Abstract
MicroRNA-21 is dysregulated in many cancers and fibrotic diseases. Since miR-21 suppresses several tumor suppressor and anti-apoptotic genes, it is considered a cancer therapeutic target. Antisense oligonucleotides are commonly used to inhibit a miRNA; however, blocking miRNA function via an antagomir is temporary, often only achieves a partial knock-down, and may be complicated by off-target effects. Here, we used transcription activator-like effector nucleases (TALENs) to disrupt miR-21 in cancerous cells. Individual deletion clones were screened and isolated without drug selection. Sequencing and quantitative RT-PCR identified clones with no miR-21 expression. The loss of miR-21 led to subtle but global increases of mRNAs containing miR-21 target sequences. Cells without miR-21 became more sensitive to cisplatin and less transformed in culture and in mouse xenografts. In addition to the increase of PDCD4 and PTEN protein, mRNAs for COL4A1, JAG1, SERPINB5/Maspin, SMAD7, and TGFBI – all are miR-21 targets and involved in TGFβ and fibrosis regulation – were significantly upregulated in miR-21 knockout cells. Gene ontology and pathway analysis suggested that cell-environment interactions involving extracellular matrix can be an important miR-21 pathogenic mechanism. The study also demonstrates the value of using TALEN-mediated microRNA gene disruption in human pathobiological studies.
Keywords: microRNA, miR-21, TALEN, gene editing, cancer, extracellular matrix, fibrosis
1. INTRODUCTION
MicroRNA-21 (miR-21) is one of the first mammalian miRNAs identified [34] and has received great attention for its involvement in cancer, cardiovascular disease, organ fibrosis, and immune response [31, 33]. miR-21 is up-regulated in virtually all cancers examined to date, and many miR-21 targets are tumor suppressors [7, 62]. The tumorigenic role of miR-21 has been tested in mouse models where shutting down miR-21 expression inhibits growth of lymphoma and lung tumors that were induced by miR-21 over-expression [21, 42]. Mouse models are useful for dissecting human diseases, but there are ample examples showing limitations of applying results obtained from mouse studies to human conditions [52]. Thus, a direct analysis of miR-21 in human cells is critical for therapeutic development.
Antisense oligonucleotides or antagomirs are perhaps the most commonly used method to inhibit a miRNA [32]. However, blocking miRNA function via an antagomir is temporary, often only achieves a partial knock-down, and may be complicated by off-target effects [28]. For example, miR-21 is up-regulated in response to cardiac injury and is consistently deregulated under various cardiovascular pathological conditions [12, 60]. When the myopathological role of miR-21 was investigated in mouse models, anti-miR-21 oligonucleotides were shown to be effective in attenuating cardiac fibrosis [56]; however, it was later found that a miR-21 knockout mouse still developed cardiac hypertrophy [48]. Thus, the investigation of a microRNA function would be more effective when the gene itself can be inactivated through direct gene editing. Three gene editing tools have recently been developed: zinc-finger nucleases, transcription activator-like effector nucleases, and RNA-guided CRISPR/cas9 nucleases [14, 24, 58]. These sequence-specific nucleases have made it possible to directly modify genes in human cells [18, 49].
Transcription activator-like effectors (TALEs) from Xanthomonas have been engineered to bind to specific DNA sequences of interest [5, 8]. The DNA binding specificity resides in 34-amino-acid repeats that can be assembled to recognize a specific DNA sequence. Fusing TALE to the nuclease domain of FokI converts the fusion protein into a TALE-nuclease (TALEN). To cut DNA, one TALEN binds upstream and another TALEN binds downstream of a target sequence so that the FokI nuclease domains can dimerize and become active. A pair of TALENs when introduced into cells will generate DNA double-stranded breaks (DSBs) at the target site; the resulted DSB can then be repaired by non-homologous end joining (NHEJ) or by homologous recombination (HR) [37]. DSB repair by NHEJ often causes insertions or deletions, resulting in targeted mutations. TALENs have been used to create site specific gene modification in plant cells, yeast, animals, and human pluripotent stem cells [24, 44].
To investigate miR-21 function in cancerous cells, we constructed 3 pairs of miR-21 targeting TALENs and used them to delete the miR-21 sequences. By analyzing single cell-derived miR-21 knockout clones, we found HeLa cells lacking miR-21 were phenotypically less transformed and more sensitive to cisplatin. We also compared the gene expression profiles of TALEN-mutagenized miR-21 disrupted clones by RNA deep sequencing. Genes and pathways that are involved in cell adhesion, extracellular matrix, and metabolism were significantly affected by the loss of miR-21. Our study indicates that alteration of cell-environment interaction may contribute to the pathogenic role of miR-21 in cancer and fibrosis, as well as demonstrates that the function of a microRNA gene can be studied in human cells using TALEN-induced gene disruption.
2. MATERIALS and METHODS
2.1. TALEN design and assembly
All TALENs were designed using TALEN Targeter 2.0 (https://TALE-nt.cac.cornell.edu/) and were assembled using the Golden Gate TALEN Kit (Addgene) as described [8]. Intermediary RVD plasmids were verified by AflII and XbaI digestions, and the complete RVD sequences were ligated into a CMV-TALEN vector and verified by a BspEI digestion. The final TALEN plasmids were confirmed by DNA sequencing using two TAL primers: 5’-CATCGCGCAATGCACTGAC and 5’-GGCGACGAGGTGGTCGTTGG.
