Abstract
Small RNA programmed Argonautes are sophisticated cellular effector platforms known to be involved in a diverse array of functions ranging from mRNA cleavage, translational inhibition, DNA elimination, epigenetic silencing, alternative splicing and even gene activation. First observed in human cells, small RNA-induced gene activation, also known as RNAa, involves the targeted recruitment of Argonaute proteins to specific promoter sequences followed by induction of stable epigenetic changes which promote transcription. The existence of RNAa remains contentious due to its elusive mechanism. A string of recent studies in C. elegans provides unequivocal evidence for RNAa's fundamental role in sculpting the epigenetic landscape and maintaining active transcription of endogenous genes and supports the presence of a functionally sophisticated network of small RNA-Argonaute pathways consisting of opposite yet complementary “yin and yang” regulatory elements. In this review, we summarize key findings from recent studies of endogenous RNAa in C. elegans, with an emphasis on the Argonaute protein CSR-1.
Keywords: Argonaute, epigenetic memory, gene expression, RNAa, RNAe
Abbreviations
- RNAa
RNA activation
- RNAe
RNA-induced epigenetic silencing
- piRNA
Piwi-interacting RNA
- RNAi
RNA interference
- WAGO
worm-specific AGO
- RISC
RNA induced silencing complex
- RDRP
RNA-dependent RNA polymerase
- LCE
lin-4 complementary element
- miRNAa
miRNA induced RNAa
- TSS
transcription start site
The gene regulatory machinery composed of small RNAs and their Argonaute protein partners have typically been synonymous with repression of gene expression given their established roles in mRNA degradation, translational suppression and heterochromatin formation.1-3 Remarkably, small RNAs also revealed an activating side4 in 2006 with the observance of stable and specific gene induction in mammalian cells following transfection with promoter-targeted synthetic small RNAs, a phenomenon known as RNA activation (RNAa).5 The underlying mechanism of RNAa remained elusive however, leading to concerns that the observed gene induction was caused by secondary effects of canonical RNA interference (RNAi). New findings from a series of recent studies in C. elegans reveal once again an activating side of the small RNA-Argonaute pathways6-11 and establish RNAa as a regulatory mechanism of endogenous gene expression.
RISC as an Epigenetic Activator
Argonautes are a family of highly conserved proteins which are classified in most organisms into 2 major clades: the AGO and PIWI clades. In C. elegans, there is an expanded family of worm-specific AGOs (WAGOs). Short RNAs depend on different Argonautes for function by forming RNA/Argonaute complexes. This requirement distinguishes Argonaute-dependent small RNA pathways from those mediated by long noncoding regulatory RNAs which function independently of Argonaute proteins. The complexes formed by short RNAs and AGOs have traditionally been referred to as the RNA-induced silencing complex (RISC) given their role as mediators of canonical RNAi.
RNA-loaded RISC typically suppresses gene expression by catalyzing the degradation of complementary mRNAs. However, when targeted to selected promoter sequences RISC can induce localized activating epigenetic marks which promote transcription (RNAa).5 RNAa was first observed in human cells following exposure to synthetic short dsRNAs designed to target gene promoters with perfect sequence complementarity.5,12 Like RNAi, RNAa is also an AGO-dependent process (predominantly AGO2 and AGO1) but displays distinct in vitro kinetics including delayed onset and persistent activity across several cell divisions. These features contrast sharply with those of RNAi and suggest that epigenetic mechanisms are involved. New findings in C. elegans now support the idea that RNAa is likely an evolutionarily conserved mechanism that utilizes such small RNA-Argonaute machinery.
