Abstract
In certain cases, a species may have access to important genetic variation present in a related species via adaptive introgression. These novel alleles may interact with their new genetic background, resulting in unexpected phenotypes. In this study, we describe a selective sweep on standing variation on the X chromosome in the mosquito Anopheles coluzzii, a principal malaria vector in West Africa. This event may have been influenced by the recent adaptive introgression of the insecticide resistance gene known as kdr from the sister species Anopheles gambiae. Individuals carrying both kdr and a nearly fixed X‐linked haplotype, encompassing at least four genes including the P450 gene CYP9K1 and the cuticular protein CPR125, have rapidly increased in relative frequency. In parallel, a reproductively isolated insecticide‐susceptible A. gambiae population (Bamako form) has been driven to local extinction, likely due to strong selection from increased insecticide‐treated bed net usage.
Keywords: adaptive introgression, Anopheles, epistasis, hybridization, insecticide resistance, kdr, malaria vector, P450
Introduction
Leaky interspecies reproductive barriers may increase genetic variability upon which selection can act, increasing the evolutionary responsiveness of a species (Stelkens et al. 2014). Adaptive introgression is thought to be most common in plants (Hedrick 2013), but clear examples are emerging in animals. Examples include warfarin resistance in mice (Song et al. 2011), coat colour in wolves (Anderson et al. 2009), wing coloration patterns in butterflies (Dasmahapatra et al. 2012; Pardo‐Diaz et al. 2012) and, more recently, insecticide resistance in African malaria mosquitoes (Clarkson et al. 2014; Norris et al. 2015).
Anopheles coluzzii is a newly described species (Coetzee et al. 2013) that is morphologically identical to Anopheles gambiae (previously referred to as the M and S forms of A. gambiae, respectively). Both are major vectors of human malaria and are sympatric throughout much of West and Central Africa (Della Torre et al. 2005; Diabaté et al. 2009). Divergence is thought to exist at least in part due to adaptation to distinct larval habitats (Gimonneau et al. 2012; Kamdem et al. 2012). The taxon A. gambiae includes two chromosomal forms, known as the Savanna and Bamako form. The two are distinguishable with respect to paracentric chromosome inversion karyotypes, are sympatric in Mali along the Niger and Senegal Rivers and are to a large extent reproductively isolated (Coluzzi et al. 1979; Toure et al. 1998; Manoukis et al. 2008). We will refer to the Savanna form as A. gambiae and use the designation Ag‐Bamako for the Bamako form.
Comparisons between A. coluzzii and A. gambiae genomes have revealed pronounced differentiation at pericentromeric regions on each chromosome (Turner et al. 2005; Lawniczak et al. 2010; White et al. 2010; Reidenbach et al. 2012). This result is consistent with a model of speciation that is not strictly dependent on allopatry, namely the ‘speciation islands’ model (Turner et al. 2005). Under this model, strong selection on genes important for reproductive isolation maintains divergence at discrete regions, while the remainder of the genome is homogenized by gene flow between sympatric populations (Turner et al. 2005; Via & West 2008; Nosil et al. 2009; Weetman et al. 2012). An alternative hypothesis poses that reduced diversity due to selection on genes within these low recombining regions may have facilitated the fixation of alternative ancestral haplotypes in these regions, resulting in ‘incidental islands’ (Lawniczak et al. 2010; Turner & Hahn 2010; White et al. 2010). The ‘incidental islands’ hypothesis argues against variable rates of gene flow as the major architect of the islands of divergence. In 2006, the insecticide resistance gene kdr and the entire 2L island were stably introgressed from A. gambiae into A. coluzzii in Selinkenyi, Mali (Clarkson et al. 2014; Norris et al. 2015). Interestingly, reproductive isolation was quickly re‐established, based on markers on X and 3L (Lee et al. 2013b).
Hybrids between A. coluzzii and A. gambiae are detected in punctuated bursts in Mali, and early‐stage hybrids are typically short lived, presumably due to reduced fitness (Lee et al. 2013b). However, cases where hybrids overcame this apparent ‘fitness bottleneck’ in nature and backcrossed with one parental strain (Uecker et al. 2014) have been reported in Guinea‐Bissau (Marsden et al. 2011), Ghana (Clarkson et al. 2014) and Mali (Norris et al. 2015). In Mali, a dramatic increase in insecticide‐treated bed net (ITN) usage starting in 2005 (Milliner 2009) likely altered the fitness landscape and promoted adaptive introgression of kdr from A. gambiae into A. coluzzii (Tripet et al. 2006; Norris et al. 2015). Kdr refers to nonsynonymous mutations in the voltage‐gated sodium channel gene (para); the most common kdr mutation in West Africa is L1014F (Ranson et al. 2011). The L1014F mutation confers resistance by altering the binding site of pyrethroid insecticides, a mechanism called target‐site resistance. Kdr has been increasing in geographical distribution and relative frequency throughout Africa, apparently in response to increased ITN use (Ranson et al. 2009, 2011; Trape et al. 2011). Genetic signatures of selection for this introgression (Clarkson et al. 2014; Norris et al. 2015) and evidence showing that A. coluzzii individuals with the introgressed kdr (kdr A. coluzzii) have increased in relative frequency since 2006 (Norris et al. 2015) suggest that this introgression is highly adaptive.
In addition to target‐site resistance, the combination of reduced cuticle penetrance (Ahmad et al. 2006; Puinean et al. 2010; Wood et al. 2010; Willis 2014) and increased activity of metabolic detoxification enzymes like cytochrome P450 genes and glutathione S‐transferases (GSTs) can also confer resistance to insecticides (Hemingway 2000; Hemingway & Ranson 2000; Müller et al. 2008; Stevenson et al. 2011). For example, a positive association between cuticle thickness and pyrethroid resistance was reported in the closely related mosquito species A. funestus (Wood et al. 2010). But, most of the identified insecticide resistance genes in A. gambiae (119 in all) are P450 genes (64%; Srivastava et al. 2010). Gene expression studies in anopheline mosquitoes have reported associations between over expression of several P450 genes and insecticide resistance, including CYP9K1 (Tene et al. 2013; Mulamba et al. 2014), CYP6P3 (Müller et al. 2008), CYP6M2 (Stevenson et al. 2011), CYP6Z1 (Nikou et al. 2003), CYP325A3 (David et al. 2005; Awolola et al. 2009) and others (Djouaka et al. 2008; McLaughlin et al. 2008). The molecular basis of DDT resistance in Drosophila has been attributed to increased copy number and cis‐regulatory variants at the P450 Cyp6g1 (Schmidt et al. 2010). Optimal insecticide resistance appears to involve the combination of multiple genes and mechanisms, including kdr (Corbel et al. 2007; Namountougou et al. 2012). For example, there is evidence that the combination of elevated P450 activity and kdr can confer a nonadditive increase in insecticide resistance (Hardstone et al. 2008). A recent report from the World Health Organization has stated that malaria vectors with both target‐site and metabolic resistance (e.g. kdr and P450) likely present the biggest threat to mosquito control efforts (WHO 2012).
We hypothesized that selection from increased ITN usage acted on multiple loci in A. coluzzii including those that have introgressed from A. gambiae in 2006 as well as on standing variation. To test this, we conducted a longitudinal study including whole‐genome sequencing and population‐scale genotyping of A. gambiae and A. coluzzii individuals collected both before and after the start of the 2006 ITN campaign in Selinkenyi, Mali. In addition, we conducted insecticide resistance bioassays to establish resistance phenotypes associated with the genotypes under study.