2.2. Cell culture and transfection
Human cervical carcinoma HeLa cells were maintained in Dulbecco's modified Eagle's medium (Corning) supplemented with 10% (v/v) fetal bovine serum (Hyclone) in a humidified incubator with 5% CO2 at 37°C. Cells were transfected at 90% confluency using lipofectamine™ 2000 (Invitrogen). A transfection mixture containing 4 μl of lipofectamine 2000, 1.6 μg of TALEN plasmid DNAs, and 100 μl of Opti-MEM was used for cells in one 12-well.
2.3. Surveyor Nuclease assay
TALEN's ability to cleave their target genomic DNA was determined using a Surveyor Nuclease assay. Briefly, genomic DNA was extracted from cells using QIAamp DNA Mini kit (Qiagen) 3 days after TALEN transfection. The targeted locus was amplified by PCR for 35 cycles using two primers: miR-21-F1: 5’-TGGGGTTCGATCTTAACAGG-3’and miR-21-R1: 5’-TTTCAAAACCCACAATGCAG-3’. The PCR products were heated at 95 °C for 10 min and cooled to 25°C with 0.3°C drop per second. Surveyor Nuclease (Transgenomic) was added and the digested sample was resolved on a 2% agarose gel. The DNA bands were quantified using Image J and the mutation rate in a cell population was calculated as 1 − (1 − fraction cleaved)1/2 [20].
2.4. Isolation and DNA analysis of mutant clones
HeLa cells transfected with miR-21 targeting TALENs were seeded 3 days post-transfection on 96-well plates at 1 cell/well. A portion of the cells from each colony appeared after 14-21 days was used to isolate genomic DNA using QuickExtract™ DNA Extraction Solution (Epicenter), while the remaining cells of each colony were maintained in culture. To detect deletions, genomic DNA was subjected to PCR using primers miR-21-F1 and miR-21-R1 and JumpStart™ Taq DNA Polymerase (Sigma) followed by agarose gel electrophoresis. Genomic DNA from candidate clones was further analyzed by Sanger sequencing of the PCR products. Selected mutant clones were further characterized by cloning the PCR fragments amplified with Hotstar HiFidelity DNA polymerase (Qiagen) into TOPO TA cloning vector (Invitrogen) and positive clones were then analyzed by Sanger sequencing.
2.5. RT-qPCR
Total RNA containing miRNA was extracted from cells using the mirVana miRNA isolation kit (Ambion). To quantify microRNAs, 0.5 μg RNA was reverse-transcribed to cDNA using miScript RT Kit II (Qiagen). Real-time PCR with 2 ng of cDNA was carried out using miScript SYBR Green PCR Kit (Qiagen) and appropriate primers (Qiagen) on MyIQ™ Real-Time PCR Detection System (Bio-Rad). The RNU6B RNA was used as control. All qPCR reactions were done in triplicate, and the relative expression levels were calculated using 2−ΔΔCt. Statistic analysis including standard deviation and p-value calculations was done following published methods [54].
2.6. RNA deep sequencing and data analysis
Total RNA with miRNA was extracted using mirVana (Ambion). For miR-Seq, RNA adaptors were ligated to the 3' and the 5' ends. After RT-PCR amplification and size selection, the cDNA fragments were sequenced on Illumina HiSeq 2000. Cluster analysis was done using Cluster 3.0 and the heatmap was generated using TreeView [15]. For RNA-Seq, poly(A)RNA was isolated (via oligo-dT) and reverse-transcribed. After fragmentation, cDNA fragments with sizes between 130-280 bp were isolated from polyacrylamide-urea gels. The isolated cDNA fragments were added a 3’ linker with a barcode and a 5’ linker. The ligated material was amplified by PCR and then put on Solexa HiSeq2000 for parallel sequencing. The 40-bp long single-ended sequence reads were mapped to the human genome (hg19) using TopHat, and the frequency of Refseq genes detected was counted with customized R scripts. The raw counts were then normalized using trimmed mean of M values (TMM) method [51] implemented in the Bioconductor package edgeR [50]. A gene was considered not expressed in the experimental conditions if the mean base coverage in all samples was less than 1, hence excluded in the expression analysis. Differentially expressed genes were identified based on the negative binomial model by comparing the two groups using edgeR, with p-value < 0.01 and fold-change > 1.5 as the cutoff values [6]. Gene enrichments were analyzed using DAVID (Database for Annotation, Visualization, and Integrated Discovery), which is a bioinformatic resource available online at http://david.abcc.ncifcrf.gov for functional interpretation of large lists of genes [26].
