Abstract
In plants, Potato spindle tuber viroid (PSTVd) replication triggers post-transcriptional gene silencing (PTGS) and RNA-directed DNA methylation (RdDM) of homologous RNA and DNA sequences, respectively. PTGS predominantly occurs in the cytoplasm, but nuclear PTGS has been also reported. In this study, we investigated whether the nuclear replicating PSTVd is able to trigger nuclear PTGS. Transgenic tobacco plants carrying cytoplasmic and nuclear PTGS sensor constructs were PSTVd-infected resulting in the generation of abundant PSTVd-derived small interfering RNAs (vd-siRNAs). Northern blot analysis revealed that, in contrast to the cytoplasmic sensor, the nuclear sensor transcript was not targeted for RNA degradation. Bisulfite sequencing analysis showed that the nuclear PTGS sensor transgene was efficiently targeted for RdDM. Our data suggest that PSTVd fails to trigger nuclear PTGS, and that RdDM and nuclear PTGS are not necessarily coupled.
Keywords: viroid, RNA silencing, DNA methylation, small RNA, tobacco
Introduction
Viroids are non-encapsidated, non-coding single-stranded (ss), 250–400 nucleotide (nt)-long circular RNA molecules, causing devastating diseases to several plant species.1 They are classified into the Pospiviroidae and Avsunviroidae families, whose members replicate in the nucleus and the chloroplast, respectively.2,3 Isolates of the Potato spindle tuber viroid (PSTVd), the Pospviroidae's type species, typically have a 359-nt circular ssRNA genome. Upon entering the plant cell, PSTVd is transported into the nucleus by the nuclear localization signal (NLS)-and bromodomain-containing VIROID RNA-BINDING PROTEIN1 (VIRP1).4–6 In the nucleus, PSTVd is replicated via the asymmetric rolling circle pathway7 by DNA-DEPENDENT RNA POLYMERASE II (POLII).8 The involvement of additional POLs and/or RNA-DIRECTED RNA POLYMERASES (RDRs) in Pospiviroidae replication can not be excluded.9 The circular monomeric ssRNA(+) is transcribed into linear multimeric ssRNA(−) that serves as the template for the production of linear multimeric ssRNA(+) that traffics into the nucleolus to be finally processed into mature unit-length circular RNAs,10,11 The mature viroid moves to neighboring cells through plasmodesmata and to distant parts of the plant through the phloem.12,13
In their hosts, viroids elicit the RNA silencing machinery (Fig. 1).14,15 The mature viroid and/or its double stranded RNA (dsRNA) replication intermediates are processed by the Dicer-like enzymes, DCL1/DCL4, DCL2 and DCL3, into 21, 22 and 24 nt viroid-derived siRNAs (vd-siRNAs).16 Similar to plant-derived siRNAs and miRNAs, vd-siRNAs are phosphorylated at their 5' and methylated at their 3' end.17 Interestingly, similar concentrations of both polarities of vd-siRNAs have been detected.9,15-20 Predominantly 21-nt vd-siRNAs are loaded onto ARGONAUTE 1 (AGO1) and target homologous transcripts for degradation, in a process termed post-transcriptional gene silencing (PTGS).21-24 Yet, similar to viral satellites, and presumably due to their secondary structure, mature viroids seem to be resistant to cleavage in some15 but not all cases.25-27 In addition to PTGS, replicating viroids trigger RNA-directed DNA methylation of cognate DNA sequences.28-32 According to the current model, AGO4-loaded 24-nt siRNAs recognize the target DNA by hybridizing with nascent POLII/POLV transcripts triggering the recruitment of de novo methyltransferases to impose cytosine methylation in every sequence context.33
Figure 1.
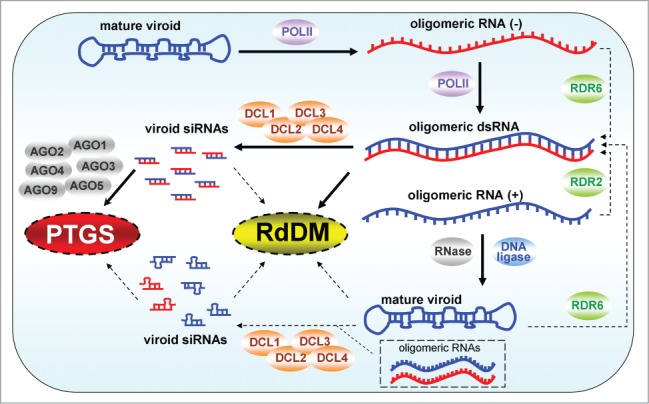
PSTVd and RNA silencing. Upon inoculation, mature PSTVd molecules enter the plant cell nucleus. During the asymmetric replication cycle, POLII transcribes oligomeric RNA of (−)-orientation from the circular viroid RNA. In the nucleoplasm, this RNA is transcribed into oligomeric (+) RNA, again by POLII. Subsequently to its transport into the nucleolus, a type III-like RNase and a DNA ligase are recruited to assemble the mature viroid molecule. Oligomeric (+) and (−) replication intermediates may form dsRNA molecules. Alternatively, dsRNA molecules may be produced by RDR6-mediated transcription of mature viroid RNA and/or of oligomeric (−) in the nucleoplasm. In addition, it cannot be excluded that in the nucleolus, dsRNA is synthesized by RDR2 transcription of oligomeric (+) RNA. All four DCLs produce vd-siRNAs. However, it is not clear if only dsRNAs or, due to the high capacity of viroid RNA to form intermolecular secondary structures, also single stranded viroid RNA, (mature viroid or oligomeric intermediates) is DCL-processed. Both polarities of vd-siRNAs could be loaded onto several AGOs and initiate PTGS in the cytoplasm and RdDM in the nucleus.