Materials and methods
Mosquito collections
Blood‐fed female mosquitoes were collected from inside human dwellings using mouth aspirators in Selinkenyi (11.700N, 8.2833W) and an adjacent (<25 km) village, Kela (11.8868N, 8.4474W), in Mali, during the rainy season (August–October). Mosquitoes were held until half‐gravid (60–70% digestion of bloodmeal), and the ovaries were removed and stored in Carnoy's solution (1 part glacial acetic acid and 3 parts 100% ethanol). The remaining carcass was stored in individual tubes containing 80% ethanol and transported to UC Davis for DNA extraction using the Qiagen Biosprint 96 system with Qiagen blood and tissue kits (Qiagen, Valencia, CA, USA). Anopheles gambiae and Anopheles coluzzii were distinguished from other Anopheline species using a diagnostic PCR developed by Scott et al. 1993 (Scott et al. 1993).
Cytogenetic analysis
To estimate the frequency of the Bamako form of A. gambaie over time, polytene chromosomes were extracted from ovarian nurse cells using the protocol described by Hunt (Hunt 1973). Chromosome banding patterns were examined using an Olympus BX‐50 phase contrast microscope. The genotypes of five chromosome inversions – 2Rj, 2Rb, 2Rc, 2Rd and 2Ru – on the right arm of chromosome 2 (2R) were scored for individual mosquitoes. Individuals that were homozygous for 2R j, c and u inversions were identified as the Bamako form (Toure et al. 1998; Lee et al. 2013a; see supplemental information).
Genotyping
To identify species and admixed individuals, we genotyped 458 mosquitoes from Selinkenyi, Mali, using the divergence island SNP (DIS) method described by Lee et al. (Lee et al. 2013b) with four additional SNPs at CYP9K1 that distinguish three major haplotypes and two additional SNPs in the para gene that distinguish L1014F and L1014S kdr mutations. Species designation was determined based on fixed SNPs on the X chromosome (Favia et al. 1997, 2001; Fanello et al. 2002; Santolamazza et al. 2004, 2008). The informative SNPs for CYP9K1 haplotypes were identified by visual inspection of paired‐end reads using the Integrated Genomics Viewer (IGV) (see Table S3 for assay design details and primer sequences). The Veterinary Genetics Laboratory at UC Davis conducted the Sequenom iPLEX SNP genotyping for this modified DIS method. CYP9K1 haplotypes were determined using phase (version 2.1 and Stephens et al. 2001; Stephens & Donnelly 2003). DIS and kdr genotypes were plotted using matplotlib (Hunter 2007) following the colour scheme used in Lee et al. (2013b) and Norris et al. (2015). The Bamako and Savanna forms of A. gambiae were determined based on genotype data and by karyotyping (see Cytogenetic analysis).
Genomic DNA library preparation and sequencing
Based on SNP genotype data, we selected 29 A. coluzzii individuals for genome sequencing: 12 pre‐2006 A. coluzzii and 17 post‐2006 A. coluzzii. In addition, we sequenced 7 A. gambiae individuals for a copy number analysis. Genomic DNA was quantified using a qubit 2.0 fluorometer (Life Technologies). DNA was cleaned and concentrated with Zymo Research DNA Clean and Concentrator kit. We used 25–50 ng of input DNA from individual mosquitoes for library construction. Genomic libraries were made with the Nextera DNA Sample Preparation Kit (Illumina) with TruSeq dual indexes (Illumina), modified to half volume. Libraries were size‐selected with Agencourt AMPure XP beads (Beckman Coulter). The concentration of finished libraries was quantified using a qubit 2.0 fluorometer. The expected library fragment size distribution was evaluated using a QIAxcel instrument (Qiagen) and Bioanalyzer 2100 (Agilent). Barcoded individual libraries were sequenced with the Illumina HiSeq2500 platform with paired‐end 100–150 bp reads at the QB3 Vincent J Coates Genomics Sequencing Laboratory at UC Berkeley (see Table S1 for raw sequence output per sample).
Genome sequence analysis
We assessed the quality of our genome sequencing reads using the fastqc software (Andrews 2010). Adaptor sequences and poor quality sequence was trimmed from the raw reads using the trimmomatic software, version 0.30 (Bolger et al. 2014), with default options. Reads were aligned to the A. gambiae reference genome (AgamP4) using BWA‐mem (Li 2013). We used the MarkDuplicates module from Picard tools to remove PCR duplicates and the genome analysis tool kit (gatk) v1.7 to realign reads around indels (McKenna et al. 2010). The resulting sorted bam (Binary sequence Alignment/Map) files, which contain sequences for each read and its mapping position, were used for analysis.
F ST between pre‐ and post‐2006 A. coluzzii was calculated in 50 kb windows with a 25‐kb step using the Weir and Cockerham estimator of F ST (–weir‐pop‐fst) in vcftools (v0.1.11). We estimated Tajima's D using vcftools (–Tajima D) for both pre‐ and post‐2006 A. coluzzii and calculated the standardized difference of D (ΔD) with the following equation adopted from Bigham et al. (Bigham et al. 2010):
where D iA is Tajima's D for a given bin for pop A, D iB is Tajima's D for a given bin for pop B, and u and SD are the mean Tajima's D and standard deviation for all bins from both populations. The step function was not available for Tajima's D with VCFtools, so a smaller window size (25 kb) was used. The data were plotted with Gaussian smoothing.
To elucidate copy number variation at the selected cyp‐l haplotype region, we analysed normalized sequencing coverage from whole‐genome sequencing data for A. gambiae (N = 7), pre‐2006 A. coluzzii (N = 12) and post‐2006 A. coluzzii (N = 17) individuals. To call individual duplicated regions, we used cnvnator (v0.3; Abyzov et al. 2011) with a bin size of 200 bp. We filtered for high‐quality calls using a t‐test P‐value threshold of 0.01, the size had to be >1 kb, and the default q0 filter was applied (calls with >50% reads with low mapping quality were ignored).
Insecticide bioassays
Gravid Anopheles mosquitoes resting inside houses were collected using mouth aspirators in Selinkenyi, Mali, in August 2014 and individually housed in a glass vial or a microtube for oviposition. Mothers were saved in 80% ethanol after oviposition. 1st instar larvae were brought to UC Kearney. Three 2nd or 3rd instar larvae from each family were preserved in 80% ethanol for DNA extraction. We extracted DNA from the mother and three larvae from each family to genotype for species and hybrid status (DIS method), kdr and CYP9K1. Using these results, families with like genotypes were combined. In total, we generated 4 colonies with the following homozygous genotypes (species: CYP9K1 haplotype: kdr status): (i) A. coluzzii:cyp‐l:kdr, (ii) A. coluzzii:cyp‐l:wt, (iii) A. coluzzii:cyp‐ll:wt and (iv) A. gambiae:cyp‐lll:kdr.
Permethrin and deltamethrin bioassays were performed on 6‐week‐old adult individuals that were first generation from the field and 1‐ to 3‐week‐old colony‐based individuals. Insecticide bottle bioassays were performed either on females or on a mix of adult males and females (see Supporting Materials) following the protocols of Brogdon & McAllister (1998). Briefly, 250 mL Wheaton bottles (Wheaton Industries, Millville, NJ, USA) were prepared by coating each with permethrin or deltamethrin dissolved in acetone at the WHO diagnostic dose (21.5 mg/mL and 12.5 mg/mL, respectively) or acetone alone (control). Bottles were left open for 1 h to evaporate residual acetone prior to bioassays. A group of 6–20 individual mosquitoes were introduced into each bottle, and the number of individuals that were nonresponsive upon disturbing the bottle (knocked down) and rotating it horizontally 360 degrees was recorded at five‐minute intervals. The time when 50% and 90% of the mosquitoes were knocked down (KD50 and KD90, respectively) within a given bottle was determined using a best fit curve. The plotted KD50 and KD90 values are the mean and standard error between replicates. Significant differences between knock‐down times was calculated using a 2‐tailed t‐test.
