Abstract
Replication of chromosomes is central to heredity. To become available for replication machinery, DNA invariably needs to dissociate from chromatin proteins. Yet, chromatin landscape must be promptly re-established during or soon after replication. Although this process underlies the epigenetic inheritance, little is known about its molecular mechanisms. This mini-review is focused on Drosophila melanogaster SUppressor of UnderReplication (SUUR) protein, which is involved both in replication and chromatin maintenance in polytene tissues. Existing data suggest that it is involved in the regulation of chromatin renewal during replication. According to this model, SUUR protein moves along the chromosomes together with the replication complex. When the replication fork enters the repressed, H3K27me3- or H3K9me3-enriched, chromatin, SUUR-containing complex slows down the replisome until these histone modifications are properly placed on the newly-synthesized DNA strands. Suggested model provides an insight into the mechanism of epigenetic information inheritance. This hypothesis could be tested by further analysis of the interplay between local enrichment of repressive histone modifications and the replication fork progression rate.
Keywords: chromatin, Drosophila, histone modifications, replication, SuUR
Abbreviations
- H3K27me3
Histone H3 tri-methylated at Lysine 27
- H3K9me3
Histone H3 tri-methylated at Lysine 9
- HP1
Heterochromatin Protein 1
- SU(VAR)3-9
Suppressor of variegation 3-9
- SUUR
Suppressor of Underreplication
- ORC
Origin Recognition Complex
- PCNA
Proliferating Cell Nuclear Antigen
- PRE
Polycomb Response Element.
Replication of the genome is essential for genetic inheritance in all organisms. Prior to division the cell needs to precisely duplicate genomic DNA and ensure its integrity so that identical DNA copies are transmitted to the daughter cells after mitosis. Consistently, DNA replication abnormalities are known to result in severe genetic disorders and cancer.
Genomic DNA in the nucleus is packaged into massive DNA/protein complexes referred to as chromosomes. Thus the replicating cell actually has to produce 2 copies of each chromosome in all their complexity, not just the DNA itself. The first level of chromosomal packaging is achieved when the DNA molecule is wound around the histone octamers. Histones, in addition to their structural function, can regulate the associated DNA sequences - certain post-translational modifications of specific amino acid residues in histone molecules contribute to gene expression and replication control.1 For example, in multicellular organisms, tri-methylation of histone H3 at lysine 27 (H3K27me3) is placed by Polycomb-group protein complexes on developmentally regulated genes that need to be repressed.2 H3K9 methylation marks repressive chromatin predominantly in the pericentric regions and is associated with HP1/SU(VAR)3-9 complex. While the core replication complex, which performs DNA synthesis, is thoroughly studied, little is known about the molecular mechanisms underlying proper inheritance of epigenetic information during replication.3,4 Yet, inability of the cell to maintain or correctly read the epigenetic information may result in cancer.5,6
Maintenance of epigenetic information throughout development requires re-establishment of histone modification marks on chromosomes after each replication cycle. During replication, the amount of histones doubles: half of them originate from the template chromosome and the other half are synthesized de novo. Original, modified histones are displaced from the template chromosome and seem to be equally distributed between the replicated copies.3,7 New histones need to be modified according to the local epigenetic state when incorporated into the chromosome. In mammals, some modifications are cast immediately after the replication fork by chromatin factors including chromatin remodelers and histone chaperones; other modifications are introduced later during chromatin maturation, when the replication fork has already passed.8
In higher eukaryotes, genomic regions replicate asynchronously - transcriptionally active regions usually replicate earlier in the S-phase of cell cycle than do repressed heterochromatic regions.9 Primarily, this replication schedule is defined by the pattern of active replication initiation sites (origins). Early origins fire at the beginning of the S-phase while others wake up later.10 This means that regions replicating the latest are those located between the 2 distant late origins. Secondly, the rate of replication fork movement itself can affect the local timing of chromosomal replication.11 Replication fork speed changes depending on the features of local chromatin environment (e.g. epigenetic state and chromosomal packaging), but the nature of this process remains elusive.
