becoming the standard of care for HCV infection, although the cost of these treatments may limit their wide-scale implementation [35]. In some trials, very high response rates have made it difficult to determine if genotype for IFNL4 variants can impacts response to DAA regimens [36], however, results from several other studies have shown that this the case [36–39]. The fixed-dose combination of ledipasvir and sofosbuvir has recently become available for treatment of HCV infection. In the phase III ION-3 trial [40], 423 previously untreated HCV genotype 1-infected patients without cirrhosis had outcome data after receiving ledipasvir/sofosbuvir for 8 or 12 weeks. The published subgroup analysis found no significant differences in SVR rates by rs12979860 genotype, however, enrollees with missing outcome data constituted a large proportion of those counted as treatment failures [40]. In an ‘efficacy analysis’ of subgroup differences in treatment based on the published data [39], we found that SVR rates varied significantly by rs12979860 genotype and exceeded 98% in individuals with the rs12979860-CC genotype (which is very strongly linked with the IFNL4-TT/TT genotype) who were treated for 8 weeks. Genotype for rs12979860 remained informative even after HCV RNA was considered. [41] Current treatment guidelines either do not endorse ledipasvir/sofosbuvir regimen for <12 weeks [42] or do so only with caution. [43] However, the results from the efficacy analysis suggest that treatment-naive patients with HCV genotype 1 and the IFNL4-TT/TT genotype are highly likely to respond to 8 weeks of ledipasvir/sofosbuvir. Thus, host genotype may be useful for personalizing treatment with DAA regimens in order to decrease treatment costs. For that purpose, IFNL4-ΔG/TT, possibly in combination with the IFNL4 Pro70Ser polymorphism, would be the most predictive variant across a range of populations.
A major strength of this study is its inclusion of AA participants in whom LD between IFNL4-ΔG/TT and rs4803217 is much weaker than in Europeans or Asians. Among AA subjects, we saw statistically significant differences between these variants in genotype associations for HCV clearance, whereas there were no significant differences between the genotypes in EA individuals. A limitation of the study is the low statistical power for some analyses. Results for comparisons of IFNL4-ΔG/TT and rs4803217 were consistent, but results for HALT-C failed to achieve statistical significance. Furthermore, although the study provides strong evidence against the hypothesis that rs4803217-G increases HCV clearance [12], our efforts to determine if there was an association in the opposite direction (i.e., that rs4803217-G is associated with impaired viral clearance) were limited by low statistical power.
In conclusion, this study provides additional evidence that the IFNL4-ΔG allele, which generates the IFN-λ4 protein, is the primary variant for impaired clearance of HCV. In the clinical setting, genotype for IFNL4-ΔG/TT will provide the best genetic prediction for HCV clearance. Future studies should examine how IFN-λ4 interferes with viral clearance and whether IFNL4-ΔG/TT plays a role in other viral infections.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the US National Institutes of Health (National Cancer Institute, Division of Cancer Epidemiology and Genetics), as well as the following grants: DA R01 013324 (D.L.T.); ALIVE cohort (G.D.K), U01-DA-036297, R01-DA-04334 and R01-DA-12568, and US National Institutes of Health grants R15 HL117199, U01 DK 065201, U54 DK 083909 (H.L.B.); R01-DA09532, R01-DA12109, R01-DA13245 and R01-DA16159 (B.R.E.); National Cancer Institute contracts N02-CP-91027 and N01-CO-12400 (B.R.E.); Substance Abuse and Mental Health Services Administration grant H79-TI12103 (B.R.E.). The VIRAHEP-C and HALT-C studies were conducted, respectively, by the VIRAHEP-C and HALT-C Investigators and supported by the National Institute of Diabetes and Digestive and Kidney Diseases. The data and samples from the VIRAHEP-C and HALT-C studies reported here were supplied by the National Institute of Diabetes and Digestive and Kidney Diseases Central Repositories. This manuscript was not prepared in collaboration with the VIRAHEP-C study group or the HALT-C study group and does not necessarily reflect the opinions or views of the VIRAHEP-C Trial and HALT-C Trial, the National Institute of Diabetes and Digestive and Kidney Diseases Central Repositories or the National Institute of Diabetes and Digestive and Kidney Diseases. Data in this manuscript were collected by the Women’s Interagency HIV Study (WIHS) Collaborative Study Group with centers (principal investigators) located at: New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); Washington DC Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); Los Angeles County/Southern California Consortium (Alexandra Levine); Chicago Consortium (Mardge Cohen); and Data Analysis Center (Stephen Gange). The WIHS is funded by the National Institute of Allergy and Infectious Diseases (U01-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The WIHS is co-funded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI grant UL1 RR024131). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the US government.