2.7. Characterization of cell phenotypes in vitro
For cell proliferation assay, HeLa cells were seeded in triplicate in 96-well plates at 3,000 cells/well in 100 μl growth medium. Cell growth was determined at indicated times by adding to each well 20 μl of tetrazolium MTS regent from CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay Kit (Promega) and measuring the 490-nm absorbance on a Bio-Tek uQuant microplate reader. For monolayer colony formation assay, HeLa cells were seeded on 10-cm culture plates at 200 cells per plate. After culturing for 21 days, plates were washed with PBS and stained with 0.05% crystal violet solution. Colonies visible to the naked eye were manually counted and the colony numbers from two separate experiments with triplicates were averaged. For soft agar colony formation assay, 2,000 HeLa cells were suspended in culture medium containing 0.35% low-melting agar and plated on a base of 0.5% agar on a 6-well plate. After culturing for 15 days, colonies were photographed and counted under 4× light microscope. The number of colonies containing more than 50 cells was determined; the average of two separate experiments of triplicate samples was calculated.
2.8. Cisplatin treatment and apoptosis assay
Cisplatin (Bristol-Myers Squibb) was added to the cell culture medium (in 96-well format) at a concentration of 1.56, 3.12, 6.25, 12.5 or 25.0 μM, and cell viability was assayed using MTS after 48 h. To measure apoptosis using Annexin-V Staining, HeLa cells (5×105 cells/well) on 6-well plates were incubated with 20 μM of cisplatin for 12 h. Cells were collected and stained using BD Pharmingen™ Annexin V: FITC Apoptosis Detection Kit II, and subjected to FACS analysis.
2.9. Mouse xenograft
Animal experiment protocols were approved by the IACUC of City of Hope. Each HeLa line was xenotransplanted into 3 immunodeficient NOD/SCID/IL2R gamma null (NSG) mice; 3 × 106 cells were inoculated subcutaneously into each mouse. Tumors were removed from mice before they reached a size of 15 mm in diameter. The tumors were fixed in formalin, sectioned, stained with hematoxylin and eosin, and examined by routine light microscopy.
3. RESULTS
3.1. TALEN-mediated disruption of the microRNA-21 gene in HeLa cells
We designed and made three pairs of TALENs, the first pair (L1-R1) targeted the seed sequence of miR-21, the second pair (L2-R2) targeted the loop of the pre-miR-21, and the third pair (L3-R3) targeted a downstream sequence of pre-miR-21 (Fig. 1A; Supplementary Fig. S1). The three TALEN pairs were tested for genomic DNA cutting by transient transfection into HeLa cells. The miR-21 region was amplified by PCR; the PCR product was denatured, re-natured, and digested with Surveyor nuclease, which cuts unpaired bases within DNA duplexes. If the genomic DNA were cut by TALENs and then repaired by non-homologous end joining (NHEJ) near the cut site, heteroduplexes formed between wild-type and a mutant (or between two different mutants) would contain mismatches and be cut into two fragments by Surveyor [20]. This was indeed the case (Fig. 1B); P1 (L1-R1) generated a lower mutation rate than P2 (L2-R2) or P3 (L3-R3) did. We also co-transfected two pairs of TALENs and found they induced higher mutation rates (Fig. 1B, P1/P3 and P2/P3). Interestingly, an additional band just below the full-length PCR product was detected in samples co-transfected with two pairs of TALENs (Fig. 1B; marked with an arrow). A similar band was also detected without Surveyor digestion, and cloning and sequencing confirmed the DNA between the two cutting sites could be deleted when two pairs of TALENs were co-transfected (Supplementary Fig. S2). Similar deletions were detected when two TALEN pairs targeting the miR-21 locus were co-transfected into the HEK293T or Ramos lymphoma cells (data not shown). Thus, disruption of miR-21 may be achieved by either using seed- or loop-cutting TALENs or by “double-digestion” with two pairs of TALENs.
Figure 1. TALEN design and miR-21 disrupted clones generated by co-transfection with the L1-R1 and L3-R3 TALEN pairs.
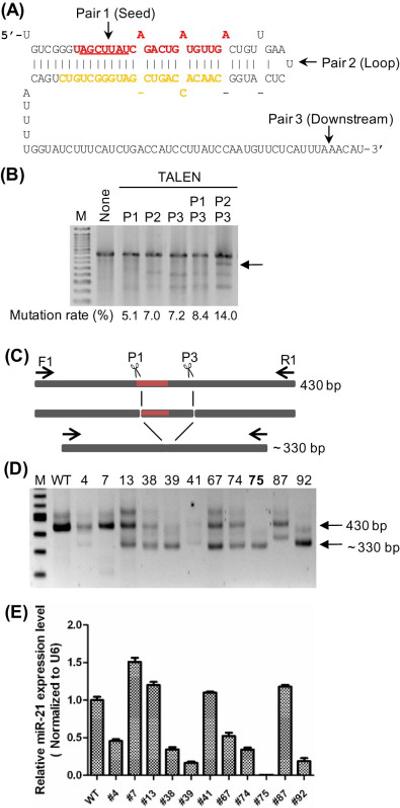
(A) TALEN cutting sites. Part of the miR-21 primary transcript is shown; miR-21 (miR-21-5p) is in red with the seed sequence (position 2 to 8) underlined and miR-21* (miR-21-3p) is in orange. The approximate cutting sites for TALEN pair 1 (L1-R1), pair 2 (L2-R2), and pair 3 (L3-R3) are labeled with arrows. (B) TALEN cutting efficiency as reflected by mutation rates. Plasmids carrying TALEN constructs were transfected into HeLa cells, genomic DNA was subjected to PCR and Surveyor digestion and run on agarose gels. M is a 50-bp marker ladder and the arrow points to the band corresponding to a deletion between two TALEN cutting sites. (C) Hypothetical NHEJ events upon TALEN P1 and P3 cutting at the miR-21 locus. The pri-miR-21 region is filled with dark red; F1 and R1 are primers used for PCR screening. (D) Genomic DNA was isolated from single cell clones (numbers on top) and subjected to PCR using F1 and R1. WT is the parental HeLa line, M is a 100-bp marker ladder, and arrows point to bands corresponding to the 430-bp full-length PCR product and the ~330-bp deletion products. (E) Mature miR-21 levels were measured by real-time RT-qPCR and normalized to the levels of the U6 snRNA. The level of miR-21 in clone #75 is significantly lower than the wild-type level with p<0.001.