The Missing “Yang” in the Small RNA-Argonaute Regulatory Network of C. elegans
The complexity of the small RNA world uncovered in C. elegans surpasses that found in all other organisms thus far. In addition to miRNAs, there is an even larger small RNA system in C. elegans which consists of Piwi-interacting RNAs (piRNAs) and their secondary small RNAs (endo-siRNAs)13 which are generated by piRNA targeting of mRNAs. One sub-type of these secondary RNA species is the 22G-RNAs so called because of their 22 nt length and a preferred 5′G residue.14 The C. elegans genome encodes for about 30,000 piRNAs which are primarily expressed in the germline and then processed into 21 nt RNAs through multiple steps (Fig. 1). These 21 nt mature piRNAs typically possess a 5′U and are thus termed 21U-RNA.15 Once bound by the Piwi protein PRG-1, 21U-RNA guides PRG-1 to mRNA sequences through imperfect base-pairing where PRG-1 further recruits EGO-1, an RNA-dependent RNA polymerase (RdRP) to amplify the silencing signal by synthesizing 22G-RNAs which are antisense to the mRNA templates (Fig. 1). This process requires DRH-3 (dicer-related helicase 3) and several additional factors.14 These 22G-RNAs can then be loaded by a WAGO (WAGO1/9/10) protein to form a 22G-RNA/WAGO complex which silences foreign sequences such as transposons, pseudogenes and aberrant transcripts either at the post-transcriptional or epigenetic level. The latter nuclear silencing mechanism has been termed RNA-induced epigenetic silencing (RNAe)14,16 which initiates transcriptional gene silencing that is then maintained across generations through the recruitment of other epigenetic factors such as H3K9 methyltransferase and heterochromatin protein 1 (HP1).17 RNAe thus serves as a surveillance mechanism to maintain the integrity of the germline genome.14
Figure 1.
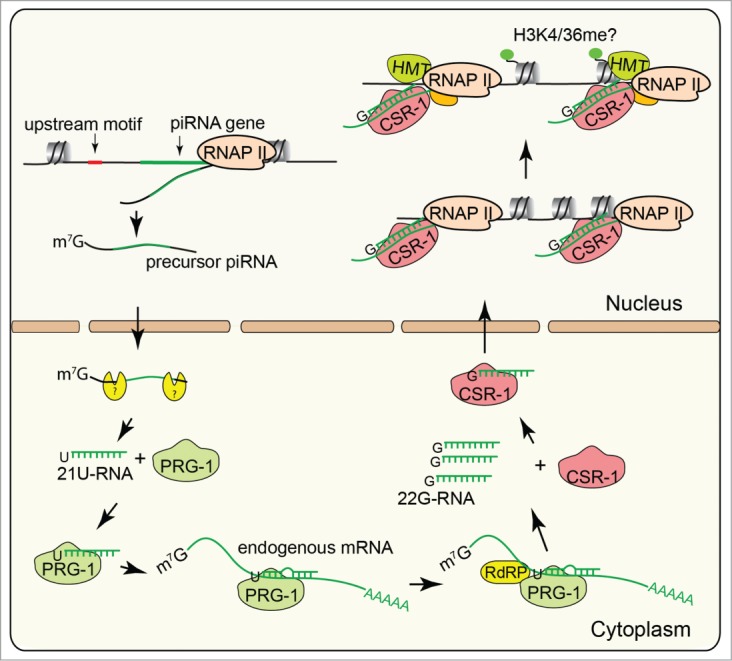
RNAa in C. elegans. piRNA genes encoded in the genome as clusters are individually transcribed in a process that requires a conserved upstream motif. The resulted piRNA precursors are then processed into mature 21U-RNAs in the cytoplasm through several undefined steps. The 21U-RNAs form a complex with PRG-1 protein and guide the complex to non-perfectly matched targets on endogenous mRNAs where PRG-1 further recruits RdRP to generate secondary 22G-RNAs. After being loaded by CSR-1, the 22G-RNAs then target CSR-1 to cognate nascent transcripts to interact with RNAP II and local chromatin where CSR-1 can further recruit histone modifying enzymes such as histone methyltransferases (HMTs) to promote epigenetic activation (H3K4me or H3K36me3).