We thank Mr. David Check for graphical assistance and Mr. Eric Boyd for assistance with data analysis.
Footnotes
Conflict of Interest: TRO’B and LP-O are inventors on patent applications filed by the National Cancer Institute for the IFNL4-ΔG/TT (rs368234815) genotype-based test and the IFN-λ4 protein. For other authors there are none to declare.
Presented, in part, at the 21th International Symposium on Hepatitis C Virus and Related Viruses, Banff, Canada, September 9, 2014
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 3.Fox BA, Sheppard PO, O’Hara PJ. The Role of Genomic Data in the Discovery, Annotation and Evolutionary Interpretation of the Interferon-Lambda Family. PLoS ONE. 2009;4:e4933. doi: 10.1371/journal.pone.0004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 6.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-[alpha] and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 8.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B Is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 10.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. The Journal of Experimental Medicine. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco S, Aparicio E, Parera M, Clotet B, Tural C, Martinez MA. IFNL4 ss469415590 variant is a better predictor than rs12979860 of pegylated interferon-alpha/ribavirin therapy failure in hepatitis C virus/HIV-1 coinfected patients. AIDS. 2014;28:133–136. doi: 10.1097/QAD.0000000000000052. [DOI] [PubMed] [Google Scholar]
- 12.McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15:72–79. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas LF, Sætrom P. Single Nucleotide Polymorphisms Can Create Alternative Polyadenylation Signals and Affect Gene Expression through Loss of MicroRNA-Regulation. PLoS Comput Biol. 2012;8:e1002621. doi: 10.1371/journal.pcbi.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–477. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Di Bisceglie AM, Shiffman ML, Everson GT, Lindsay KL, Everhart JE, Wright EC, et al. Prolonged therapy of advanced chronic hepatitis C with low-dose peginterferon. The New England journal of medicine. 2008;359:2429–2441. doi: 10.1056/NEJMoa0707615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee WM, Dienstag JL, Lindsay KL, Lok AS, Bonkovsky HL, Shiffman ML, et al. Evolution of the HALT-C Trial: pegylated interferon as maintenance therapy for chronic hepatitis C in previous interferon nonresponders. Control Clin Trials. 2004;25:472–492. doi: 10.1016/j.cct.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Aka PV, Kuniholm MH, Pfeiffer RM, Wang AS, Tang W, Chen S, et al. Association of the IFNL4-ΔG allele with impaired spontaneous clearance of hepatitis C virus. Journal of Infectious Diseases. 2014;209:350–354. doi: 10.1093/infdis/jit433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlahov D, Anthony JC, Munoz A, Margolick J, Nelson KE, Celentano DD, et al. The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr. 1991;109:75–100. [PubMed] [Google Scholar]
- 20.Watters JK, Bluthenthal RN, Kral AH. HIV seroprevalence in injection drug users. JAMA. 1995;273:1178. [PubMed] [Google Scholar]
- 21.Tseng FC, O’Brien TR, Zhang M, Kral AH, Ortiz-Conde BA, Lorvick J, et al. Seroprevalence of hepatitis C virus and hepatitis B virus among San Francisco injection drug users, 1998 to 2000. Hepatology. 2007;46:666–671. doi: 10.1002/hep.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacon MC, von Wyl V, Alden C, Sharp G, Robison E, Hessol N, et al. The Women’s Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clinical and diagnostic laboratory immunology. 2005;12:1013–1019. doi: 10.1128/CDLI.12.9.1013-1019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissner EG, Wu D, Osinusi A, Bon D, Virtaneva K, Sturdevant D, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. The Journal of Clinical Investigation. 2014;124:0–0. doi: 10.1172/JCI75938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoofnagle JH, Wahed AS, Brown RS, Howell CD, Bellw SH. Early Changes in Hepatitis C Virus (HCV) Levels in Response to Peginterferon and Ribavirin Treatment in Patients with Chronic HCV Genotype 1 Infection. Journal of Infectious Diseases. 2009;199:1112–1120. doi: 10.1086/597384. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford: Oxford University Press; 2003. [Google Scholar]