The molecular basis of symptom development in viroid-infected plants is still elusive.34 It is reasonable to assume that host genes sharing sequence homology with viroids are transcriptionally or post-transcriptionally silenced. In this view: (1) expression of a hairpin RNA (hpRNA) transgene encoding a nearly full length sequence of PSTVd in tomato induced PSTVd disease symptom-like phenotypes.24 (2) In Nicotiana species, an artificial miRNA corresponding to the PSTVd virulence modulating region directed PTGS of a soluble inorganic pyrophospahatase gene and led to the development of abnormal phenotypes.35 (3) In PSTVd-infected tomato, the accumulation of several endogenous miRNAs is suppressed, suggesting vd-siRNA-induced downregulation of the respective pre-miRNAs.36 (4) Of those genes that are downregulated early in PSTVd infection in tomato, 2 are involved in gibberellin and jasmonic acid biosynthesis and contain binding sites for vd-siRNAs in their respective ORFs.37 (5) In cucumber plants, Hop stunt viroid (HSVd) infection induced dynamic changes in the DNA methylation of rRNA genes.38 Whether viroids lead to PTGS and RdDM of other host genes is still elusive. It is also not clear whether viroid-induced PTGS is solely a cytoplasmic process39,40 or whether it may also take place in the nucleus.41,42 The nuclear co-localization of plant DCLs with the replication and maturation of PSTVd, would render the replicating and abundantly siRNA-producing viroid a promising candidate to trigger nuclear PTGS. In this study, however, we showed that PSTVd infection indeed triggered RdDM and cytoplasmic PTGS but failed to trigger nuclear PTGS in Nicotiana tabacum.
Results
Generation of GFPint98 and its introduction into tobacco plants
In order to generate a nuclear PTGS sensor construct, we reasoned that the target transcript should be retained in the nucleus. After splicing of an intron-containing pre-mRNA, the polyadenylated mRNA is exported to the cytoplasm while the intronic lariat is retained in the nucleus.43 Thus, a chimeric intron was produced, where a 98 bp PSTVd subfragment (serving as target site for vd-siRNAs) was inserted in plus (int98(+)) and in minus (int98(−)) orientation into the Solanum lycopersicum RNA-DIRECTED RNA POLYMERASE 1 (SeRDR1) intron 3, respectively.44,45 We inserted the 98 bp subfragment in both orientations to exclude that any insensitivity to nuclear PTGS would be due to the either plus-only or minus-only polarity of the vd-siRNAs (Fig. 1). Subsequently, int98(+) and int98(−) were inserted into the GFP5 cDNA46 producing the nuclear PTGS sensor constructs GFPint98(+) and GFPint98(−), respectively (Fig. 2). GFPint89(+)/(−) constructs were cloned into the expression cassette of the pPCV702SM binary vector and the corresponding plasmids were introduced into Agrobacterium tumefaciens, that was then used to transform Nicotiana tabacum (Nt) plants. In order to show that in PSTVd-infected tobacco plants, vd-siRNAs with the potential to target the 98-nt PSTVd sequence are accumulating, the transgenic tobacco Nt-GFP98(+) line, harbouring the full length GFP5 cDNA fused to the 98 bp PSTVd subfragment22 (Fig. 2) was employed. The polyadenylated GFP98(+) mRNA is transported into the cytoplasm, and upon PSTVd infection, it is silenced by vd-siRNAs.22 Thus, transcripts of the GFP98 transgene constructs could be considered as an ideal cytoplasmic PTGS sensor.
Figure 2.
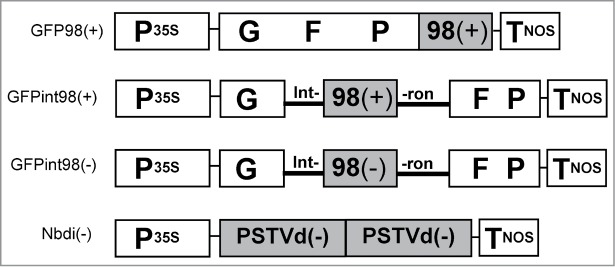
Schematic representation of transgenes. P35S: Cauliflower mosaic virus 35S promoter; TNOS: nopaline synthase (NOS) terminator; GFP: green fluorescent protein; intron: S. lycopersicum intron 3; 98: 98 bp PSTVd subfragment: PSTVd: full-length PSTVd cDNA; (+): plus orientation; (−): minus orientation.
The 98 bp PSTVd subfragment did not abolish intron functionality
Plant introns do not have absolute requirements for branch sites and polypyrimidine tracts. An A/U-rich base composition, a minimal size of 70-nt and the presence of 5´-GU and 3´-AG splice sites are sufficient for intron functionality.47–49 Thus, chimeric introns should be functional in plants. However, secondary structures within introns could potentially inhibit pre-mRNA splicing in dicotyledons.50 Viroids and long viroid subfragments, as for example the 98-nt PSTVd RNA, form extensive secondary structures. Nevertheless, Northern blot analysis showed that spliced GFP mRNA (797-nt) was highly abundant indicating that the 98-nt PSTVd RNA did not significantly interfere with the splicing process. Some unspliced GFPint98(+)/(−) pre-mRNA (1365-nt) was also detected at low levels (Fig. 4), reflecting most probably the steady state levels investigated by Northern blot analysis.
Figure 4.
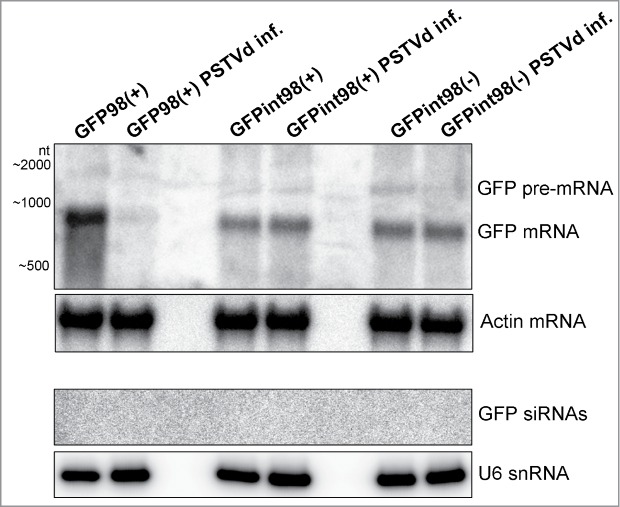
Northern blot analysis for pre-mRNA processing and degradation. Total RNA (7 µg) was extracted from PSTVd-infected and non-infected plants and subjected to long and small RNA analysis using the full-length GFP cDNA as a hybridization probe. Hybridization took place at 64°C and the blot was exposed for 24 hrs to phosporimager screen. Actin and U6 hybridization of the stripped membranes served as loading controls.