Given the large number of piRNAs that do not require strict base-pairing for target binding, any mRNA could theoretically be targeted by piRNAs for silencing.18 Indeed, 50% of 22G-RNAs found in C. elegans target endogenous mRNAs.14 This raises the question: how are ‘self’ RNAs distinguished from ‘non-self’ RNAs and protected from RNAe-mediated silencing? Interestingly, 22G-RNAs antisense to “self” RNAs interact with a different Argonaute protein, CSR-1 (chromosome segregation and RNAi deficiency 1), to form the 22G-RNA/CSR-1 complex which then binds nascent transcripts in a sequence-specific manner to associate with local chromatin19 (Fig. 1). Although CSR-1 has been demonstrated to possess slicer activity in vitro,20 there is no evidence to suggest that in vivo it cleaves the transcripts which it binds.19 It is therefore plausible that the 22G-RNA/CSR-1 pathway is responsible for marking and protecting “self” RNAs from being silenced by the 22G-RNA/WAGO pathway, as has been suggested by Beth et al.7 Previous studies have already shown that perturbation of this pathway causes defects in chromosome segregation, histone pre-mRNA processing and sterility.6,19,21 New studies7-9,22 have now proved that the 22G-RNA/CSR-1 system can do more than just passively defending “self” RNAs; it can actively promote their expression via epigenetic mechanisms as discussed below (Fig. 1).
RNAa in C. elegans
In the work by Wedeles et al.,9 the authors first observed that CSR-1 interacts with RNA polymerase II (RNAP II) and is recruited to its target gene loci via nascent transcripts. To further assess the functions of CSR-1, they then employed an in vivo RNA tethering assay in which a gfp transcript containing phage lambda box b RNA hairpins (gfp::boxb) is expressed under the control of a germline promoter, and a fusion CSR-1 protein containing a phage lambda N anti-termination protein fragment (CSR-1::λN) is also expressed in the same cells. In this system, CSR-1 protein can be specifically tethered to the gfp transcript due to the ability of the Lambda N peptide to recognize box b hairpin RNA.23 The authors found that in germline, tethering CSR-1 to gfp can stably protect the gfp transgene from piRNA-induced silencing. Moreover, tethering CSR-1 to an already silenced gfp can reactivate its expression in trans. Even more interesting is that in the continued presence (a few generations) of gfp::boxb and CSR-1::λN in the same strain, cdk-1:gfp, an allele silenced by RNAe (referred to as RNAe allele), could be activated although no CSR-1 is tethered to its mRNA. As suggested by the authors, it is possible that small RNAs derived from the gfp::boxb allele target CSR-1 to and activate the expression of cdk-1;fgp. Together, these analyses revealed that the CSR-1 pathway functions to maintain active germline gene expression by opposing the piRNA surveillance mechanism.
Using a different genetic approach, Seth et al.7 first examined the role of CSR-1 in the process of epigenetically transmitted and RNA-induced transactivation (also named as RNAa). A previous study by the same group has found that when a worm strain carrying an RNAe transgene, gfp::cdk-1, which is silenced via the RNAe pathway as described above, is crossed with a strain carrying oma-1::gfp transgene which is resistant to RNAe, the latter transgene could activate the expression the RNAe transgene gfp::cdk-1 in the F1 progeny.17 In the new study, the authors found that oma-1::gfp RNAa allele failed to activate the gfp::ckd-1 RNAe allele when CSR-1 is inactivated by either RNAi or genetic mutation, indicating that CSR-1 is required for RNAa.7
To find out whether RNAa could stably activate an RNAe transgene, the authors set up a series of crossing experiments to establish double-transgenic lines expressing 2 transgenes with either RNAa or RNAe activity, then separated the 2 transgenes by outcrossing to a wild-type strain. They found that conversion of a silent RNAe allele to a permissive state requires multiple generations of continued exposure to RNAa. For example, the expression of gfp::cdk-1, the RNAe transgene, was activated in the presence of oma-1::gfp, the RNAa transgene, but disappeared when the RNAa transgene was crossed away in the next generation. When they cultured the double-transgenic strain oma-1::gfp;gfp::cdk-1 for over 10 and 30 generations before separating the 2 transgenes, they observed respectively one full generation and 10 generations of RNAe gene expression in the absence of RNAa allele. These data suggest that RNAa can counteract the effect of RNAe and be epigenetically transferred across generations, albeit with diminishing efficacy.