Temporal shift in replication between active and repressed genomic regions implies that chromatin state must affect the replication time. However, very little is known about the chromatin factors that can regulate replication time. One of these factors was found in Drosophila.12,13
In Drosophila melanogaster polytene chromosomes, late-replicating heterochromatic regions often fail to complete replication and remain under-replicated (Fig. 1).14 Mutation in the Suppressor of Underreplication (SuUR) gene results in full polytenization of under-replicated regions in chromosome arms and partial polytenization of pericentric regions.13 This means that under-replication is an active process, which specifically requires functional SUUR protein. SUUR is associated with heterochromatin and acts as a weak position effect modifier.15 In other aspects SuUR mutants are viable, fertile and show no overt changes in gene expression and Origin Recognition Complex (ORC) binding.11,13 It is worth mentioning that SuUR is a fast-evolving gene that was found only in Diptera.16
Figure 1.
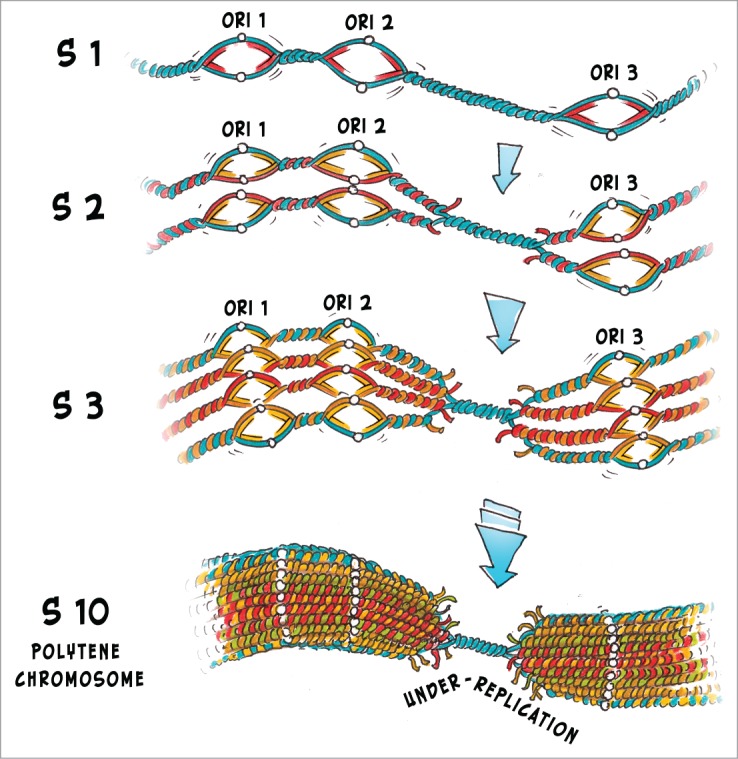
Replication and under-replication in polytene chromosome. Endocycles (cell cycles lacking mitosis) in salivary gland cells start early in Drosophila melanogaster embryogenesis. Replication begins at multiple sites - origins of replication (ORI 1, ORI 2 and ORI 3 on the scheme). By the end of the first S-phase (S1) the amount of DNA doubles along the most of the chromosome, apart from the heterochromatic regions where replication forks fail to collide before the next endocycle begins. This local under-replication occurs for 2 main reasons: (i) larger distance between ORI2 and ORI 3, because in heterochromatin origins are depleted; and (ii) slower replication progression through the repressed regions due to chromatin compaction. During the next S-phases (S2 and S3) the number of DNA strands increases and under-replication becomes dramatic. After 10 endocycles (S10) the polytene chromosome consisting of about 1000 DNA strands is formed. The under-replicated heterochromatic regions appear as specific constrictions on the polytene chromosomes.
Despite the low evolutionary conservation, recent studies revealed several important features of SuUR. First, SUUR protein interacts with the replication machinery: it co-purifies with PCNA (Proliferating Cell Nuclear Antigene), the sliding clamp component of the replisome17; there are some indications that CDC45, the component of eukaryotic replicative helicase, could also be a part of the common complex with SUUR.18 Second, SUUR protein has a seemingly paradoxical function - it actively slows down replication fork progression.11 Third, genomic regions with restored polytenization in SuUR mutants, demonstrate the depletion of the H3K27me3 histone mark levels compared with wild-type.11 Here we present a hypothesis that explains these and other observations by connecting SUUR protein function to replication-coupled chromatin renewal.