Viroid infection failed to trigger nuclear PTGS of GFPint98 transcripts
Mechanical inoculation-mediated viroid infection does not exclude the presence of non-infected cells in systemic leaves.51 In order to achieve PSTVd infection in presumably every cell, ‘Nt-Nbdi(−) plants were genetically crossed with Nt-GFP98(+) and Nt-GFPint98(+)/(−). Nt-Nbdi(−) plants carry an infectious, dimeric full-length PSTVd cDNA transgene in (−)-orientation (Fig. 2).52 As a mock control, the Nt-GFP98(+) and Nt-GFPint98(+)/(−) plants were crossed with Nt-WT. For each crossing, the presence of transgenes was validated by PCR (data not shown) and viroid infection was validated by Northern blot analysis. In infected plants, the mature 359-nt PSTVd RNA as well as the 21–24-nt vd-siRNAs were detected (Fig. 3).
Figure 3.
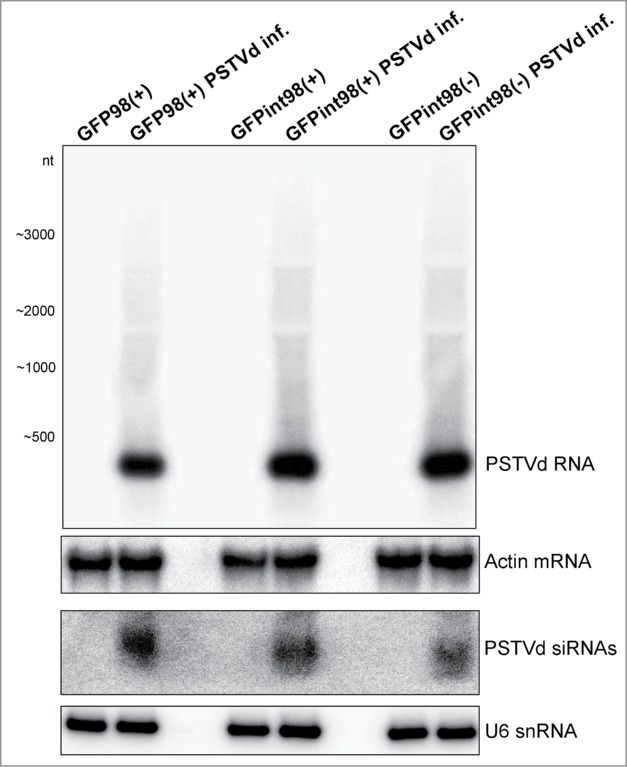
Northern blot analysis for validation of PSTVd replication. Total RNA (2 µg) was extracted from PSTVd-infected and non-infected plants and subjected to long and small RNA analysis using the full-length PSTVd cDNA as a hybridization probe. Actin and U6 hybridization of the stripped membranes served as loading controls. Hybridization took place at 64°C and the blot was exposed for 12 hrs to phosporimager screen. At these hybridization conditions, PSTVd probe did not hybridize with GFP98(+) or GFPint98(+)/(−).
In order to investigate whether vd-siRNAs targeted the nuclear PTGS sensor constructs, Northern blot analysis was conducted using the full-length GFP cDNA as a hybridization probe (Fig. 4). Our data showed that, in PSTVd-infected GFPint98(+)/(−) plants, neither the spliced GFP mRNA (797-nt) nor the unspliced GFPint98(+)/(−) pre-mRNA (1365-nt) were targeted for cleavage (Fig. 4). In plants, cleaved transcripts may be further processed by RDR6 into dsRNA and subsequently by DCLs into secondary siRNAs. Thus, silencing may spread to 5´ and 3´ direction, in a process termed transitive silencing.53–56 In the case that nuclear vd-siRNA-targeting of the int98 sequence occurred prior to splicing, transitive silencing into the GFP sequence could not be ruled out. Thus, it was important to show that GFP mRNA accumulated to the same degree in PSTVd-infected and non-infected GFPint98(+)/(−) plants. The failure to detect secondary GFP siRNAs, at least under the applied conditions, may underpin that no transitive silencing process was activated (Fig. 4). In contrast, the mRNA of the cytoplasmic PTGS sensor GFP98(+) was efficient silenced by vd-siRNAs. However, also in this case, secondary 21–24-nt GFP siRNAs were not detected which was in agreement with previous data, where viroid sequences were proposed to inhibit RDR6 processing and initiation of transitivity.22
In order to monitor the RNA levels of the post-spliced int98(+)/(−) lariat (568-nt) in PSTVd-infected plants, Northern blot analysis was performed using the 3' region of the intron (-ron) as a hybridization probe (Fig. 5). Our data revealed that in the PSTVd-infected plants, also the int98(+)/(−) splicing products were not targeted for degradation. Moreover, intronic secondary 21–24-nt siRNAs were not detectable (Fig. 5). These data suggest that vd-siRNAs fail to trigger nuclear PTGS of intronic sequences.
Figure 5.
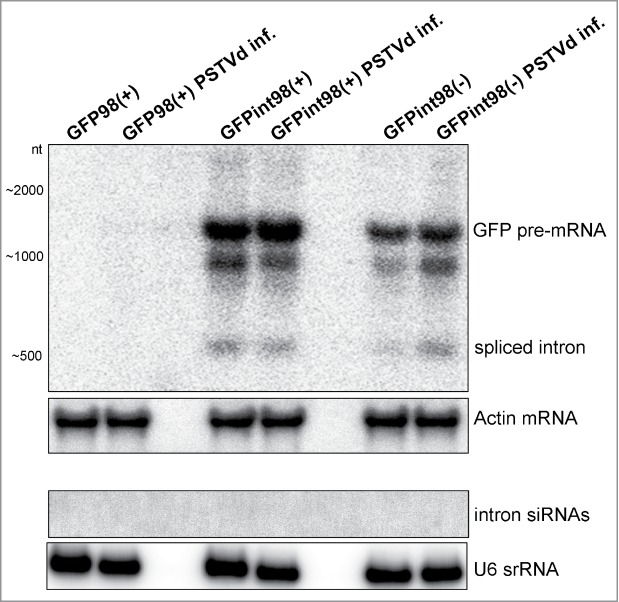
Northern blot analysis for int98(+)/(−) spliced intronic lariat degradation. Total RNA (7 µg) was extracted from PSTVd-infected and non-infected plants and subjected to long and small RNA analysis using the 3´ part of S. lycopersicum intron (‘-ron’) as a hybridization probe. Hybridization took place at 64°C and the blot was exposed for 72 hrs to phosporimager screen. Actin and U6 hybridization of the stripped membranes served as loading controls. An unexpected band of unknown nature of approximate 1000-nt in size was detected below the 1365-nt GFP pre-mRNA. We suggest that this RNA is most probably reflecting an intermediate splicing product.