Results
Temporal dynamics of species composition
In 2005, the President's Malaria Initiative initiated a major ITN campaign in Mali (Flaxman et al. 2010; WHO 2013). To explore the potential relationship between this anthropogenic selection pressure on the relative fitness of Anopheles coluzzii, Anopheles gambiae and Ag‐Bamako, we plotted their relative abundance at our study site (Selinkenyi) based on adult mosquito collections starting in 1980 through 2014 (Fig. 1). These data were gathered from Touré et al. (Toure et al. 1998) and our own collections, some of which have been published (Lee et al. 2013a,b). For our analysis, we used only wet season collections (May–October). During the 25 years prior to 2005, the frequencies of A. gambiae, Ag‐Bamako and A. coluzzii in the population were remarkably stable, representing approximately 10%, 25% and 65%, respectively, at Selinkenyi (Figs 1 and 2). In 2006, we observed a punctuated burst of A. coluzzii x A. gambiae hybrids, including 16 F1s and 9 recombinants, N = 124 (Fig. 2). By 2010, early‐stage hybrid genotypes were no longer detected, but A. coluzzii individuals with kdr and the physically linked 2L island from A. gambiae were common (Fig. 2). The A. coluzzii population increased in relative frequency from approximately 65% pre‐2006 to 88% in 2014 (N = 179), likely due to the increased representation of the kdr‐introgressed A. coluzzii genotype (80%; N = 155). This gain in relative abundance of A. coluzzii is proportional to the decline of Ag‐Bamako from 25% in pre‐2006 to 0% in 2014 (N = 179, Fig. 1).
Figure 1.
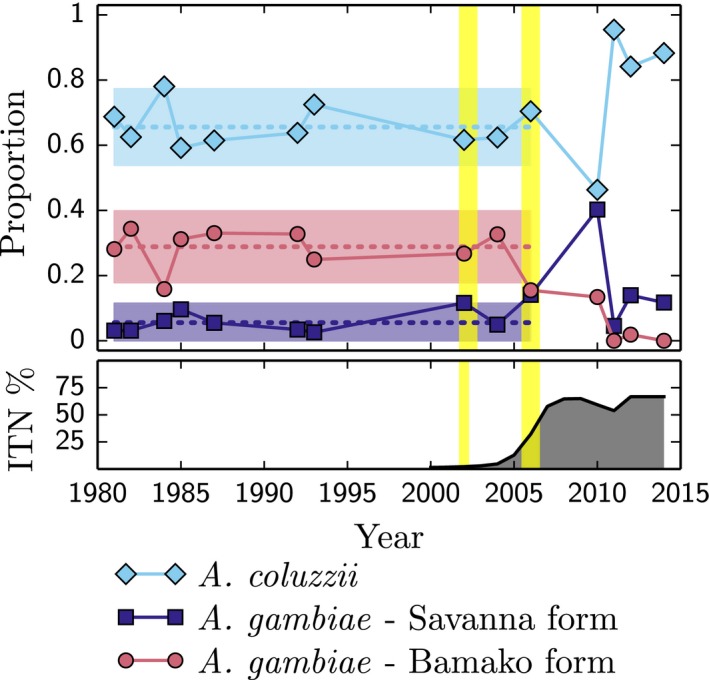
Temporal Dynamics of Species Composition at Selinkenyi, Mali. Shown is the relative abundance of the Savanna and Bamako Forms of Anopheles gambiae and Anopheles coluzzii collected from the town of Selinkenyi, Mali, between the years 1980 through 2014. Data prior to 1991 were taken from Toure et al. 1998;. Data from 1991 through 2010 were collected by us and reported in Lee et al. 2013a,b. Data since 2010 are new. The yellow vertical lines mark when F1 hybrids were observed. The bottom graph displays the estimated proportion of the population sleeping under insecticide‐treated bed nets (ITN). ITN usage data was taken from WHO (2013).
Figure 2.
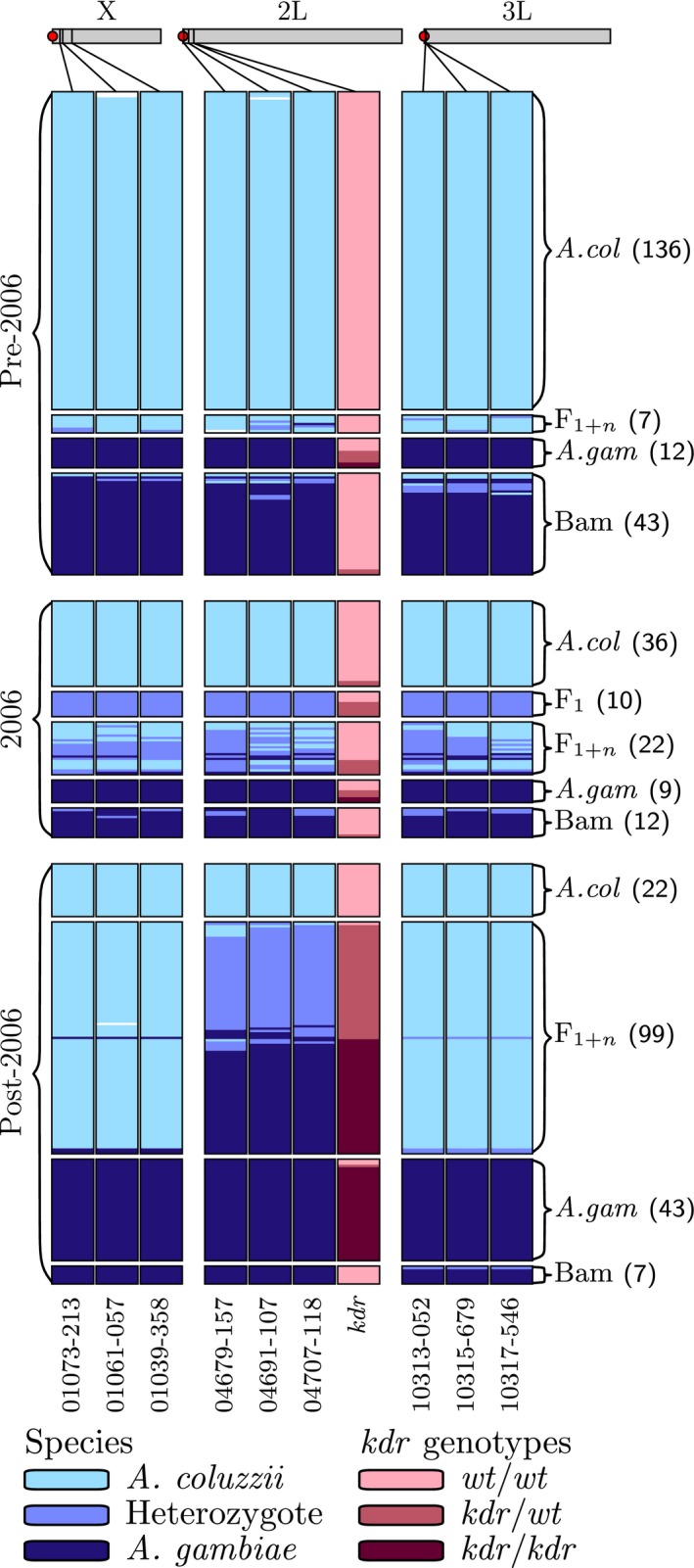
A Longitudinal Survey of Divergence Island SNP (DIS) and kdr Genotype Frequencies at Selinkenyi, Mali. Data are organized into three time periods: Top panel – pre‐2006 (includes data for 2002 and 2004); centre panel – 2006; bottom panel – post‐2006 (2010 and 2012). Individual SNPs are organized by chromosome location: X chromosome (N = 3), chromosome 2L (N = 4) and chromosome 3L (N = 3). Individual SNP identifiers are provided at the bottom of each column. Details for each DIS can be found in Lee et al. (2013a) and for the kdr SNP in Norris et al. (2015). Light blue = homozygous for Anopheles coluzzii DIS, medium blue = DIS heterozygote and dark blue = homozygous for Anopheles gambiae DIS. Dark red = homozygous kdr resistant (kdr), medium red = kdr heterozygote and pink = kdr susceptible (wt) homozygote. The chromosomal forms of A. gambiae are represented as follows: Bamako = Bam and Savanna = A. gam. F1 = first‐generation hybrid and F1+n = backcross genotypes. Sample sizes for each genotype are given in parentheses.