We then used “double-digestion” by co-transfecting HeLa cells with TALEN pairs L1-R1 and L3-R3 (Fig. 1C) and isolated single cell clones. A screen of 92 clones by genomic DNA-PCR identified 11 clones carrying deletions in the miR-21 locus (Fig. 1D); one of the deletion clones, #75, appeared not to contain any intact miR-21 allele. Seven deletion-carrying clones expressed a low amount of miR-21 as determined by quantitative RT-PCR, and clone #75 expressed less than 0.1% of the wild type level (Fig. 1E). We further characterized the miR-21 locus of clones #38, #39, #75, and #92 by cloning the PCR products and sequencing. An intact pre-miR-21 sequence was detected in clones #38, #39, and #92, while in clone #75 only disrupted pre-miR-21 sequence was found (Supplementary Fig. S3). While most clones we characterized have three miR-21 alleles, we only recovered two different alleles in clones #75 and #92; these two clones could either lose one of the three alleles or have two identical alleles through recombination [4].
To further verify the loss of miR-21 expression, we sequenced small RNAs from the parental line (WT), clone #39, and clone #75 by next generation sequencing. A heatmap comparing the expression of the top 50 highly expressed microRNAs in WT showed that both miR-21-5p and miR-21-3p were decreased in clone #39 but depleted in clone #75, while the other 48 microRNAs (except for miR-143-3p) were largely unaffected (Fig. 2A). Although the expression of miR-143-3p was higher in clone #75, it was actually lower in clone #39 as compared to the parental line. By measuring miR-143-3p in other miR-21 disrupted clones, we did not observe a strong correlation between these two miRNAs (data not shown). Thus, the TALEN-mediated mutagenesis specifically disrupted the miR-21 expression.
Figure 2. Expression of microRNA and poly(A)RNA in miR-21 disrupted clones.
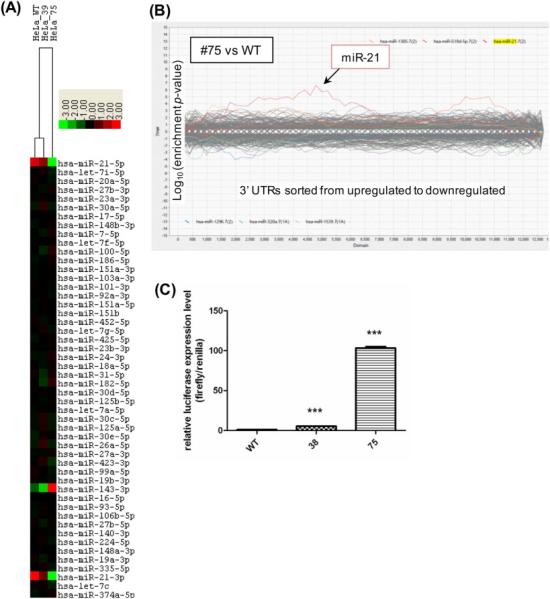
(A) A heatmap of the miR-Seq data from HeLa WT and disrupted clones #39 and #75. The 50 most abundant microRNAs were ranked and clustered by centering on the row-wise mean log(2) value [17]. A scale of 8-fold deviation from the mean is shown. (B) Sylamer analysis of gene expression changes. Poly(A) RNA was quantified by deep sequencing and the expression changes between miR-21 disrupted clones #75 and the WT were analyzed using Sylamer. Gene expression changes were ranked from increase to decrease (left to right) and enrichment p-value was determined for all possible 7-mers in the genes’ 3’UTR. Genes carrying a miR-21 target sequence, miR-21 7(2) or miR-21 7(1A), were over-represented in the up-regulated region in both cases. (C) Expression of a luciferase reporter with a miR-21 target sequence in clones #38 and #75. A miR-21 reporter plasmid carrying a firefly luciferase with 3 copies of the miR-21 complementary sequence and a control Renilla luciferase (our unpublished) was transfected into the cells. Luciferase activity was determined 48 h after transfection and normalized to the level detected in the WT cells; *** represents a significant difference in luciferase reporter expression with p≤0.001.