According to this model, SUUR protein is involved in the process of chromatin re-assembly during chromosome replication (Fig. 2): it moves together with the replication complex and controls incorporation of proper repressive histone modifications into the nascent chromatin. During replication of the repressed chromatin (enriched with H3K27 or H3K9 methylated histones), SUUR pauses or slows down the replication complex thereby allowing some yet unknown mechanism to faithfully reproduce histone marks on the newly replicated DNA strands. This control mechanism would decrease the replication kinetics in heterochromatin and could explain the phenomenon of under-replication (Fig. 2A).
Figure 2.
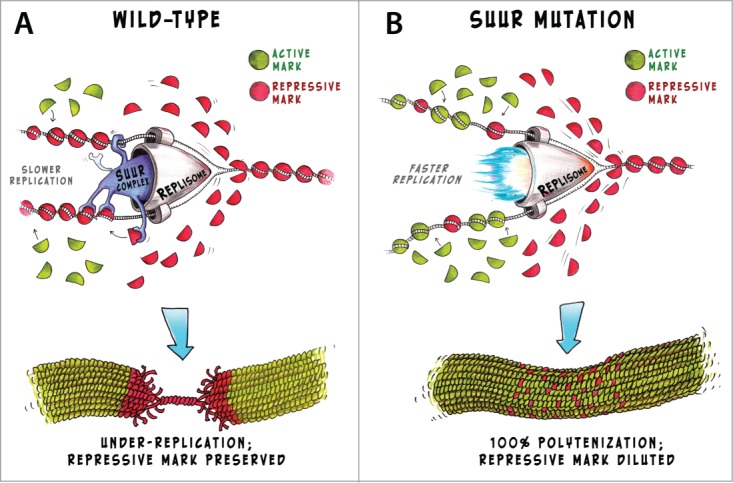
Suggested role of SUUR in replication and heterochromatin maintenance. In wild-type, SUUR protein moves through the chromatin together with the replisome (A). We suggest that SUUR controls the re-assembly of repressive chromatin on the daughter DNA strands thus preserving the chromatin state after the replication. This process is time-consuming and replication complex moves through the large heterochromatin blocks slowly. In polytene chromosomes this eventually results in under-replication. In SUUR mutants this control is turned off (B) and replisome progresses faster leading to complete polytenization of heterochromatin. However the repressive histone modifications are gradually diluted in SuUR mutants, as newly synthesized DNA strands incorporate histone marks randomly.
The absence of the functional SUUR protein would turn off the renewal mechanism controlling H3K27me3 and H3K9me3 incorporation into the nascent chromatin. This could account for 2 major effects observed in SuUR mutants. First, with the renewal process switched off, replication forks progress through the repressed chromatin faster. Ultimately this results in complete polytenization of these regions in polytene chromosomes.13 Second, the decrease in H3K27me3 histones observed in SuUR mutants11 is, in fact, a result of gradual dilution of H3K27me3 mark in repressed chromatin regions (Fig. 2B). These effects are the most prominent manifestations of SUUR protein loss. In contrast, the excess of SUUR protein in the cell would result in the enhanced control of chromatin renewal and could lead to even slower rate of replication. Indeed, in flies bearing extra copies of normal SuUR gene under-replication is more severe than in wild-type animals.12
Suggested model could explain other reported effects of SuUR mutation. SuUR mutation is considered to be a weak suppressor of position effect variegation.15 Position effect is observed in genetic systems where reporter gene is translocated into heterochromatic environment and becomes partially repressed. One classic example involves the white gene, normally responsible for the red eye phenotype, placed into pericentric heterochromatin of the X chromosome by chromosomal inversion. Flies bearing this inversion display mosaic red/white eye pigmentation due to random repression of white gene.19 SuUR mutant background increases the proportion of red eye facets in these flies.15 Again, this could be a result of the dilution of repressive histone marks in SuUR mutants leading to a more permissive chromatin composition and facilitating the white gene transcription. Notably, SUUR protein involvement in repression appears indirect, as tethering of SUUR to an artificial chromosomal site does not lead to inactivation of adjacent reporter gene.20
One cannot rule out the possibility that SUUR protein could participate in the histone modification process, but available experimental data do not support this suggestion. If this were the case, we should assume that SUUR participates in both H3K9 and H3K27 methylation pathways, 2 very distinct processes that are performed by different protein complexes. Also, the experimental data revealed that SuUR mutants do not show decrease in H3K9me2 histone mark and its pattern is unaffected in the chromocenter.21 Thus, replication-mediated action of SUUR is a more plausible scenario.