Sequence differentiation between pre‐ and post‐2006 Anopheles coluzzii
The kdr‐introgressed A. coluzzii genotype first appeared in 2006 in Selinkenyi. Since then, it has outcompeted wt A. coluzzii, reaching 97% in 2014 (N = 159). To elucidate additional introgressed regions (other than 2L) and/or selection on standing variation elsewhere in the A. coluzzii genome, we sequenced the genomes of 12 pre‐2006 A. coluzzii (kdr freq. = 0) and 17 post‐2006 A. coluzzii (kdr freq. = 0.56). All genomic libraries for this study were prepared from single, field‐collected adult females and were sequenced at a median depth of 14× (Table S1, Supporting information). To identify major differentiated regions between pre‐2006 and post‐2006 A. coluzzii genomes, we calculated F ST and a relative Tajima's D statistic [∆D, standardized difference of D (Fig. 3; see Methods)]. In brief, negative ∆D values may be indicative of a selective sweep after 2006, while positive ∆D values could be due to an enrichment of heterozygotes (e.g. due to balancing selection) after 2006. This analysis revealed two prominent F ST peaks, including the expected kdr locus within the speciation island on chromosome 2L (Fig. 3). Interestingly, we observed positive ∆D values at this pericentromeric 2L region.
Figure 3.
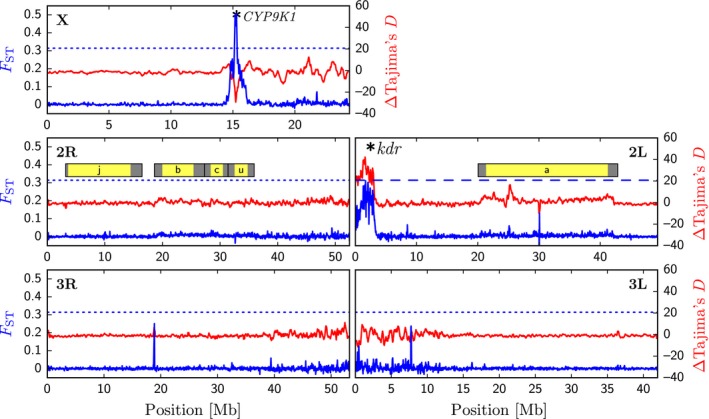
Sequence divergence between pre‐ and post‐2006 Anopheles coluzzii. Panels represent each of the three chromosomes (X, 2 and 3), as indicated by labels on the top outside corner of each box. FST were calculated in 50 kb windows with 25 kb steps comparing 12 ‘pure’ (pre‐2006) A. coluzzii and 17 kdr Anopheles coluzzii (post‐2006). FST is displayed in blue (with Gaussian smoothing). The 99.9% threshold for FST is indicated with a dashed line. The standardized difference of D (∆D) is a relative Tajima's D statistic (Bigham et al. 2010) shown in red. Negative ∆D values indicate fewer haplotypes than expected in post‐2006 samples vs. pre‐2006. The positions of common inversions are shown for reference and indicated in yellow with breakpoints highlighted in grey. Genes of interest are labelled with a star.
The second differentiated region had high F ST and negative ∆D values that were centred at approximately 15.24 Mb on the X chromosome. To approximately estimate the size of the region under selection in post‐2006 A. coluzzii, we examined a zoomed‐in region from 14.6 Mb to 16 Mb with F ST at higher resolution (5 kb windows and 1 kb steps). Peak F ST values (>95%ile) span a 156‐kb region (Fig. S1, Supporting information) including 4 genes: an uncharacterized gene (AGAP012997), a P450 gene (CYP9K1), a terminal gap gene (Tailless) and a cuticular protein (CPR125). We detected 30 nonsynonymous mutations among these genes. However, only one (I221T; rs5558865), located in AGAP012997, was associated with the selective sweep (Table S2, Supporting information). This gene has four known paralogs (AGAP013173, AGAP000816, AGAP000817 AGAP013424) located nearby (within 200 kb), but a well‐characterized orthologue was not identified. Although we can rule out nonsynonymous mutations in the remaining three genes associated with the selective sweep, it is possible that adaptive regulatory variation at these genes is the target of selection. Thus, CYP9K1, Tailless and CPR125 remain important candidate genes under selection.
Visual inspection of long paired‐end reads using Integrative Genomics Viewer (IGV) revealed haplotype‐specific SNPs in the transcription start sites, intron and 3′UTR of CYP9K1 and Tailless. To estimate the frequency of the highly selected haplotype in the populations, we used a custom Sequenom iPLEX genotyping assay to sequence 6 SNPs that span the CYP9K1 3′UTR and the Tailless gene, including two SNPs in the CYP9K1 5′ transcription start site (−47 bp, −100 bp). In parallel, we genotyped kdr and species‐specific SNPs in the speciation islands on each chromosome (Supplemental Data). Using this multiplex genotyping data, we identified three common haplotypes at the CYP9K1 region (see methods) and estimated haplotype frequencies by species and collection year. Because CYP9K1 is a primary candidate gene, we refer to the haplotypes as cyp‐l, cyp‐ll and cyp‐lll (Table 1). The genotype at synonymous Tailless SNPs were perfectly correlated with CYP9K1 genotypes (r 2 = 1; N = 27 A. coluzzii and N = 21 A. gambiae from 2010), indicating that this highly selected haplotype spans at least CYP9K1 and Tailless, but appears to commonly span approximately 156 kb in A. coluzzii collected in 2012 (Fig. S1, Supporting information).
Table 1.
Frequency of CYP9K1 haplotypes in pre‐ and post‐2006 individuals. To rule out the possibility of introgression of the cyp‐l haplotype from Anopheles gambiae, we genotyped 5 informative SNPs that span CYP9K1 and Tailless (+9.4 kb) to assess the frequency of the cyp‐l haplotype in Anopheles coluzzii and A. gambiae pre‐ and post‐2006. Note that cyp‐l was common in A. coluzzii (4–23%) and not detected in A. gambiae prior to the 2006 hybridization event. Also, cyp‐ll was more common that cyp‐l pre‐2006
| Frequency of CYP9K1 haplotypes, % | ||||||||
|---|---|---|---|---|---|---|---|---|
| A. coluzzii | A. gambiae | |||||||
| 2002 | 2004 | 2010 | 2014 | 2002 | 2004 | 2010 | 2014 | |
| cyp‐l | 4 | 23 | 85 | 99 | 0 | 0 | 0 | 9 |
| cyp‐ll | 46 | 31 | 4 | <1 | 0 | 0 | 0 | 0 |
| cyp‐lll | 48 | 39 | 12 | 0 | 100 | 97 | 100 | 87 |
| cyp‐other | 2 | 7 | 0 | <1 | 0 | 3 | 0 | 4 |
| N | 26 | 35 | 26 | 158 | 13 | 18 | 21 | 21 |
Distinguishing Introgression from selection on standing variation
To determine whether the differentiated locus on the X chromosome was due to introgression, like the previously described introgression of kdr and 2L island on chromosome 2 (Norris et al. 2015), we assessed the frequency of the cyp haplotypes and kdr in the A. coluzzii and A. gambiae specimens collected from 2002 to 2014 using the iPLEX SNP genotyping assay described above (Table 1). In total, we genotyped 87 A. coluzzii and 52 A. gambiae individuals. The cyp‐l haplotype has increased in frequency in A. coluzzii from 4% in 2002 to 99% in 2014. The cyp‐lll haplotype was the predominant haplotype (>89%) in A. gambiae in all years. The cyp‐l haplotype was not detected in A. gambiae prior to 2014 (N = 52).