3.2. Up-regulation of genes containing miR-21-targeted sequences in a miR-21 knockout clone
To investigate whether there would be a general increase of mRNAs carrying a predicted miR-21 target sequence, we deep-sequenced used the poly(A)+RNAs from WT, clone #39, and clone #75, and used the Sylamer algorithm to analyze the RNA-Seq data. In a Sylamer analysis, genes are ranked from most up-regulated to most down-regulated, and the occurrence bias (hypergeometric significance) of a specific sequence in the 3’ UTR of each gene batch within this ranked list is then calculated [59]. By comparing clone #75 and the parental line using Sylamer for every possible 7-mer, it appeared that predicted miR-21 target sequences were over-represented in the up-regulated portion of the expression ranking (Fig. 2B). We also transfected a miR-21 luciferase reporter into the parental line, clone #38, and clone #75, and found that the de-repression of the reporter expression was miR-21 dosage dependent: clone #38 with partial miR-21 deficiency increased the reporter activity by 5 fold, while miR-21 depleted clone #75 increased the reporter activity by nearly 100 fold (Fig. 2C). Thus, the expressing gene enrichment and the reporter de-repression both indicated that disruption of the miR-21 gene by TALEN could result in global up-regulation of miR-21 target genes.
3.3. Phenotypical characterization of miR-21 knockout clones in culture and in mouse xenografts
To aid in the phenotypical analysis, we isolated additional miR-21 knockout HeLa clones. We took clone #38, which still has one functional miR-21 allele (Supplementary Fig. S3), and performed a second round of mutagenesis using TALENs. Since the remaining, functional miR-21 allele (38-A) did not have an intact L3-R3 binding site, we decided to use L2-R2 that cuts the loop region to disrupt the gene (Fig. 1A). We also included a single-stranded oligodeoxynucleotide (ssODN) for homologous recombination to precisely delete the mature miR-21 sequence (Fig. 3A). We screened 164 single cell derived clones and found 1 clone (#38124) had a precise deletion of the miR-21 sequence (Fig. 3B), and 2 NHEJ-derived deletion clones (#3837 and #3838) (Supplementary Fig. S3). Quantitative RT-PCR showed that all three clones did not express mature miR-21 (Fig. 3C). MicroRNA deep sequencing of clones #38 and #3837 confirmed a depletion of miR-21-5p and miR-21-3p in clone #3837 (Fig. 3D). The expression of miR-143-3p was not significantly different between #38 and #3837.
Figure 3. miR-21 completely disrupted clones derived from clone #38 using TALEN L2-R2.
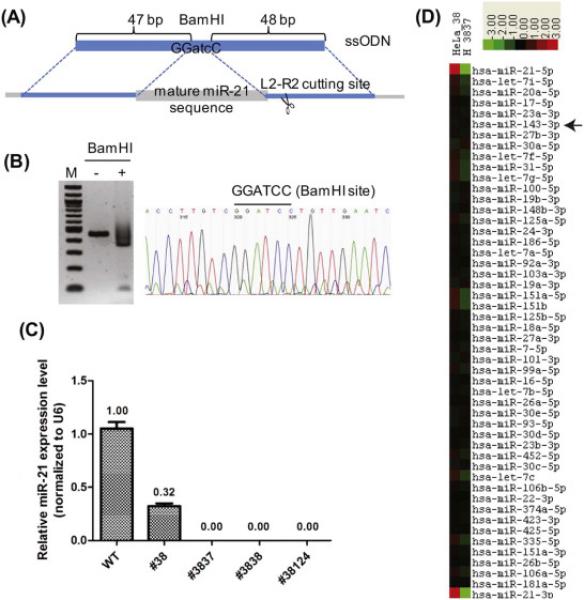
(A) A homologous recombination event between the single-stranded oligodeoxynucleotides (ssODN) and the miR-21 genomic locus will delete the miR-21 sequence and replace with 3 bases of atc to generate a BamHI site. (B) Genomic DNA from clone #38124 was subjected to PCR and BamHI digestion (left) and Sanger sequencing (right). (C) Mature miR-21 levels were measured by RT-qPCR. (D) A heatmap of the miR-Seq data from clones #38 and #3837. The 49 most abundant microRNAs plus miR-21-3p were ranked and clustered by centering on the row-wise mean log(2) value [15, 17]. A scale of 8-fold deviation from the mean is shown.
Although miR-21 is up-regulated in many cancers, it is not clear how loss of miR-21 would affect the highly transformed HeLa cells. We measured the cell growth rates using the metabolic-activity-based MTS assay and found that all four miR-21 knockout clones are less proliferative (Fig. 4A). We also performed a monolayer colony formation assay and found that miR-21 knockout cells were much less clonogenic than the parental line (Fig. 4B). By performing a soft agar colony formation assay, we found the miR-21 knockout cells had decreased anchorage-independent growth (Fig. 4C). Thus, proliferation of HeLa cells indeed was hindered by the loss of miR-21.
Figure 4. Analysis of cell proliferation of miR-21 disrupted clones.