Viroid infection efficiently triggered RdDM of GFPint98 transgene
DsRNA-induced PTGS is tightly related to RdDM57 and replicating viroids trigger both mechanisms.22,32 So far, and at least in the case of transgenes, PTGS is always associated with RdDM of transgene coding regions.22,56–64 RdDM seems be interwoven with PTGS in a mutual-reinforcing loop, but whether RdDM acts upstream or downstream of PTGS or whether one is a prerequisite of the other is not clear. We were thus interested to investigate whether the failure to post-transcriptionally silence the GFPint98(+)/(−) transgenes was, nevertheless, accompanied by RdDM. In order to analyze the 98 bp PSTVd subfragment methylation status at a single-base resolution, bisulfite sequencing was performed. Treatment of DNA with sodium bisulfite results in the conversion of non-methylated cytosine to uracil and during PCR amplification uracil is replaced by thymine. Thus, in the case of non-methylated cytosines, sequencing of PCR products from bisulfite-treated DNA reveals thymines instead of cytosines. DNA from non-infected and PSTVd-infected GFPint98(+) was subjected to bisulfite sequencing and the upper strand of the 98 bp viroid DNA region was analyzed. In non-infected plants, cytosine conversion of this strand was complete (Fig. 6), indicating that it was not methylated, at all, and that it did not form secondary structures that affect bisulfite conversion. In contrast, in the infected plants, the 98 bp subfragment of the GFPint98(+) transgene was densely methylated in all sequence contexts (84% mCG, 85% mCHG and 51% mCHH) (Fig. 6), indicating ongoing de novo DNA methylation.30 Our data thus show that viroid infection efficiently triggered RdDM of intronic sequences but failed to target the corresponding transcripts for cleavage.
Figure 6.
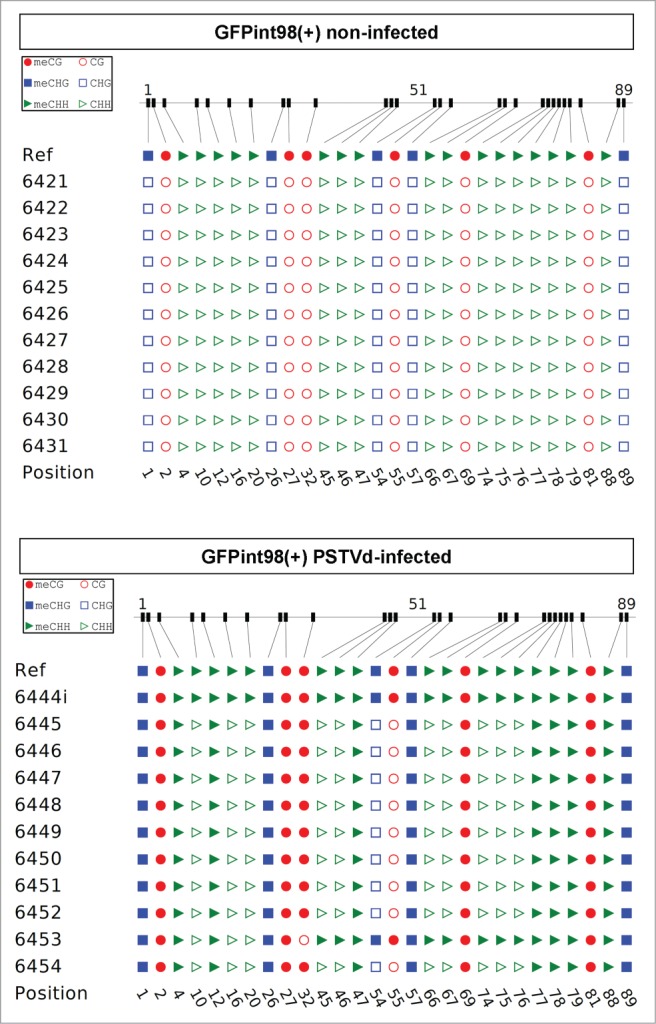
Bisulfite sequencing analysis for the methylation status of the 98 bp region of the GFPint98(+) transgene. DNA was extracted from non-infected (upper panel) and PSTVd-infected (crossed with Nt-Nbdi) (lower panel) Nt-GFPint98(+) plants and subjected to bisulfite sequencing. In each case, 10–15 clones were sequenced and data were interpreted with CyMate software.88 ‘Ref’ corresponds to the reference sequence (not bisulfite treated). Open symbols denote unmethylated and filled symbols methylated cytosines, respectively. Circles, squares and triangles symbolize cytosines in CG, CHG and CHH context, respectively.