Yet another study in Drosophila revealed that activity of Polycomb Group proteins (largely responsible for H3K27 methylation) peaks in early S-phase, when the regions enriched with H3K27me3 are not yet replicated. As suggested, this leads to the accumulation of histones bearing this modification that are later being used as a supply during the replication of the repressed regions at the end of the S-phase.22 According to our model, SUUR protein may function as one of the factors required for this process.
The suggested mechanism of repressed chromatin renewal has important parallels with the analogous processes in mammals. For example, an extensive study in human cells revealed that in contrast to other histone modifications, H3K9me3 and H3K27me3 are inherited via the incorporation of the histones from the template chromatin (parental histones) to the nascent chromatin.8 The mechanism that distinguishes H3K9me3 and H3K27me3 from the rest of the histone modifications during replication remains unknown.
It is known that the abundance of histone modifications at the particular genomic location oscillates during the cell cycle. As a rule, histone modifications are first diluted after the replication and are subsequently restored more or less rapidly, prior to the next cell cycle (for review see3). Yet, a slightly different scenario was observed for H3K27me3 histone mark in Drosophila cells. In early S-phase, Polycomb Group proteins bound to PREs exhibit enhanced activity producing extra amount of H3K27me3 histones. This excess of repressive marks would then be used in the late S-phase, when repressed regions are replicated. This mechanism keeps the level of H3K27me3 above a certain threshold and thus precludes the unauthorized gene activation.22 These studies imply that H3K27 methylation pattern is rather maintained via local over-methylation of parental histones followed by their dilution, than by rapid “on-the-run” placement of methylation marks into the nascent chromatin.
It is presently unclear what mechanism regulates incorporation of parental histones with their marks to the newly assembled chromatin. If the suggested role of SUUR in chromatin replication is confirmed, it would give a clue to the regulation of this process, at least for H3K27me3 and likely for H3K9me3.
Fast evolution of SuUR gene impedes straightforward identification of proteins with the homologous function in other organisms by protein similarity searches.16 However, it is unlikely that chromatin-dependent regulation of replication fork progression is a phenomenon restricted to Diptera. In mammals and other eukaryotes, SUUR function may well be performed by a different protein. Given that other players in this process could be more conserved, identification of SUUR protein interactors at the replication fork could help unravel the puzzling nature of chromatin-renewal mechanism.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Andrey Gortchakov for English proofs. Figures were prepared by Olga Posukh.
Funding
This work was supported by Russian Science Foundation grant 14-14-00934, Russian Foundation for Basic Research grants 15-04-05301, 12-04-33080, 12-04-31777, 12-04-31128, 12-04-00160 and 15-04-02264.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41-5; PMID:10638745; http://dx.doi.org/ 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 2.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 2007; 8:9-22; PMID:17173055; http://dx.doi.org/ 10.1038/nrg1981 [DOI] [PubMed] [Google Scholar]
- 3.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 2012; 13:153-67; PMID:22358331; http://dx.doi.org/ 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- 4.Budhavarapu VN, Chavez M, Tyler JK. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 2013; 6:32; PMID:24225278; http://dx.doi.org/ 10.1186/1756-8935-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra M, Bohlander SK. Disturbing the histone code in leukemia: translocations and mutations affecting histone methyl transferases. Cancer Genet 2015; 208(5):192-205; PMID:25592767; http://dx.doi.org/19234478 10.1016/j.cancergen.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 6.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther 2015; 150:1-22; PMID:25578037; http://dx.doi.org/19234478 10.1016/j.pharmthera.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 7.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 2009; 10:192-206; PMID:19234478; http://dx.doi.org/ 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- 8.Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 2014; 16:281-93; PMID:24561620; http://dx.doi.org/ 10.1038/ncb2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol 2002; 14:377-83; PMID:12067662; http://dx.doi.org/ 10.1016/S0955-0674(02)00326-5 [DOI] [PubMed] [Google Scholar]