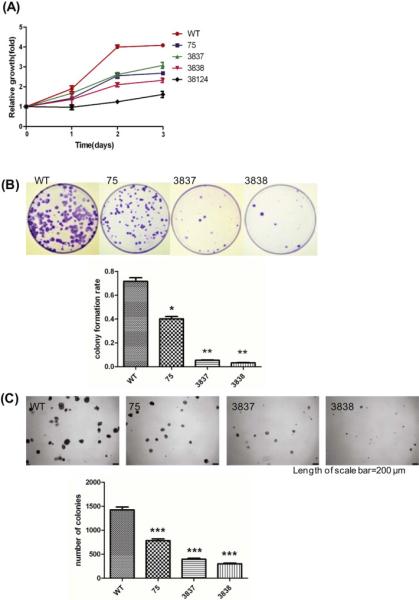
(A) Cell viability was measured using tetrazolium MTS assay. (B) Clonogenic ability was measured using monolayer colony formation assay; * is p≤0.05, ** is p≤0.01. (C) Anchorage-independent growth was measured using soft agar colony formation assay; *** is p≤0.001.
We also investigated whether loss of miR-21 would affect chemotherapeutic sensitivity of the cells. miR-21 knockout clones and the parental line were treated with cisplatin at concentrations from 1.5 μM to 25.0 μM, and cell viability was determined using the MTS assay. The miR-21 knockout clones were less viable than the parental line (Fig. 5A). By performing Annexin V staining of cells treated with or without cisplatin, we found the miR-21 knockout cells showing a greater cisplatin sensitivity had a higher apoptotic rate, particularly in the early apoptotic cell population, which were Annexin V positive but propidium iodine negative – WT at 1.8%, #75 at 7.8%, #3837 at 16.4%, and #3838 at 17.9% (Fig. 5B). Thus, HeLa cells lacking miR-21 expression were more apoptotic and sensitive to cisplatin.
Figure 5. Cisplatin sensitivity in vitro and mouse xenografts of miR-21 disrupted cells.
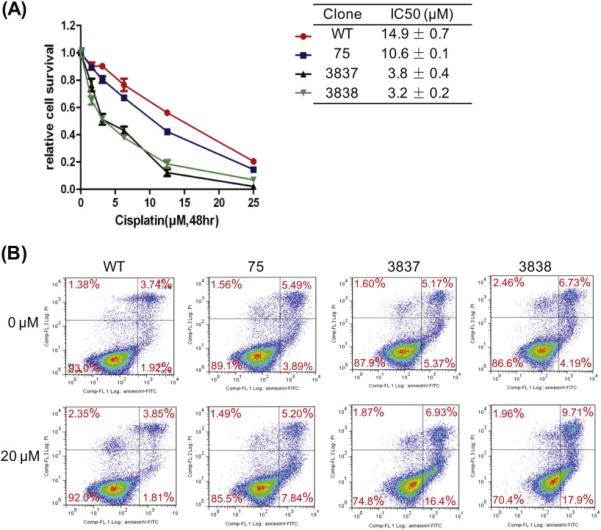
(A) Cells were treated with cisplatin at various concentrations for 48 h and the cell viability was measured using MTS assay. (B) Cells were treated with cisplatin for 12 h, stained with propidium iodine and Annexin V, and subjected to FACS analysis. Significant increases in early apoptotic cells present in the lower-right quadrant were observed in clones #75, #3837, and #3838 particularly with cisplatin treatment.
We further characterized tumorigenic properties of miR-21 knockout cells by xenotransplantation into immunodeficient NSG mice. Cells from a miR-21 knockout clone (#75), a partially disrupted clone (#39), and the parental line (WT) were xenotransplanted, and the appearance of tumor growth and size were followed in situ. The tumors were eventually excised from mice and analyzed by H&E staining. Although the tumor size was not significantly different (data not shown), the xenograft derived from clone #75 appeared to be less proliferative than the xenograft derived from WT or clone #39: the cell density was lower, the nucleoli were less prominent, and there were fewer mitotic figures (Supplementary Fig. S4). The xenograft derived from clone #75 also appeared to have fewer apoptotic bodies and more matrixes between cells. A combination of a lower mitotic rate, less apoptosis, and more extracellular matrix in clone #75 might explain why its xenografts had a similar size as those from WT and clone #39. Taken together, these results indicated that miR-21 knockout cells were less proliferative in vivo as in vitro.
3.4. Alteration of gene expression in miR-21 knockout cells
To find out what genes or pathways might be affected upon depletion of miR-21 in HeLa cells, we compared the RNA-Seq data of 4 TALEN-mutagenized single-cell clones: partially disrupted clones #39 and #38, and completely disrupted clones #75 and #3837. After filtering out non-expressing genes, we identified 200 up-regulated genes and 122 down-regulated genes in the two knockout clones as compared to the two disrupted clones that still expressed considerable amounts of miR-21 (Supplementary Table S1 and Table S2, respectively). We used DAVID (Database for Annotation, Visualization, and Integrated Discovery [16]) to analyze the up-regulated gene set for cellular processes affected in the miR-21 knockout clones. Genes involved in cell-environment interactions such as cell adhesion, extracellular matrix (ECM), and growth factor and integrin binding were enriched (Table 1). DAVID analysis of the down-regulated gene set indicated that genes involved in metabolic pathways such as reduction-oxidation and fatty acid/lipid synthesis were enriched (Table 1).
Table 1.