Discussion
Plant RNA viruses replicating in the cytoplasm serve as triggers and targets of PTGS.65,66 In Nicotiana benthamiana, the exons but not the introns of the phytoene desaturase (PDS) endogene could be efficiently targeted by virus-induced gene silencing (VIGS) suggesting that, rather than nuclear pre-mRNAs, cytosolic mature mRNAs are targeted by PTGS.40 Most plant miRNAs are generated in the nucleus but are exported to the cytoplasm where they trigger PTGS of target mRNAs through transcript cleavage and/or translational inhibition.67,68 These data led to the view that PTGS is exclusively a cytoplasmic process.39 In support of this assumption, AGO1-containing processing bodies (P-bodies) and AGO7-containing siRNA bodies in Arabidopsis thaliana are localized in the cytoplasm.69,70
However, recent data have demonstrated that also nuclear PTGS takes place in plants. In soybean, siRNAs originating from an inverted repeat (IR) corresponding to the FAD2–1A desaturase intron efficiently silenced FAD2–1A expression, suggesting that siRNAs targeted the nuclear pre-mRNA rather than the cytosolic mRNA.41,42 However, it cannot be excluded that non-spliced FAD2–1A transcripts were transported into the cytoplasm, at first, and then targeted by IR-derived siRNAs. In a more recent report, several miRNAs appeared to target the introns of pre-mRNAs in the nucleus of rice cells.71 Whether RNA degradation of intron-containing pre-mRNA is a post-transcriptional or co-transcriptional process is not clear. Co-transcriptional gene silencing (CTGS) has been well documented in Neurospora crassa72 but conclusive data on CTGS in plants are lacking. Interestingly, in Arabidopsis, the flowering-time regulators FCA and FPA target aberrant RNA for nuclear silencing, most probably, in a co-transcriptional manner.73
Immunofluorescence analysis revealed that the functionally redundant Arabidopsis DCL1, DCL2, DCL3 and DCL4 co-localize in the nuclear periphery.74 DCL-produced small RNAs (sRNAs) are exported to the cytoplasm, where they are loaded onto AGO proteins. While AGO1 loaded with 21-nt siRNAs seems to operate predominantly in the cytoplasm, at least AGO4 loaded with 24-nt siRNAs is transported into the nucleus.75 Similar to AGO1, AGO4 exhibits the characteristic DDH motif which is essential for RNA cleavage.76 In current RdDM models, 24-nt siRNA-loaded AGO4 is considered to hybridize with nascent POLII/POLV transcripts, presumably cleaving them to enable RDR2 to copy the cleaved transcript into dsRNA.33,77,78
The above observations suggest that in plants, nuclear PTGS may be more common than previously assumed. Yet, it is striking that vd-siRNAs of the nucleus-replicating PSTVd failed to trigger nuclear PTGS. We could exclude, (1) that the vd-siRNAs were not produced, (2) that they were not PTGS-competent and (3) that the PTGS machinery itself was negatively affected by environmental conditions such as light or temperature79,80 since GFP98(+) transcripts were efficiently targeted for degradation. We thus conclude that PSTVd infection indeed fails to trigger nuclear PTGS of intronic sequences in plants. Why nuclear PTGS takes place in some cases41,42,71 while not in others,40 is not clear. Though, it is reasonable to assume that the nature of sRNAs which are produced in various biosynthetic pathways (MIR gene, IR gene, VIGS, viroid replication) may greatly differ in terms of size, structure, chemical composition/modification. Importantly, different AGO proteins appeared to have various impacts on silencing processes. In Arabidopsis, 9 functional AGOs were detected and some of them have very specific functions, e.g. AGO7 was reported to specifically bind miR390 which is essential for TAS3 mRNA processing.21,81 Since loading of sRNAs onto different AGO proteins is dependent on their sizes, 5' nucleotides, structures and probably also on their biosynthesis pathway one may speculate that processing of PSTVd RNA into sRNAs does not include the production of nuclear PTGS-competent vd-siRNAs. Interestingly, in N. benthamiana vd-siRNAs seem to associate with all AGOs except AGO6, AGO7 and AGO10.82
Both, Pospiviroidae and Avsunviroidae infections result in the production of abundant siRNAs.20,83 Yet, in contrast to viruses, viroids do not encode any suppressors of RNA silencing. Thus, one may hypothesize that for reasons of fitness, viroids must replicate in an organelle (nucleus and chloroplast, respectively) where they avoid attack by the host silencing machinery. In tomato, the host VirP1 protein ensures PSTVd's transport into the nucleus.6 Deep sequencing of sRNA from viroid-infected tomato plants suggested that both, mature viroid and dsRNA viroid replication intermediates are processed by nuclear DCLs into vd-siRNAs.15,18,19,37 Nevertheless, vd-siRNAs were reported to accumulate in the cytoplasm but not in the nucleus of tomato cells.84 In our case, the cytoplasmic restriction of vd-siRNAs may explain the inability of vd-siRNAs to trigger nuclear PTGS of GFPint98(+)/(−). Yet, it fails to explain how the GFPint98(+) transgene was targeted for dense RdDM in the nucleus. Challenging the siRNA-guided RdDM models, several lines of evidence suggested that siRNAs are involved in the production and amplification of but are not representing the actual RdDM-guide RNA. Thus, RdDM may be triggered by double-stranded PSTVd replication intermediates even in the absence of nuclear vd-siRNAs85,86 (Fig. 1). Alternatively, and in accordance with siRNA-guided RdDM models, it cannot be excluded that AGO4-loaded vd-siRNAs which were not detectable under the experimental conditions applied in previous studies,84 are re-directed into the nucleus and are involved in RdDM.75 In any case, our data suggest that these AGO4-loaded 24-nt vd-siRNAs are unable to trigger nuclear PTGS. Collectively, our data show that viroid replication leads to RdDM and cytoplasmic PTGS but not nuclear PTGS.
Materials and Methods
Plasmid construction
The plasmid pPCV702SM-GFP9822 served as a PCR template. The 98 fragment corresponds to bases 292–359 and 1–30 of PSTVd strain KF440–2 (accession number X58388). For int98(+) the primer pair 5´-AGC TTC TAG AAC TCC GCT TTT-3´/5´-GAA GAT CTT TGA ACC ACA GGA-3´ and for int98(−) the primer pair 5´-AGC TTT TAG ATC TCC GCT TTT-3´/5´-GTC TAG ATT TGA ACC ACA GGA-3´ were used. PCR products were cleaved with XbaI and BglII and ligated into the XbaI/BglII-cleaved pPCV702SM-GFPintron plasmid.44 The resulting plasmids, pPCV702SM-GFPint98(+) and pPCV702SM-GFPint98(−), were introduced into Agrobacterium tumefaciens GV3101. The generated GVGFPint98(+) and GVGFPint98(−) strains were used for leaf-disc transformation of N. tabacum as previously described by Horsch et al.87
RNA extraction and Northern blot analysis
Total RNA was extracted from N. tabacum leaves with PeqGold TriFast (PeqLab, www.peqlab.de). For long RNA molecule analysis, 7 µg of total RNA were separated on a 1.2% agarose/formaldehyde gel while for small RNA molecule analysis 25 µg of total RNA were separated on a 20% Tris-Borate-EDTA (TBE)-acrylamide gel (Anamed, www.anamed-gele-com). Capillary blotting of long RNAs, electro-blotting of small RNAs, and hybridization with radiolabelled probes were performed as previously described.58 In the cases of GFP and PSTVd probes, full-length cDNAs were used. For the intron (‘-ron’) probe, the 410 bp fragment amplified by PCR with the primers 5´-TCT AAA GGC ATA TTG AAA TAC-3´ and 5´-TCA TCA AAG TTT TAA CAC TTG-3´ was used. The mRNA of the N. tabacum actin gene (GQ339768.1) served as a loading control for cytoplasmic RNA. For actin RNA detection a 592 bp long cDNA fragment was amplified in a reverse-transciptase PCR reaction using the primers 5'-ATG TAT GTT GCT ATT CAG GCT GTC C-3' and 5'-CCT TAA TCT TCA TGC TGC TAG GAG C-3' and used as a probe. The U6 snRNA was used as a loading control for nuclear RNA. In order to detect U6 snRNA, the end-labeled 5´-AGG GGC CAT GCT AAT CTT CTC-3 oligo, was used as a probe.58
DNA extraction and bisulfite sequencing analysis
Genomic DNA was extracted from N. tabacum leaves with DNEasy Plant mini kit (Qiagen, www.qiagen.com) and subjected to bisulfite treatment and PCR as previously described44 using the primers 5´-TTA TGG TGT TYA ATG YTT TTY AAG ATA-3´ and 5´-TTT CCT CCC ATA ART TAG TRT ARA ATA T-3´. Bisulfite PCR products were cloned into the pJET vector (ThermoScientific, www.thermoscientificbio.com), 10–15 clones were sequenced for each experiment and the CyMate software88 was used for interpretation of data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the grants of the Sixth Research Framework Programs of the European Union, Project LSHG-CT-2006–037900 (SIROCCO) and by the German Research Foundation (DFG) (grant: Wa1019/8–1).