Insecticide resistance bioassays
To determine if elevated insecticide resistance is associated with the increase in the relative frequency of kdr:cyp‐I A. coluzzii individuals, we performed insecticide resistance bioassays (see methods) on 4 recently derived mosquito colonies representing the following homozygous genotypes (species: CYP9K1 haplotype: kdr status): (i) A. coluzzii:cyp‐l:kdr, (ii) A. coluzzii:cyp‐l:wt, (iii) A. coluzzii:cyp‐ll:wt and (iv) A. gambiae:cyp‐lll:kdr. Some genotypes (e.g. A. gambiae:cyp‐lll:wt and A. coluzzii:cyp‐ll:kdr) were not evaluated because they were not present in our collection. The cyp‐l:kdr A. coluzzii individuals were significantly more resistant than cyp‐l:wt A. coluzzii (t‐test; P = 0.01) and cyp‐lll:kdr A. gambiae (t‐test; P < 0.0001) at both KD50 and KD90 (Fig. 4). The cyp‐l:wt A. coluzzii (N = 40) genotype is trending towards slightly less resistance than cyp‐ll:wt A. coluzzii (N = 7), but more replicates are needed to assess significance (dashed bar; Fig. 4). For external reference, cyp‐l:kdr A. coluzzii was also highly resistant compared the cyp‐ll:wt A. coluzzii MOPTI colony (MRA‐763; 0% vs. 100% KD at 30 min; N = 20, 19). Resistance was not limited to permethrin as cyp‐l:kdr A. coluzzii were also more resistant than cyp‐lll:kdr A. gambiae under deltamethrin exposure (12.5 μg/bottle; 0% vs. 85% KD at 30 min; N = 20, 20). The bioassays in Fig. 4 were performed on a mix of male and females due to sample size limitations with some genotypes. As there may be gender‐specific differences in KD times, more bioassays are needed to accurately quantify the relative contributions of the three CYP9K1 haplotypes to insecticide resistance with or without kdr.
Figure 4.
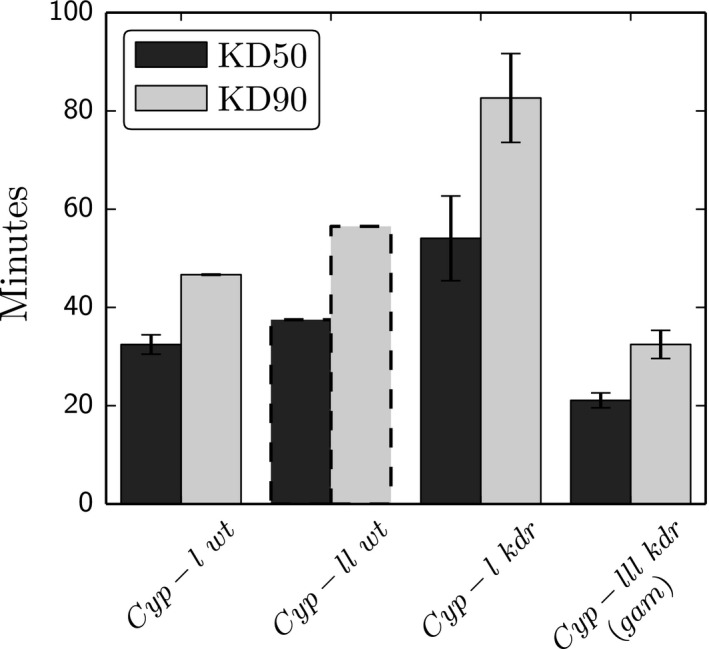
Insecticide resistance bioassay results. KD 50 is the estimated time (minutes) when 50% of the mosquitoes in a given insecticide (permethrin) coated bottle were nonresponsive (knocked down) after holding the bottle horizontally and rotating the bottle 360 degrees. These mixed gender assays were first‐generation offspring from field‐collected females. Shown are mean KD 50 (black) and KD 90 (light grey) for specific genotypes with standard error. The CYP9K1 haplotype (cyp‐l, ll or lll) and kdr status are noted below each bar. Note the small sample size of cyp‐ll:wt (dashed bar; one replicate of N = 7).
Analysis of copy number variation
To test for copy number variation specifically at the CYP9K1 region in (i) A. gambiae, (ii) pre‐2006 A. coluzzii and (iii) post‐2006 A. coluzzii, we examined relative sequencing depth across the genome in 200 bp bins for each individual (see Methods). Using this approach, we did not detect multiple copies at the CYP9K1 region in pre‐2006 A. coluzzii or post‐2006 A. coluzzii. However, a 21‐kb region, including CYP9K1 in A. gambiae, had normalized read depth of 2.5 (t‐test P‐value <0.0001, N = 7), indicating a duplication at this region in A. gambiae (Table S2, Supporting information). As A. gambiae is nearly fixed for cyp‐lll, individual paralogs within its genome are likely not confounding our genotyping results.
Discussion
Adaptive introgression of kdr
Using fixed SNP markers within the pericentromeric ‘speciation islands’ on X, 2L and 3L, a longitudinal study identified punctuated bursts of F1 hybrids between Anopheles coluzzii and Anopheles gambiae in Selenkenyi, Mali (Lee et al. 2013b). In 2006, local ITN use dramatically increased coincident with a particularly large burst of hybrids (Fig. 1). By 2010, linkage disequilibrium (LD) between the X and 3L islands was re‐established, but the entire 2L island was lost, replaced by the A. gambiae island containing kdr (Fig. 2). It has been hypothesized that increased ITN usage altered the fitness landscape, resulting in a relative fitness increase of normally unfit hybrid genotypes (Norris et al. 2015). The subsequent increase in relative abundance of A. coluzzii (Fig. 1) and elevated insecticide resistance of kdr A. coluzzii individuals (Fig. 4) indicate that this introgression event is adaptive. The burst of typically unfit hybrids detected in 2006 (Lee et al. 2013b) likely produced myriad mixed genotypes upon which strong selection could act. Ultimately, only A. coluzzii (based on fixed X chromosome markers) that had the 2L introgression (with kdr) from A. gambaie and the A. coluzzii 3L island persisted in the population, resulting in the re‐establishment of LD between the X and 3L islands. This is an important observation because the maintenance of LD between unlinked loci in the face of gene flow is a critical requirement for divergence with gene flow (i.e. the speciation islands model).