DAVID analysis of up-regulated or down-regulated genes in miR-21 knockout HeLa cells
| Up Cluster Representative Term | Enrichment Score | Representative Genes |
|---|---|---|
| Cell adhesion | 8.16 | PCDHA6, PCDHA7, PCDHA8, PCDHA2, TNFRSF12A, PCDHA3, TNC, PCDHA4, IGFBP7, PCDHA5, ITGA11, ITGB5, NEDD9, CDH2, PCDHA1, PCDHAC2, PCDHAC1, CD97, CTGF, TGFBI, PCDHA10, PCDHA11, PCDHA12, PCDHA13, NEGR1, FN1, COL15A1, CERCAM, COL4A6, AMIGO2, ITGA6, ITGA5, CYFIP2 |
| Extracellular matrix | 4.58 | COL4A2, LTBP1, COL4A1, LTBP2, TNC, NTN4, COL15A1, OLFML2A, COL5A3, ECM1, COL5A1, COL4A6, CPZ, TIMP1, CTGF, TGFBI, THSD4, COL1A1, TFPI2, FN1 |
| Cell motion and migration | 3.78 | TNFRSF12A, PLXNA2, TGFBR1, ITGA11, CDH2, CERCAM, COL5A1, PLAUR, EPHB2, CD97, ITGA6, ITGA5, CTGF, SERPINB5, DNER, SEMA3A, ETV4, DCLK1, RUNX3, FN1 |
| Growth factor binding | 3.41 | FGFBP3, LTBP1, COL4A1, LTBP2, CTGF, HTRA1, IGFBP7, TGFBR1, COL1A1, COL5A1, CRIM1 |
| Cell differentiation | 2.62 | NOTCH3, SMAD7, TGFBR1, RCAN1, ABCA1, SEMA3A, JAG1, ABCG1, KLF4, EPHB2, MBP, THY1 |
| EGF/Notch signaling | 2.34 | NOTCH3, CD97, LTBP1, LTBP2, TNC, DNER, PTGS1, JAG1, ADAM19, MAML3, FOXC2 |
| Integrin binding | 2.06 | ITGA6, CTGF, ITGA5, TGFBI, COL5A1, THY1 |
| Down Cluster Representative Term | Enrichment Score | Representative Genes |
|---|---|---|
| Aldo/keto reductase and NADP | 2.92 | AKR1C3, AKR1C2, AKR1C4, KCNAB2, FASN, AKR1C1, FDFT1 |
| GPI-anchor | 2.52 | CPM, ALPPL2, FOLR1, NTNG1, PSCA, ALPP |
| Steroid metabolic process | 2.34 | SOAT1, AKR1C2, AKR1C4, SQLE, HMGCS1, LSS, IDI1, AKR1C1, SREBF2, FDFT1 |
| Lipid and fatty acid synthesis | 1.76 | ELOVL2, HMGCS1, FASN, FADS2, LSS, IDI1, FDFT1 |
Five of the up-regulated genes are validated miR-21 targets: COL4A1, JAG1, SERPINB5/Maspin, SMAD7, and TGFBI (Supplementary Table S1); interestingly, they have all been implicated in TGFβ regulation and playing a role in fibrosis [13, 39, 45, 46, 67]. TGFBI as a secreted protein interacts with collagen and integrins and enhances cell interactions. Overexpression of TGFBI contributes to apoptotic cell death and higher TGFBI levels are associated with better prognosis of lung cancer [47]. Two up-regulated genes are predicted miR-21 targets: FMN1 and NEGR1. The fmn1 mRNA has been reported to be up-regulated in miR-21 knockout mice after fibrosis-related kidney injury [9]. It is possible that many miR-21 targets were not highly upregulated at the RNA level but up-regulated at the protein level [27]. By using immuno-blot to analyze two well-characterized miR-21 regulated genes, PDCD4 [3] and PTEN [43], we observed a significant increase of PDCD4 but a subtle increase of PTEN in miR-21 knockout clones #75, #3837, and #3838 (Supplementary Fig. S5). There are more than 30 miRNAs have been validated to target PTEN [55], so depletion of one microRNA may not drastically derepress PTEN [69]. Conversely, many highly over-expressed mRNAs detected in the knockout cells were probably not direct targets of miR-21, but might represent downstream events. Taken together, the RNA-Seq data indicated that HeLa cells lacking miR-21 increased expression of genes related to cell-environment interactions and decreased expression of genes related to the redox regulation.
4. DISCUSSION
MicroRNA-21 is of great interest given it is highly expressed and plays a pathological role in many cancers and fibrotic organs. We show in this report that human cell lines without a functional miR-21 can be readily obtained using TALE-Nucleases, and miR-21 expression is necessary to support robust cell proliferation and to augment cisplatin resistance. We also show that genes involved in cell-environment interaction or metabolism are deregulated, and that miR-21-targeted mRNAs involved in TGFβ and fibrosis regulation are up-regulated in miR-21 null cells.
Antisense oligonucleotides or antagomirs have been widely used in investigating the function and therapeutic potentials of microRNAs [38]. The advantage of using antagomirs is that it is relatively simple to design, synthesize, and deliver; however, inhibition by antagomirs is almost always incomplete and off-target effects are difficult to control [28]. For instance, inhibiting miR-21 using antisense oligonucleotides results in profound increase in HeLa cell growth [11], while a similar antagomir approach causes a complete opposite effect of profound suppression of HeLa cell proliferation [66]. Strategies to inactivate microRNA genes using gene-editing nucleases such as TALEN are being developed more recently [30]. The advantage of nuclease-mediated knockout is that the microRNA of interest can be completely depleted so the outcome would likely be more consistent. For instance, knocking out miR-21 using TALEN in two different cell lines, HeLa in this study and HEK293 in two others [57, 63], all resulted in decrease in cell proliferation. However, the utility of this approach still needs to be further investigated [64].