In another study also from the Mello group, Conine et al. further examined the functional role of RNAa mediated by the CSR-1/22G-RNA pathway in spermatogenesis in C. elegans. The group has previously shown that ALG3/4 Argonautes (paralogous AGO-clade members) can engage 26G-RNAs antisense to mRNAs expressed during spermatogenesis.24 In the new study, they showed that the primary function of the ALG3/4 pathway is to promote the transcription of spermiogenesis genes; as evidenced by increased H3K4me2 at their promoters, a chromatin modification associated with active transcription.8 Both ALG3/4 double and CSR-1 single mutants resulted in partial temperature-dependent sterility, suggesting these 2 systems function in the same pathway. Indeed, csr-1, alg-3 and alg-4 triple mutant males showed defects in spermatogenesis leading to complete sterility at 25°C. They further showed that the CSR-1/22G-RNA pathway shares most target genes with the ALG3/4 26G-RNA pathway, and promotes expression of ALG-3/4 target mRNAs. Despite their similar target genes and function, ALG-3/4 proteins are expressed only during spermatogenesis, whereas CSR-1 protein is expressed beyond spermatogenesis and remains abundant in sperms. They then asked whether the 2 pathways might cooperate to propagate epigenetic memory of male-specific gene expression from one generation to the next via RNAa. By repeated backcrossing heterozygous hermaphrodites to homozygous mutant males, they found that the crossing led to a progressive loss of fertility which can be rescued by mating to wild-type males. These data suggest that the CSR-1/22G-RNA mediated RNAa pathway is capable of transmitting a transgenerational epigenetic signal.
RNAa mediated by the 22G-RNA/CSR-1 pathway operates not only in germ cells but also in somatic cells as shown in the study by Cecere et al.10 By profiling nascent transcripts using global run-on sequencing (GRO-seq) in wild-type worms and a strain hypomorphic for csr-1, the authors found that 22G-RNA target genes were significantly enriched among genes whose transcription was decreased in the csr-1 hypomorphic strain compared to wild-type worms. These genes are represented by genes highly expressed at the particular developmental stages of worms (later L3 and early L4) used in the analysis, including genes expressed ubiquitously and germline-specifically. The downregulation of these genes appears to be 22G-RNA dependent since they are also downregulated in a drh-3 mutant strain which depleted 22G-RNAs. Interestingly, transcripts antisense to those downregulated genes were globally increased in both csr-1 and drh-3 mutants, and are genes that do not express or express low at later L3 and early L4 stages. They further showed that CSR-1 is enriched in euchromatic regions and interacts with RNAP II in an RNA-dependent manner. These findings suggest that 22G-RNA targets CSR-1 to nascent sense transcripts where CSR-1 interacts with RNAP II to stabilize its transcription while restricting RNAP II transcription in the opposite direction. Together, this study not only corroborates the observations from the Claycomb and Mello groups but also suggests that the CSR-1/22G-RNA pathway functions to globally reinforce the transcription of actively transcribed genes expressed at certain developmental stages. How CSR-1 promotes RNAP II transcription of its target genes remains to be elucidated.