Introgression vs. selection on standing variation
For an initial assessment of sequence differentiation between pre‐ and post‐2006 A. coluzzii, we analysed F ST in sliding windows (see Methods). This approach revealed a prominent differentiated region at the expected kdr locus and linked 2L island and a second region at approximately 15.24 Mb on the X chromosome, centred at CYP9K1. To reveal signatures of recent selection in the A. coluzzii genome, we used ∆D, a relative Tajima's D statistic (see Methods). The relationship between F ST and ∆D was different between the X and 2L regions. At the known introgression on 2L, there was a positive relationship, with elevated F ST and ∆D. The positive Tajima's D trend in post‐2006 A. coluzzii is likely due to the increased representation of individuals that were heterozygous for the 3 Mb island (10/17 were heterozygous for kdr). In addition, model‐based estimates predict that the acquisition of a selected allele from a differentiated population (like kdr from A. gambiae) can result in elevated Tajima's D values at sequences linked to the selected allele (Santiago & Caballero 2005).
On the X, ∆D and F ST were negatively associated. The dip in ∆D in this case was due, at least in part, to the near fixation of a haplotype (cyp‐l hereafter) in post‐2006 A. coluzzii, which spans at least four genes including AGAP013173, CYP9K1, Tailless and CPR125. The cyp‐l haplotype was present in A. coluzzii at 23% in 2004 (N = 35) and was not detected in A. gambiae prior to 2014 (N = 52), indicating that selection acted upon standing variation within A. coluzzii (Table 1). Continued population sampling is needed to assess whether the cyp‐l A. gambiae genotype detected in 2014 will be selected for in the population and warrant further study. We hypothesize that selection has acted upon cis‐regulatory variation at CYP9K1 because (i) CYP9K1 has been shown to be upregulated in resistant anopheline mosquitoes (Tene et al. 2013; Mulamba et al. 2014), (ii) the dramatic increase in the cyp‐l haplotype was coincident with the increase in ITN usage in 2006, and (iii) bioassay results confirm that kdr alone cannot account for the level of resistance observed for kdr:cyp‐l A. coluzzii genotype. That said, the nonsynonymous mutation in AGAP013173 and regulatory variation at Tailless or CPR125 should also be considered good candidates for selection.
Evidence for synergistic epistasis
In 2004, the A. coluzzii population was kdr susceptible (N = 61) and variation at the CYP9K1 locus was composed of three major haplotypes: cyp‐l (23%), cyp‐ll (31%) and cyp‐lll (39%) (N = 35; Table 1). After 2006, the cyp‐I:kdr genotype appears to be approaching fixation in A. coluzzii and the cyp‐II genotype with the kdr introgression (cyp‐ll:kdr) was never observed in this species. This is despite the fact that the frequency of cyp‐ll (31%) was higher than cyp‐l (23%) in pre‐2006 A. coluzzii. Insecticide resistance bioassays indicate that the cyp‐l A. coluzzii with the kdr introgression is more resistant than any other genotype tested, including A. gambiae (cyp‐lll) with kdr (Fig. 4). This result supports the hypothesis that selection for insecticide resistance is contributing to the observed changes in the modern A. coluzzii genome and altering long‐standing species dynamics in Selenkenyi, Mali (Fig. 1). This is also evidence that resistance in A. coluzzii is more complex than kdr alone as A. gambiae has kdr, but is less resistant and has not increased in relative frequency as of 2014 estimates (Fig. 1). Bioassay results comparing kdr‐susceptible colonies (or families) with homozygous cyp‐I or cyp‐II genotypes indicate that cyp‐I may not offer increased resistance compared to cyp‐II in the absence of kdr (Fig. 4). So why did the cyp‐l haplotype dramatically increase in relative frequency? One hypothesis is that the exclusive selection for cyp‐l in the presence of kdr is due to an allele‐specific interaction between kdr and cyp‐I resulting in a nonadditive increase in resistance. Synergistic epistasis between P450 genes and kdr has been described previously in Culex mosquitoes (Hardstone et al. 2008), further supporting the possibility that CYP9K1 is the target of selection within the cyp‐l haplotype. Another hypothesis is that alleles in the cyp‐l haplotype were selected for because they were the most compatible with the A. gambaie alleles in the 2L island in nature. We also cannot rule out the possibility that selection for the cyp‐l haplotype could have been independent of the 2L introgression and kdr (no epistasis).
Understanding the genetic basis for the potential interaction between kdr and CYP9K1 is important because the World Health Organization recently suggested that insecticide resistance via the combination of kdr and elevated P450 activity represents the biggest threat to vector control for malaria in Africa (WHO 2012). The latest ITNs add piperonyl‐butoxide (PBO), a general P450 inhibitor, to better combat complex insecticide resistance (e.g. PermaNet® 3.0). Functional verification of the nonsynonymous mutation in AGAP013173 as well as regulatory variation in the four candidate genes within the cyp‐l haplotype is an important next step beyond this study to confirm that selection is acting on CYP9K1 and/or other linked loci. For example, allele‐specific gene expression assays between pairwise hybrids of the cyp‐l, cyp‐ll and cyp‐lll haplotypes would be ideal to estimate the effects of the 5′ proximal SNPs on gene expression.
We described evidence for elevated copy number at the CYP9K1 region exclusively in A. gambiae. Variation in P450 copy number appears common between species (Good et al. 2014). A population‐scale analysis of copy number variation at the CYP9K1 region in A. gambiae (e.g. via qPCR) would reveal whether the detected duplication is fixed in the population and whether higher copy genotypes exist. It would be interesting if the lack of cyp‐l in A. gambiae is partially compensated for by an A. gambiae ‐specific increase in copy number, but bioassay data indicate that cyp‐lll:kdr A. gambiae is significantly less resistant to insecticide than cyp‐l:kdr A. coluzzii (Fig. 4). Thus, this copy number variation may be unrelated to insecticide resistance. Metabolic studies would also be very informative as CYP9K1 is closely related to, but not among the several P450 genes that have been proven to metabolize pyrethroid insecticides in vitro (Müller et al. 2008; Stevenson et al. 2011).
Intraspecific mating fidelity and evolutionary responsiveness
Anopheles gambiae, Ag‐Bamako and A. coluzzii have fairly consistently represented 10%, 25% and 65%, respectively, of the mosquito population at Selinkenyi, based on collection data from 1980 to 2006 (Fig. 1). Ag‐Bamako appears to be part of an adaptive radiation in the A. gambiae species complex via its adaptation in the larval stage to riverine rock pools (Toure et al. 1998; Manoukis et al. 2008). The time since divergence between Ag‐Bamako and A. gambiae is thought to be much more recent than that between A. gambiae and A. coluzzii (Taylor et al. 2001; Slotman et al. 2006). Ag‐Bamako is identified primarily by the presence of the j inversion on chromosome 2R and is A. gambiae ‐like on the X chromosome. Based on karyotype and genotype data, we demonstrate that the longstanding species dynamics between these three populations has changed following the start of the major ITN campaign in 2006 (Fig. 1). In 2002 and 2004, A. coluzzii and Ag‐Bamako populations were kdr susceptible (N = 142 and 43, respectively), whereas the kdr frequency in A. gambiae was approximately 50% (N = 12). In post‐2006 samples, leaky reproductive barriers between A. gambiae and A. coluzzii and strong selection resulted in the stable introgression of kdr into A. coluzzii. The kdr A. coluzzii genotype is now significantly more resistant to insecticides (Fig. 4) and has increased in relative frequency in the population from 65% to 97% in 2014 (N = 179; Fig. 1). We suggest that the brief increase in the relative frequency of A. gambiae in the population after 2006 may be due to the presence of relatively unfit early‐stage mixed A. coluzzii genotypes. Unlike A. gambiae and A. coluzzii, reproductive isolation appears to be nearly complete between Ag‐Bamako and the other two taxa in Mali (Toure et al. 1998; Powell et al. 1999; Manoukis et al. 2008). The high mating fidelity in Ag‐Bamako appears to have prevented the acquisition of kdr from either A. coluzzii or A. gambiae (N = 7; Fig. 2), which may be responsible for its steady decline towards local extinction in Selinkenyi, Mali. Thus, unstable reproductive barriers resulting in adaptive introgression of kdr appear to have enabled A. coluzzii to adapt to a rapid environmental change (i.e. increased ITN use) and even out‐compete Ag‐Bamako, a population associated with the donor species, A. gambiae.