References
- 1.Hammann C, Steger G. Viroid-specific small RNA in plant disease. RNA Biol 2012; 9:809–19; PMID:22617880; http://dx.doi.org/ 10.4161/rna.19810. [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Gas ME, Molina D, Hernandez C, Daros JA. Analysis of viroid replication. Methods Mol Biol 2008; 451:167–83; PMID:18370255; http://dx.doi.org/ 10.1007/978-1-59745-102-4_12. [DOI] [PubMed] [Google Scholar]
- 3.Flores R, Gas ME, Molina-Serrano D, Nohales MA, Carbonell A, Gago S, De la Pena M, Daros JA. Viroid replication: rolling-circles, enzymes and ribozymes. Viruses 2009; 1:317–34; PMID:21994552; http://dx.doi.org/ 10.3390/v1020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gozmanova M, Denti MA, Minkov IN, Tsagris M, Tabler M. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res 2003; 31:5534–43; PMID:14500815; http://dx.doi.org/ 10.1093/nar/gkg777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalantidis K, Denti MA, Tzortzakaki S, Marinou E, Tabler M, Tsagris M. Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J Virol 2007; 81:12872–80; PMID:17898061; http://dx.doi.org/ 10.1128/JVI.00974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maniataki E, Martinez de Alba AE, Sagesser R, Tabler M, Tsagris M. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 2003; 9:346–54; http://dx.doi.org/ 10.1261/rna.2162203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNA's. Science 1984; 223:450–5; http://dx.doi.org/ 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 8.Schindler I, Mühlbach H. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci 1992; 84:221–9; http://dx.doi.org/ 10.1016/0168-9452(92)90138-C. [DOI] [Google Scholar]
- 9.Di Serio F, Martinez de Alba AE, Navarro B, Gisel A, Flores R. RNA-dependent RNA polymerase 6 delays accumulation and precludes meristem invasion of a viroid that replicates in the nucleus. J Virol 2010; 84:2477–89; PMID:20015979; http://dx.doi.org/ 10.1128/JVI.02336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabler M, Tsagris M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci 2004; 9:339–48; PMID:15231279; http://dx.doi.org/ 10.1016/j.tplants.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Tsagris EM, Martinez de Alba AE, Gozmanova M, Kalantidis K. Viroids. Cell Microbiol 2008; 10:2168–79; PMID:18764915; http://dx.doi.org/ 10.1111/j.1462-5822.2008.01231.x. [DOI] [PubMed] [Google Scholar]
- 12.Ding B. Viroids: self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip Rev RNA 2009; 1:362–75; http://dx.doi.org/ 10.1002/wrna.22. [DOI] [PubMed] [Google Scholar]
- 13.Ding B, Itaya A, Zhong X. Viroid trafficking: a small RNA makes a big move. Curr Opin Plant Biol 2005; 8:606–12; PMID:16181802; http://dx.doi.org/ 10.1016/j.pbi.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Itaya A, Folimonov A, Matsuda Y, Nelson RS, Ding B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol Plant Microbe Interact 2001; 14:1332–4; PMID:11763132; http://dx.doi.org/ 10.1094/MPMI.2001.14.11.1332. [DOI] [PubMed] [Google Scholar]
- 15.Itaya A, Zhong X, Bundschuh R, Qi Y, Wang Y, Takeda R, Harris AR, Molina C, Nelson RS, Ding B. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J Virol 2007; 81:2980–94; PMID:17202210; http://dx.doi.org/ 10.1128/JVI.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dadami E, Boutla A, Vrettos N, Tzortzakaki S, Karakasilioti I, Kalantidis K. DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on Potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol Plant 2012; 6(1):232–4; PMID:23100483. [DOI] [PubMed] [Google Scholar]
- 17.Martin R, Arenas C, Daros JA, Covarrubias A, Reyes JL, Chua NH. Characterization of small RNAs derived from Citrus exocortis viroid (CEVd) in infected tomato plants. Virology 2007; 367:135–46; PMID:17559901; http://dx.doi.org/ 10.1016/j.virol.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Li R, Gao S, Hernandez AG, Wechter WP, Fei Z, Ling KS. Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PloS one 2012; 7:e37127; PMID:22623984; http://dx.doi.org/ 10.1371/journal.pone.0037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machida S, Yamahata N, Watanuki H, Owens RA, Sano T. Successive accumulation of two size classes of viroid-specific small RNA in potato spindle tuber viroid-infected tomato plants. J Gen Virol 2007; 88:3452–7; PMID:18024916; http://dx.doi.org/ 10.1099/vir.0.83228-0. [DOI] [PubMed] [Google Scholar]
- 20.Papaefthimiou I, Hamilton A, Denti M, Baulcombe D, Tsagris M, Tabler M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res 2001; 29:2395–400; PMID:11376158; http://dx.doi.org/ 10.1093/nar/29.11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci 2008; 13:350–8; PMID:18508405; http://dx.doi.org/ 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Vogt U, Pelissier T, Putz A, Razvi F, Fischer R, Wassenegger M. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J 2004; 38:107–18; PMID:15053764; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02029.x. [DOI] [PubMed] [Google Scholar]
- 23.Voinnet O. Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility. Curr Opin Plant Biol 2008; 11:464–70; PMID:18583181; http://dx.doi.org/ 10.1016/j.pbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Wang MB, Bian XY, Wu LM, Liu LX, Smith NA, Isenegger D, Wu RM, Masuta C, Vance VB, Watson JM, et al.. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci U S A 2004; 101:3275–80; PMID:14978267; http://dx.doi.org/ 10.1073/pnas.0400104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbonell A, Martinez de Alba AE, Flores R, Gago S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008; 371:44–53; PMID:18028975; http://dx.doi.org/ 10.1016/j.virol.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 26.Kasai A, Sano T, Harada T. Scion on a stock producing siRNAs of potato spindle tuber viroid (PSTVd) attenuates accumulation of the viroid. PloS one 2013; 8:e57736; PMID:23469061; http://dx.doi.org/ 10.1371/journal.pone.0057736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwind N, Zwiebel M, Itaya A, Ding B, Wang MB, Krczal G, Wassenegger M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol Plant Pathol 2009; 10:459–69; PMID:19523100; http://dx.doi.org/ 10.1111/j.1364-3703.2009.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalakouras A, Dadami E, Zwiebel M, Krczal G, Wassenegger M. Transgenerational maintenance of transgene body CG but not CHG and CHH methylation. Epigenetics 2012; 7:1071–8; PMID:22863736; http://dx.doi.org/ 10.4161/epi.21644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalakouras A, Moser M, Krczal G, Wassenegger M. A chimeric satellite transgene sequence is inefficiently targeted by viroid-induced DNA methylation in tobacco. Plant Mol Biol 2010; 73:439–47; PMID:20364297; http://dx.doi.org/ 10.1007/s11103-010-9631-6. [DOI] [PubMed] [Google Scholar]