miRNAa in C. elegans
Naturally occurring miRNAs may also have the ability to induce RNAa of genes whose promoters contain imperfect binding sites for the miRNAs,25 a phenomenon which can be referred to as miRNA-induced RNAa (miRNAa). Until very recently, miRNAa has only been observed in cultured mouse and human cells.25-27
Turner et al. now showed that lin-4, the first miRNA discovered in C. elegans and important for the timing of stem cell fate decisions,28 can activate its own transcription by binding to its own promoter.11 By analyzing the promoter sequence of lin-4, the authors found a putative lin-4 complementary element (LCE) conserved in the lin-4 promoters of several nematode species. Deletion of the LCE in the lin-4-GFP reporter transgenic animals resulted in decreased transcriptional activity of the lin-4 promoter suggesting that the LCE is a functional cis-regulatory element potentially trans-regulated by lin-4. To test this idea, the authors went on to cross the GFP reporter line into a strain with mutated lin-4 that had no production of endogenous wild-type lin-4. Compared to the wild-type strain, the mutant animals had 3 times less GFP transcriptional activity with decreased RNAP II association with lin-4 promoter. Furthermore, overexpression of lin-4 in the lin-4-GFP reporter animals resulted in increased lin-4 transcriptional activity compared to control animals. These results point to a model in which mature lin-4 functions as a trans-acting factor and binds to the conserved LCE in its promoter to enhance promoter transcription by recruiting RNAP II and possibly additional protein factors. Therefore, the work by Turner et al. provided the first example of miRNAa that functions in vivo and to regulate transcripts other than mRNA. Future experiments are needed to identify the Argonaute proteins and chromatin factors involved in this process and the prevalence of miRNAa in C. elegans. Similar positive feedback loop regulation has been demonstrated with another well-known miRNA, let-7,29 although a non-transcriptional mechanism that leads to enhanced maturation of let-7 seems to be responsible for the regulation.29
Potential RNAa Regulatory Networks in Human Cells
Accumulating evidence supports an intricate interplay between the small RNA machinery and chromatin in mammalian cells.30 In analogy to C. elegans RNAe and RNAa, this additional layer of epigenetic regulation may constitute an epigenetic memory transmitted by small RNAs across cell divisions and may play important roles in cell physiology and disease. For example, cancer cells may exploit RNAa to sustain active expression of genes necessary for growth and survival. A recent study of genome-wide AGO-chromatin interaction carried out in human cancer cells by our group suggests that AGO1 interacts with RNAP II and binds to thousands of promoters of actively transcribed genes.31 Similar to the effects of CSR-1 binding on chromatin in C. elegans22 the impact of AGO1-promoter interactions is largely positive on gene expression.31 The interactions appear to be small RNA/miRNA dependent since they can be affected by perturbing miRNA biogenesis31 Apart from miRNAs, small RNAs that are equivalent to the C. elegans 22G-RNAs and capable of mediating AGO1 binding have yet to be identified in mammalian cells. We speculated that nascent promoter transcripts may directly recruit AGOs to chromatin,30 given the spatial overlapping of AGO1 binding sites on chromosomes31 and the origin of previously identified promoter transcripts known as TSSa-RNAs (transcription start site-associated RNAs)32 or tiRNAs (transcription-initiation RNAs).33 In this regard, a very recent study34 has identified a new class of non-canonical miRNAs which are originated from the transcription start site (TSS) of hundreds of protein coding genes as single stranded transcripts and form hairpin structures which is processed by Dicer into 21–24 nt miRNAs termed TSS-miRNAs. Importantly, TSS-miRNAs are bound by AGO2,34 suggesting that they could potentially be involved in RNAa by mediating AGO-chromatin interactions.
Conclusions
Initially, the finding that the RNAi machinery is also involved in gene upregulation appeared to be counterintuitive. A cascade of recent C. elegans studies corroborates this concept by showing concrete evidence for the existence of RNAa in worm germline and somatic cells. Collectively, these studies demonstrate a positive role for an endogenous Argonaute pathway in gene regulation. While the detailed molecular mechanisms for how 22G-RNA/CSR-1 complex activates target genes await to be solved, it will be very interesting to examine whether similar piRNA-initiated RNAa pathway exists in mammalian cells and whether other small RNAs such as TSS-miRNAs could target Argonautes to chromatin loci to maintain active expression of gene pathways.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a grant from the National Institutes of Health (1R01GM090293–0109 to Li L-C).