Conclusion
Our results indicate that extant kdr A. coluzzii populations in Mali are highly resistant to insecticides (both permethrin and deltamethrin) and have increased in relative frequency in the presence of increased ITN usage. We hypothesize that this elevated insecticide resistance is due to interactions between the introgressed kdr allele and allele/s within the cyp‐l haplotype on the X chromosome, which was already present in the A. coluzzii population. Thus, surveys of insecticide resistance in malaria vectors may benefit from assessing the population frequencies of both kdr and the cyp‐l haplotype. That said, it is also possible that increased ITN use drove the introgression of the A. gambiae 2L island containing kdr, and then, variable genetic incompatibilities between A. gambiae alleles in the 2L island and alleles in the CYP9K1 region resulted in fixation of the most amenable haplotype (cyp‐l). We also cannot rule out selection from other environmental changes, for example climate change or new pathogens. The remarkable adaptive radiation of the A. gambiae mosquito complex and leaky reproductive barriers between species may underlie their resilience to rapid environmental changes and ultimately their persistence through prehistory.
B.J.M., Y.L. and G.C.L. designed research; B.J.M., Y.L. and T.C.C. performed the data analysis; G.C.L., A.F. and A.J.C. carried out field collections; A.F. and A.J.C. performed the cytogenetic analysis; B.J.M. and K.B. performed the bioassays; B.J.M., Y.L., A.J.C., L.C.N. and G.C.L. wrote the paper.
Data accessibility
Illumina sequencing data were deposited in SRA at NCBI under accession number SRP063464. Variant data has been deposited in Dryad (http://dx.doi.org/10.5061/dryad.f3dn2). Other data associated with this study are available in supporting information and in the PopI open online database (https://popi.ucdavis.edu/PopulationData/OpenProjects/AgKDR/).
Supporting information
Table S1 Genome sequencing reads per sample.
Table S2 Candidate nonsynonymous mutations.
Table S3 Sequenome iPLEX primer design.
Fig. S1 Estimating the size of the cyp‐l haplotype.
Data S1 Haplotype estimates from genotype data at the CYP9K1 region.
Data S2 Bioassay data.
Data S3 Copy number analysis using cnvnator (v0.3).
Acknowledgements
We would like to thank Catelyn Neiman and Allison M. Weakley and Youki Yamasaki for assisting with DNA extraction and mosquito rearing, Julia Malvick at the UC Davis Veterinary Genetics Laboratory for performing iPLEX SNP genotyping, Stephanie Seifert and Dr. Rebecca Trout Fryxell for their assistance in field collection in Selinkenyi, Mali, in 2010 and three anonymous reviewers for helpful comments on this manuscript. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R21AI117174, D43TW007390, R01AI078183 and T32AI074550. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Abyzov A, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Research, 21, 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Denholm I, Bromilow RH (2006) Delayed cuticular penetration and enhanced metabolism of deltamethrin in pyrethroid‐resistant strains of Helicoverpa armigera from China and Pakistan. Pest Management Science, 62, 805–810. [DOI] [PubMed] [Google Scholar]
- Anderson TM, Vonholdt BM, Candille SI et al (2009) Molecular and evolutionary history of melanism in North American gray wolves. Science, 323, 1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FASTQC. A quality control tool for high throughput sequence data. 2010.
- Awolola TS, Oduola OA, Strode C et al (2009) Evidence of multiple pyrethroid resistance mechanisms in the malaria vector Anopheles gambiae sensu stricto from Nigeria. Transactions of the Royal Society of Tropical Medicine and Hygiene, 103, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D et al (2010) Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics, 6, e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogdon WG, McAllister JC (1998) Simplification of adult mosquito bioassays through use of time‐mortality determinations in glass bottles. Journal of the American Mosquito Control Association, 14, 159–164. [PubMed] [Google Scholar]
- Clarkson CS, Weetman D, Essandoh J et al (2014) Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nature Communications, 5, 4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R et al (2013) Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa, 3619, 246–274. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Di Deco MA (1979) Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Transactions of the Royal Society of Tropical Medicine and Hygiene, 73, 483–497. [DOI] [PubMed] [Google Scholar]
- Corbel V, N'Guessan R, Brengues C et al (2007) Multiple insecticide resistance mechanisms in Anopheles gambiae and Culex quinquefasciatus from Benin, West Africa. Acta Tropica, 101, 207–216. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD et al (2012) Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature, 487, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J‐P, Strode C, Vontas J et al (2005) The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic‐based insecticide resistance in malaria vectors. Proceedings of the National Academy of Sciences of the United States of America, 102, 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Torre A, Tu Z, Petrarca V (2005) On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochemistry and Molecular Biology, 35, 755–769. [DOI] [PubMed] [Google Scholar]
- Diabaté A, Dao A, Yaro AS et al (2009) Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae . Proceedings of the Royal Society of London B: Biological Sciences, 276, 4215–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouaka RF, Bakare AA, Coulibaly ON (2008) Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics, 9, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della TA (2002) Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR‐RFLP. Medical and Veterinary Entomology, 16, 461–464. [DOI] [PubMed] [Google Scholar]
- Favia G, della Torre A, Bagayoko M et al (1997) Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Molecular Biology, 6, 377–383. [DOI] [PubMed] [Google Scholar]
- Favia G, Lanfrancotti A, Spanos L (2001) Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Molecular, 10, 19–23. [DOI] [PubMed] [Google Scholar]
- Flaxman AD, Fullman N, Otten MW et al (2010) Rapid scaling up of insecticide‐treated bed net coverage in Africa and its relationship with development assistance for health: a systematic synthesis of supply, distribution, and household survey data. PLoS Medicine, 7, e1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimonneau G, Pombi M, Dabire RK, Diabate A (2012) Behavioural responses of Anopheles gambiae sensu stricto M and S molecular form larvae to an aquatic predator in Burkina Faso. Parasit Vectors, 5, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good RT, Gramzow L, Battlay P et al (2014) The molecular evolution of cytochrome P450 genes within and between Drosophila species. Genome Biology and Evolution, 6, 1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardstone MC, Leichter CA, Scott JG (2008) Multiplicative interaction between the two major mechanisms of permethrin resistance, kdr and cytochrome P450‐monooxygenase detoxification, in mosquitoes. Journal of Evolutionary Biology, 22, 416–423. [DOI] [PubMed] [Google Scholar]
- Hedrick PW (2013) Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology, 22, 4606–4618. [DOI] [PubMed] [Google Scholar]
- Hemingway J (2000) The molecular basis of two contrasting metabolic mechanisms of insecticide resistance. Insect Biochemistry and Molecular Biology, 30, 1009–1015. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H (2000) Insecticide resistance in insect vectors of human disease. Annual Review of Entomology, 45, 371–391. [DOI] [PubMed] [Google Scholar]
- Hunt RH (1973) A cytological technique for the study of Anopheles gambiae complex. Parassitologia, 15, 137–139. [PubMed] [Google Scholar]
- Hunter JD (2007) Matplotlib: A 2D graphics environment. Computing in Science & Engineering, 9, 90–95. [Google Scholar]
- Kamdem C, Tene Fossog B, Simard F et al (2012) Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae . PLoS ONE, 7, e39453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MKN, Emrich SJ, Holloway AK et al (2010) Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science, 330, 512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Collier TC, Sanford MR et al (2013a) Chromosome inversions, genomic differentiation and speciation in the African malaria mosquito Anopheles gambiae . PLoS ONE, 8, e57887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Norris LC et al (2013b) Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae . Proceedings of the National Academy of Sciences of the United States of America, 110, 19854–19859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM. arXiv.org.