- 27.de Castellarnau M, Aparicio E, Parera M, Franco S, Tural C, Clotet B, et al. Deciphering the Interleukin 28B Variants That Better Predict Response to Pegylated Interferon-α and Ribavirin Therapy in HCV/HIV-1 Coinfected Patients. PLoS ONE. 2012;7:e31016. doi: 10.1371/journal.pone.0031016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedergnana V, Abdel-Hamid M, Guergnon J, Mohsen A, Le Fouler L, Theodorou I, et al. Analysis of IL28B Variants in an Egyptian Population Defines the 20 Kilobases Minimal Region Involved in Spontaneous Clearance of Hepatitis C Virus. PLoS ONE. 2012;7:e38578. doi: 10.1371/journal.pone.0038578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terczyńska-Dyla E, Bibert S, Duong FHT, Krol I, Jørgensen S, Collinet E, et al. 75: A variant of IFNλ4 displaying decreased activity is associated with better hepatitis C virus clearance and reduced ISG expression. Cytokine. 2014;70:45–46. [Google Scholar]
- 30.Galmozzi E, Aghemo A. Nonsynonymous variant Pro70Ser (rs117648444) in IFNL4 gene identifies carriers of the rs368234815 ΔG allele with higher HCV RNA decline during the first 4 weeks of pegylated interferon and ribavirin therapy in HCV-1 patients. Journal of Clinical Virology. 2014;59:274–275. doi: 10.1016/j.jcv.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Key FM, Peter B, Dennis MY, Huerta-Sánchez E, Tang W, Prokunina-Olsson L, et al. Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 (IFNL4) PLoS Genet. 2014;10:e1004681. doi: 10.1371/journal.pgen.1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, et al. Evolutionary genetic dissection of human interferons. The Journal of Experimental Medicine. 2011;208:2747–2759. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-λ4: the paradoxical new member of the interferon lambda family. Journal of Interferon & Cytokine Research. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamming OJ, Terczyńska-Dyla E, Vieyres G, Dijkman R, Jørgensen SE, Akhtar H, et al. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. The EMBO Journal. 2013;32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoofnagle JH, Sherker AH. Therapy for Hepatitis C — The Costs of Success. New England Journal of Medicine. 2014;370:1552–1553. doi: 10.1056/NEJMe1401508. [DOI] [PubMed] [Google Scholar]
- 36.Stättermayer AF, Scherzer T, Beinhardt S, Rutter K, Hofer H, Ferenci P. Review article: genetic factors that modify the outcome of viral hepatitis. Alimentary Pharmacology & Therapeutics. 2014 doi: 10.1111/apt.12717. n/an/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meissner EG, Bon D, Prokunina-Olsson L, Tang W, Masur H, O’Brien TR, et al. IFNL4-ΔG genotype is associated with slower viral clearance in hepatitis C, genotype-1 patients treated with sofosbuvir and ribavirin. Journal of Infectious Diseases. 2014;209:1700–1704. doi: 10.1093/infdis/jit827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeuzem S, Soriano V, Asselah T, Bronowicki J-P, Lohse AW, Mullhaupt B, et al. Faldaprevir and Deleobuvir for HCV Genotype 1 Infection. New England Journal of Medicine. 2013;369:630–639. doi: 10.1056/NEJMoa1213557. [DOI] [PubMed] [Google Scholar]
- 39.O’Brien TR, Lang Kuhs KA, Pfeiffer RM. Subgroup differences in response to 8 weeks of ledipasvir/sofosbuvir for chronic hepatitis C. Open Forum Infectious Diseases. 2014 doi: 10.1093/ofid/ofu110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, et al. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. New England Journal of Medicine. 2014;370:1879–1888. doi: 10.1056/NEJMoa1402355. [DOI] [PubMed] [Google Scholar]
- 41.O’Brien TR, Pfeiffer RM. Reply: Subgroup Differences in Response to 8 Weeks of Ledipasvir/Sofosbuvir for Chronic Hepatitis C. Open Forum Infectious Diseases. 2015:2. doi: 10.1093/ofid/ofv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panel AIHG. Hepatitis C Guidance: AASLD-IDSA Recommendations for Testing, Managing, and Treating Adults Infected with Hepatitis C Virus. Hepatology. 2015 doi: 10.1002/hep.27950. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 43.EASL Recommendations on Treatment of Hepatitis C. Journal of Hepatology. 2015;63:199–236. doi: 10.1016/j.jhep.2015.03.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.