- 30.Pelissier T, Thalmeir S, Kempe D, Sanger HL, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res 1999; 27:1625–34; PMID:10075993; http://dx.doi.org/ 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelissier T, Wassenegger M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA 2000; 6:55–65; http://dx.doi.org/ 10.1017/S135583820099201X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenegger M, Heimes S, Sanger HL. An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J 1994; 13:6172–7; PMID:7813454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev 2014; 15:394–408; PMID:24805120; http://dx.doi.org/ 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 34.Owens RA, Hammond RW. Viroid pathogenicity: one process, many faces. Viruses 2009; 1:298–316; PMID:21994551; http://dx.doi.org/ 10.3390/v1020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eamens AL, Smith NA, Dennis ES, Wassenegger M, Wang MB. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 2013; 450-451:266–77; http://dx.doi.org/ 10.1016/j.virol.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Diermann N, Matousek J, Junge M, Riesner D, Steger G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol Chem 2010; 391:1379–90; PMID:21087089; http://dx.doi.org/ 10.1515/bc.2010.148. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Shibuya M, Taneda A, Kurauchi T, Senda M, Owens RA, Sano T. Accumulation of Potato spindle tuber viroid-specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology 2011; 413:72–83; PMID:21353278; http://dx.doi.org/ 10.1016/j.virol.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 38.Martinez G, Castellano M, Tortosa M, Pallas V, Gomez G. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res 2013; 42:1553–62; PMID:24178032; http://dx.doi.org/ 10.1093/nar/gkt968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baulcombe D. RNA silencing in plants. Nature 2004; 431:356–63; PMID:15372043; http://dx.doi.org/ 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell 1998; 10:937–46; PMID:9634582; http://dx.doi.org/ 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. Posttranscriptional gene silencing in nuclei. Proc Natl Acad Sci U S A 2011; 108:409–14; PMID:21173264; http://dx.doi.org/ 10.1073/pnas.1009805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mroczka A, Roberts PD, Fillatti JJ, Wiggins BE, Ulmasov T, Voelker T. An intron sense suppression construct targeting soybean FAD2-1 requires a double-stranded RNA-producing inverted repeat T-DNA insert. Plant Physiol 2010; 153:882–91; PMID:20424004; http://dx.doi.org/ 10.1104/pp.110.154351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res 1992; 20:5345–50; PMID:1437551; http://dx.doi.org/ 10.1093/nar/20.20.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dalakouras A, Moser M, Zwiebel M, Krczal G, Hell R, Wassenegger M. A hairpin RNA construct residing in an intron efficiently triggered RNA-directed DNA methylation in tobacco. Plant J 2009; 60:840–51; PMID:19702668; http://dx.doi.org/ 10.1111/j.1365-313X.2009.04003.x. [DOI] [PubMed] [Google Scholar]
- 45.Schiebel W, Pelissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger HL, Wassenegger M. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 1998; 10:2087–101; PMID:9836747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci U S A 1997; 94:2122–7; PMID:9122158; http://dx.doi.org/ 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodall GJ, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 1989; 58:473–83; PMID:2758463; http://dx.doi.org/ 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- 48.Goodall GJ, Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol 1990; 14:727–33; PMID:2102851; http://dx.doi.org/ 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- 49.Goodall GJ, Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J 1991; 10:2635–44; PMID:1868837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu HX, Goodall GJ, Kole R, Filipowicz W. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5' but not the 3' splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J 1995; 14:377–88; PMID:7835348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harders J, Lukacs N, Robert-Nicoud M, Jovin TM, Riesner D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J 1989; 8:3941–9; PMID:2591366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi Y, Pelissier T, Itaya A, Hunt E, Wassenegger M, Ding B. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 2004; 16:1741–52; PMID:15194818; http://dx.doi.org/ 10.1105/tpc.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. Twenty-two-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci U S A 2010; 107:15269–74; PMID:20643946; http://dx.doi.org/ 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci U S A 2012; 109:2461–6; PMID:22308502; http://dx.doi.org/ 10.1073/pnas.1200169109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 2007; 13:1268–78; http://dx.doi.org/ 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 2002; 14:857–67; PMID:11971140; http://dx.doi.org/ 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 1999; 11:2291–301; PMID:10590159; http://dx.doi.org/ 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dadami E, Dalakouras A, Zwiebel M, Krczal G, Wassenegger M. An endogene-resembling transgene is resistant to DNA methylation and systemic silencing. RNA Biol 2014; 11:934–41; PMID:25180820; http://dx.doi.org/ 10.4161/rna.29623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dadami E, Moser M, Zwiebel M, Krczal G, Wassenegger M, Dalakouras A. An endogene-resembling transgene delays the onset of silencing and limits siRNA accumulation. FEBS Lett 2013; 18:706–10; http://dx.doi.org/ 10.1016/j.febslet.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 60.Dalakouras A, Moser M, Boonrod K, Krczal G, Wassenegger M. Diverse spontaneous silencing of a transgene among two Nicotiana species. Planta 2011; 234:699–707; PMID:21617990; http://dx.doi.org/ 10.1007/s00425-011-1433-9. [DOI] [PubMed] [Google Scholar]