- Manoukis NC, Powell JR, Toure MB et al (2008) A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae . Proceedings of the National Academy of Sciences of the United States of America, 105, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden CD, Lee Y, Nieman CC (2011) Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Molecular Ecology, 20, 4983–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Research, 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LA, Niazi U, Bibby J et al (2008) Characterization of inhibitors and substrates of Anopheles gambiae CYP6Z2. Insect Molecular Biology, 17, 125–135. [DOI] [PubMed] [Google Scholar]
- Milliner J (2009) Net Mapping Project. Alliance for Malaria Prevention, President's Malaria Initiative, United States Agency for International Development, Washington, District of Columbia. [Google Scholar]
- Mulamba C, Irving H, Riveron JM et al (2014) Contrasting Plasmodium infection rates and insecticide susceptibility profiles between the sympatric sibling species Anopheles parensis and Anopheles funestus s.s: a potential challenge for malaria vector control in Uganda. Parasites & Vectors, 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Warr E, Stevenson BJ et al (2008) Field‐caught permethrin‐resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genetics, 4, e1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namountougou M, Simard F, Baldet T et al (2012) Multiple insecticide resistance in Anopheles gambiae s.l. populations from Burkina Faso, West Africa. PLoS ONE, 7, e48412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikou D, Ranson H, Hemingway J (2003) An adult‐specific CYP6 P450 gene is overexpressed in a pyrethroid‐resistant strain of the malaria vector, Anopheles gambiae . Gene, 318, 91–102. [DOI] [PubMed] [Google Scholar]
- Norris LC, Main BJ, Lee Y et al (2015) Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide‐treated bed nets. Proceedings of the National Academy of Sciences of the United States of America, 112, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz‐Barrientos D (2009) Divergent selection and heterogeneous genomic divergence. Molecular Ecology, 18, 375–402. [DOI] [PubMed] [Google Scholar]
- Pardo‐Diaz C, Salazar C, Baxter SW et al (2012) Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genetics, 8, e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Petrarca V, della Torre A, Caccone A, Coluzzi M (1999) Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia, 41, 101–113. [PubMed] [Google Scholar]
- Puinean AM, Foster SP, Oliphant L et al (2010) Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae . PLoS Genetics, 6, e1000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H, Abdallah H, Badolo A et al (2009) Insecticide resistance in Anopheles gambiae: data from the first year of a multi‐country study highlight the extent of the problem. Malaria Journal, 8, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H, N'Guessan R, Lines J et al (2011) Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends in Parasitology, 27, 91–98. [DOI] [PubMed] [Google Scholar]
- Reidenbach KR, Neafsey DE, Costantini C et al (2012) Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biology and Evolution, 4, 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago E, Caballero A (2005) Variation after a selective sweep in a subdivided population. Genetics, 169, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, della Torre A, Caccone A (2004) Short report: a new polymerase chain reaction‐restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. The American Journal of Tropical Medicine and Hygiene, 70, 604–606. [PubMed] [Google Scholar]
- Santolamazza F, Calzetta M, Etang J et al (2008) Distribution of knock‐down resistance mutations in Anopheles gambiae molecular forms in west and west‐central Africa. Malaria Journal, 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B et al (2010) Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6 g1 locus. PLoS Genetics, 6, e1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH (1993) Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. The American Journal of Tropical Medicine and Hygiene, 49, 520–529. [DOI] [PubMed] [Google Scholar]
- Slotman MA, Mendez MM, Torre AD et al (2006) Genetic differentiation between the BAMAKO and SAVANNA chromosomal forms of Anopheles gambiae as indicated by amplified fragment length polymorphism analysis. The American Journal of Tropical Medicine and Hygiene, 74, 641–648. [PubMed] [Google Scholar]
- Song Y, Endepols S, Klemann N et al (2011) Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Current Biology, 21, 1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava H, Sharma M, Dixit J, Das A (2010) Evolutionary insights into insecticide resistance gene families of Anopheles gambiae . Infection, Genetics and Evolution, 10, 620–628. [DOI] [PubMed] [Google Scholar]
- Stelkens RB, Brockhurst MA, Hurst GDD, Greig D (2014) Hybridization facilitates evolutionary rescue. Evolutionary Applications, 7, 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. American Journal of Human Genetics, 73, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics, 68, 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Bibby J, Pignatelli P et al (2011) Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochemistry and Molecular Biology, 41, 492–502. [DOI] [PubMed] [Google Scholar]
- Taylor C, Touré YT, Carnahan J et al (2001) Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics, 157, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tene BF, Poupardin R, Costantini C et al (2013) Resistance to DDT in an urban setting: common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS ONE, 8, e61408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure YT, Petrarca V, Traore SF et al (1998) The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia, 40, 477–511. [PubMed] [Google Scholar]
- Trape J‐F, Tall A, Diagne N et al (2011) Malaria morbidity and pyrethroid resistance after the introduction of insecticide‐treated bednets and artemisinin‐based combination therapies: a longitudinal study. The Lancet Infectious Diseases, 11, 925–932. [DOI] [PubMed] [Google Scholar]
- Tripet F, Wright J, Lanzaro G (2006) A new high‐performance PCR diagnostic for the detection of pyrethroid knockdown resistance kdr in Anopheles gambiae . The American Journal of Tropical Medicine and Hygiene, 74, 658–662. [PubMed] [Google Scholar]
- Turner TL, Hahn MW (2010) Genomic islands of speciation or genomic islands and speciation? Molecular Ecology, 19, 848–850. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV (2005) Genomic islands of speciation in Anopheles gambiae . PLoS Biology, 3, e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uecker H, Setter D, Hermisson J (2014) Adaptive gene introgression after secondary contact. Journal of Mathematical Biology, 70, 1523–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, West J (2008) The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Molecular Ecology, 17, 4334–4345. [DOI] [PubMed] [Google Scholar]
- Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ (2012) Gene flow‐dependent genomic divergence between Anopheles gambiae M and S forms. Molecular Biology and Evolution, 29, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ (2010) Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae . Molecular Ecology, 19, 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2012) Global Plan for Insecticide Resistance Management in Malaria Vectors. World Health Organization Press, Geneva. [Google Scholar]
- WHO (2013) World Health Organization. Country Profile: Mali. World Health Organization Press, Geneva: Available at: http://www.who.int/countries/mli/en/. [Google Scholar]
- Willis JH (2014) Temporal and spatial expression of cuticular proteins of Anopheles gambiae implicated in insecticide resistance or differentiation of M/S incipient species. Parasites & Vectors, 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood O, Hanrahan S, Coetzee M, Koekemoer L, Brooke B (2010) Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus . Parasites & Vectors, 3, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Genome sequencing reads per sample.
Table S2 Candidate nonsynonymous mutations.
Table S3 Sequenome iPLEX primer design.
Fig. S1 Estimating the size of the cyp‐l haplotype.
Data S1 Haplotype estimates from genotype data at the CYP9K1 region.
Data S2 Bioassay data.
Data S3 Copy number analysis using cnvnator (v0.3).
Data Availability Statement
Illumina sequencing data were deposited in SRA at NCBI under accession number SRP063464. Variant data has been deposited in Dryad (http://dx.doi.org/10.5061/dryad.f3dn2). Other data associated with this study are available in supporting information and in the PopI open online database (https://popi.ucdavis.edu/PopulationData/OpenProjects/AgKDR/).