References
- 1. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID: 9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 2. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843-54; PMID: 8252621; http://dx.doi.org/ 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 3. Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833-7; PMID: 12193640; http://dx.doi.org/ 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 4. Garber K. Genetics. Small RNAs reveal an activating side. Science 2006; 314:741-2; PMID: 17082428; http://dx.doi.org/ 10.1126/science.314.5800.741a [DOI] [PubMed] [Google Scholar]
- 5. Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 2006; 103:17337-42; PMID: 17085592; http://dx.doi.org/ 10.1073/pnas.0607015103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avgousti DC, Palani S, Sherman Y, Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J 2012; 31:3821-32; PMID: 22863779; http://dx.doi.org/ 10.1038/emboj.2012.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr., Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 2013; 27:656-63; PMID: 24360782; http://dx.doi.org/ 10.1016/j.devcel.2013.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr, Yates JR, 3rd, Mello CC. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 2013; 155:1532-44; PMID: 24360276; http://dx.doi.org/ 10.1016/j.cell.2013.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell 2013; 27:664-71; PMID: 24360783; http://dx.doi.org/ 10.1016/j.devcel.2013.11.016 [DOI] [PubMed] [Google Scholar]
- 10. Cecere G, Hoersch S, O'Keeffe S, Sachidanandam R, Grishok A. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat Struct Mol Biol 2014; PMID: 24681887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner MJ, Jiao AL, Slack FJ. Autoregulation of lin-4 microRNA transcription by RNA activation (RNAa) in C. elegans. Cell Cycle 2014; 13:772-81; PMID: 24398561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 2007; 3:166-73; PMID: 17259978; http://dx.doi.org/ 10.1038/nchembio860 [DOI] [PubMed] [Google Scholar]
- 13. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 2003; 13:807-18; PMID: 12747828; http://dx.doi.org/ 10.1016/S0960-9822(03)00287-2 [DOI] [PubMed] [Google Scholar]
- 14. Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. . Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 2009; 36:231-44; PMID: 19800275; http://dx.doi.org/ 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. . PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 2008; 31:67-78; PMID: 18571452; http://dx.doi.org/ 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J 2012; 31:3422-30; PMID: 22850670; http://dx.doi.org/ 10.1038/emboj.2012.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012; 150:65-77; PMID: 22738726; http://dx.doi.org/ 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr., Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 2012; 150:78-87; PMID: 22738724; http://dx.doi.org/ 10.1016/j.cell.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. . The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 2009; 139:123-34; PMID: 19804758; http://dx.doi.org/ 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 2007; 26:5007-19; PMID: 18007599; http://dx.doi.org/ 10.1038/sj.emboj.7601910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006; 127:747-57; PMID: 17110334; http://dx.doi.org/ 10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- 22. Cecere G, Grishok A. A nuclear perspective on RNAi pathways in metazoans. Biochim Biophys Acta 2014; 1839:223-33; PMID: 24361586; http://dx.doi.org/ 10.1016/j.bbagrm.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 2004; 10:1518-25; PMID: 15337849; http://dx.doi.org/ 10.1261/rna.7131604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2010; 107:3588-93; PMID: 20133686; http://dx.doi.org/ 10.1073/pnas.0911685107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 2008; 105:1608-13; PMID: 18227514; http://dx.doi.org/ 10.1073/pnas.0707594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 2012; 40:1695-707; PMID: 22053081; http://dx.doi.org/ 10.1093/nar/gkr934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 2013; 41:10086-109; PMID: 23999091; http://dx.doi.org/ 10.1093/nar/gkt777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997; 88:637-46; PMID: 9054503; http://dx.doi.org/ 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- 29. Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 2012; 486:541-4; PMID: 22722835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li LC. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 2013; 9:45-52; PMID: 24149777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet 2013; 9:e1003821; PMID: 24086155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science 2008; 322:1849-51; PMID: 19056940; http://dx.doi.org/ 10.1126/science.1162253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. . Tiny RNAs associated with transcription start sites in animals. Nat Genet 2009; 41:572-8; PMID: 19377478; http://dx.doi.org/ 10.1038/ng.312 [DOI] [PubMed] [Google Scholar]
- 34. Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 2014; 156:920-34; PMID: 24581493; http://dx.doi.org/ 10.1016/j.cell.2014.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]