- 10.Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect-part II. Curr Opin Genet Dev 2009; 19:142-9; PMID:19345088; http://dx.doi.org/ 10.1016/j.gde.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sher N, Bell GW, Li S, Nordman J, Eng T, Eaton ML, Macalpine DM, Orr-Weaver TL. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res 2012; 22:64-75; PMID:22090375; http://dx.doi.org/ 10.1101/gr.126003.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Volkova EI, Alekseyenko AA, Andreyeva EN, Makarevich GF, Boldyreva LV, Nanayev RA, et al.. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 2003; 111:377-98; PMID:12644953; http://dx.doi.org/ 10.1007/s00412-002-0218-0 [DOI] [PubMed] [Google Scholar]
- 13.Belyaeva ES, Zhimulev IF, Volkova EI, Alekseyenko AA, Moshkin YM, Koryakov DE. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc Natl Acad Sci U S A 1998; 95:7532-7; PMID:9636184; http://dx.doi.org/ 10.1073/pnas.95.13.7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhimulev IF, Belyaeva ES, Makunin IV, Pirrotta V, Semeshin VF, Alekseyenko AA, Belyakin SN, Volkova EI, Koryakov DE, Andreyeva EN, et al.. Intercalary heterochromatin in Drosophila melanogaster polytene chromosomes and the problem of genetic silencing. Genetica 2003; 117:259-70; PMID:12723705; http://dx.doi.org/ 10.1023/A:1022912716376 [DOI] [PubMed] [Google Scholar]
- 15.Belyaeva ES, Boldyreva LV, Volkova EI, Nanayev RA, Alekseyenko AA, Zhimulev IF. Effect of the Suppressor of Underreplication (SuUR) gene on position-effect variegation silencing in Drosophila melanogaster. Genetics 2003; 165:1209-20; PMID:14668376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurlova AA, Makunin IV, Kolesnikova TD, Posukh OV, Belyaeva ES, Zhimulev IF. Conservation of domain structure in a fast-evolving heterochromatic SUUR protein in drosophilids. Genetics 2009; 183:119-29; PMID:19596903; http://dx.doi.org/ 10.1534/genetics.109.104844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolesnikova TD, Posukh OV, Andreyeva EN, Bebyakina DS, Ivankin AV, Zhimulev IF. Drosophila SUUR protein associates with PCNA and binds chromatin in a cell cycle-dependent manner. Chromosoma 2013; PMID:23149855 [DOI] [PubMed] [Google Scholar]
- 18.Nordman JT, Kozhevnikova EN, Verrijzer CP, Pindyurin AV, Andreyeva EN, Shloma VV, Zhimulev IF, Orr-Weaver TL. DNA copy-number control through inhibition of replication fork progression. Cell Rep 2014; 9:841-9; PMID:25437540; http://dx.doi.org/ 10.1016/j.celrep.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 2013; 5:a017780; PMID:23906716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokholkova GV, Koryakov DE, Pindyurin AV, Kozhevnikova EN, Belyakin SN, Andreyenkov OV, Belyaeva ES, Zhimulev IF. Tethering of SUUR and HP1 proteins results in delayed replication of euchromatic regions in Drosophila melanogaster polytene chromosomes. Chromosoma 2015; 124(2):209-20; PMID:25398563; http://dx.doi.org/21340745 10.1007/s00412-014-0491-8 [DOI] [PubMed] [Google Scholar]
- 21.Koryakov DE, Walther M, Ebert A, Lein S, Zhimulev IF, Reuter G. The SUUR protein is involved in binding of SU(VAR)3-9 and methylation of H3K9 and H3K27 in chromosomes of Drosophila melanogaster. Chromosome Res 2011; 19:235-49; PMID:21340745; http://dx.doi.org/ 10.1007/s10577-011-9193-8 [DOI] [PubMed] [Google Scholar]
- 22.Lanzuolo C, Lo Sardo F, Diamantini A, Orlando V. PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLoS Genet 2011; 7:e1002370; PMID:22072989; http://dx.doi.org/ 10.1371/journal.pgen.1002370 [DOI] [PMC free article] [PubMed] [Google Scholar]