- 61.Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 2001; 11:747–57; PMID:11378384; http://dx.doi.org/ 10.1016/S0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 62.Lunerova-Bedrichova J, Bleys A, Fojtova M, Khaitova L, Depicker A, Kovarik A. Trans-generation inheritance of methylation patterns in a tobacco transgene following a post-transcriptional silencing event. Plant J 2008; 54:1049–62; PMID:18315537; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03475.x. [DOI] [PubMed] [Google Scholar]
- 63.Vermeersch L, De Winne N, Nolf J, Bleys A, Kovarik A, Depicker A. Transitive RNA silencing signals induce cytosine methylation of a transgenic but not an endogenous target. Plant J 2013; 74:867–79; PMID:23480471; http://dx.doi.org/ 10.1111/tpj.12172. [DOI] [PubMed] [Google Scholar]
- 64.Haque AK, Yamaoka N, Nishiguchi M. Cytosine methylation is associated with RNA silencing in silenced plants but not with systemic and transitive RNA silencing through grafting. Gene 2007; 396:321–31; PMID:17521830; http://dx.doi.org/ 10.1016/j.gene.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Brigneti G, Martin-Hernandez AM, Jin H, Chen J, Baulcombe DC, Baker B, Jones JD. Virus-induced gene silencing in Solanum species. Plant J 2004; 39:264–72; PMID:15225290; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02122.x. [DOI] [PubMed] [Google Scholar]
- 66.Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. Virus-induced gene silencing in plants. Methods 2003; 30:296–303; PMID:12828943; http://dx.doi.org/ 10.1016/S1046-2023(03)00037-9. [DOI] [PubMed] [Google Scholar]
- 67.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 2006; 20:759–71; PMID:16600909; http://dx.doi.org/ 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 68.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009; 136:669–87; PMID:19239888; http://dx.doi.org/ 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 69.Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J 2012; 31:1704–13; PMID:22327216; http://dx.doi.org/ 10.1038/emboj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maldonado-Bonilla LD. Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci 2014; 5:201; PMID:24860588; http://dx.doi.org/ 10.3389/fpls.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meng Y, Shao C, Ma X, Wang H. Introns targeted by plant microRNAs: a possible novel mechanism of gene regulation. Rice (N Y) 2013; 6:8; PMID:24280590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell 2006; 125:873–86; PMID:16751098; http://dx.doi.org/ 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 73.Baurle I, Smith L, Baulcombe DC, Dean C. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 2007; 318:109–12; http://dx.doi.org/ 10.1126/science.1146565. [DOI] [PubMed] [Google Scholar]
- 74.Pontes O, Vitins A, Ream TS, Hong E, Pikaard CS, Costa-Nunes P. Intersection of small RNA pathways in Arabidopsis thaliana sub-nuclear domains. PloS One 2013; 8:e65652; PMID:23776518; http://dx.doi.org/ 10.1371/journal.pone.0065652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 2012; 46:859–70; PMID:22608924; http://dx.doi.org/ 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 76.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 2006; 443:1008–12; PMID:16998468; http://dx.doi.org/ 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 77.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 2011; 12:483–92; PMID:21779025; http://dx.doi.org/ 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 78.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev 2010; 11:204–20; PMID:20142834; http://dx.doi.org/ 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kotakis C, Vrettos N, Kotsis D, Tsagris M, Kotzabasis K, Kalantidis K. Light intensity affects RNA silencing of a transgene in Nicotiana benthamiana plants. BMC Plant Biol 2010; 10:220; PMID:20939918; http://dx.doi.org/ 10.1186/1471-2229-10-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, Lakatos L, Banfalvi Z, Burgyan J. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 2003; 22:633–40; PMID:12554663; http://dx.doi.org/ 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell 2006; 127:565–77; PMID:17081978; http://dx.doi.org/ 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 82.Minoia S, Carbonell A, Di Serio F, Gisel A, Carrington JC, Navarro B, Flores R. Specific ARGONAUTES bind selectively small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J Virol 2014; PMID:25100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez de Alba AE, Flores R, Hernandez C. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J Virol 2002; 76:13094–6; PMID:12438638; http://dx.doi.org/ 10.1128/JVI.76.24.13094-13096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Denti MA, Boutla A, Tsagris M, Tabler M. Short interfering RNAs specific for potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J 2004; 37:762–9; PMID:14871315; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02001.x. [DOI] [PubMed] [Google Scholar]
- 85.Dalakouras A, Dadami E, Wassenegger M. Viroid-induced DNA methylation in plants. BioMol Concepts 2013; 4:557–65; PMID:25436756; http://dx.doi.org/ 10.1515/bmc-2013-0030. [DOI] [PubMed] [Google Scholar]
- 86.Dalakouras A, Wassenegger M. Revisiting RNA-directed DNA methylation. RNA biology 2013; 10:453–5; PMID:23324611; http://dx.doi.org/ 10.4161/rna.23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horsch R, Fry J, Hoffmann N, Eichholz D, Rogers S, Fraley R. A simple and general method for transferring genes into plants. Science 1985; 227:1229–31; http://dx.doi.org/ 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 88.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 2007; 51:526–36; PMID:17559516; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]