MicroRNA-21 has been knocked out in human colorectal cancer cell lines using adeno-associated viruses to transduce a neomycin marker flanked by miR-21 homologous sequences [61]. The procedure requires two rounds of drug selection and LoxP-Cre recombinase-mediated deletion in order to obtain complete knockout clones. By using sequence-specific TALENs, complete miR-21 knockout clones can be isolated without drug selection and in one step [63] (and this study). TALENs have been used to disrupt the miR-155, miR-146a, and miR-125b genes, but the study did not characterize the knockout lines created [25]. TALENs in combination with a homologous template carrying a selectable marker have also been used to disrupt the miR-21 gene in HEK293 cells, which result in cell growth reduction in culture [57]. Our study went further to characterize miR-21 knockout clones for gene expression and cell proliferation changes in vitro and in vivo.
MicroRNA biogenesis is often studied using minigene constructs in transfected cells [68] or using relatively short synthetic RNA substrates in cell-free extracts [36]. Using TALEN – or other sequence-specific nucleases – to generate genomic mutant alleles would be quite valuable in evaluating microRNA processing from its native transcript. For example, in our HeLa clone #3838 a deletion of 11 base pairs between miR-21-5p and miR-21-3p, without deleting any nucleotide in either microRNA, completely abolished the biogenesis of miR-21 (Supplementary Fig. S3). Conversely, in HeLa clone #39, a deletion of 7 base pairs immediately upstream of miR-21-5p did not abolish the expression of miR-21 from that allele (Supplementary Fig. S3).
Homozygous miR-21 knockout mice are viable indicating that miR-21 is not essential for cell growth [40, 48]. The fact that viable miR-21 knockout clones can be isolated from RKO and DLD1 colorectal cancer cells [61], HeLa cervical adenocarcinoma cells (this study), HEK293 transformed human embryonic kidney cells [57, 63], and Ramos Burkitt's lymphoma cells (our unpublished results), suggests that miR-21 expression is possibly not essential for many human cancer cell lines. However, the growth of pre-B cell lymphoma resulted from miR-21 overexpression is abolished when miR-21 expression is turned off [42]. Our results indicated that miR-21 enhances robust growth in vitro and aggressiveness in vivo in highly transformed HeLa cells. HeLa cells are widely used as a cancer cell model in general and as a cervical adenocarcinoma cell model in particular [41]. In addition, the complete genome sequence of HeLa has recently been determined, making it an ideal cellular model for cancer study [2, 35]. Cisplatin remains a backbone of therapy for early and late-stage cervical cancer [19], so it is of clinical interest that knocking out miR-21 enhances sensitivity of HeLa to cisplatin. Thus, our study further advances the idea that miR-21 may be explored as a safe adjuvant therapeutic target [10, 65].
We compared the 200 up-regulated genes in our miR-21 knockout HeLa cells with the 118 up-regulated genes in the miR-21 knockout colon cancer cell lines [61]; only 4 genes appear on both lists (CTGF, ECM1, EEF1A2, and HAS2). Three of the 4 overlapped genes are involved in cell-environment interactions: CTGF is connective tissue growth factor, ECM1 is extracellular matrix protein 1, and HAS2 is hyaluronan synthase 2. We have also isolated TALEN-induced miR-21 knockout Ramos lymphoma clones and analyzed gene expression changes by RNA-Seq; there were 101 up-regulated genes and none overlapped with the up-regulated genes in miR-21 knockout HeLa or colon cancer cell lines (our unpublished). The lack of overlaps in gene expression changes among the miR-21 knockout clones from various human cell lines suggests that different cells may have different ways to respond to the loss of miR-21.
Knockout of miR-21 in HeLa cells leads to up-regulation of genes enriched in extracellular matrix, which include genes encoding collagen, integrin, and fibronectin (Supplementary Tables S1 and S3). There are 5 validated miR-21 targets up-regulated in the knockout HeLa cervical cancer cells: COL4A1, JAG1, SERPINB5, SMAD7, and TGFBI (this study), 3 validated targets in knockout colon cancer cells: BTG2, CDC25A, and TGFB1 [61], and 2 validated targets in knockout Ramos lymphoma cells: ACTA2 and MEF2C (our unpublished). Although there are no overlaps, all these genes are implicated in TGFβ regulation and fibrosis. NEGR1, a putative miR-21 target, was also found to be up-regulated in our study. The NEGR1 protein localizes to cell-cell contact areas of the cell membrane and is down-regulated in many cancerous tissues. NEGR1 is regulated by the TGFβ signaling, and overexpressing NEGR1 inhibits cell growth while suppressing NEGR1 leads to increase of cell migration and invasion [23, 29, 53]. Thus, NEGR1 may represent a novel tumor-suppressor target for miR-21.
The role of miR-21 as a regulator of ECM formation has also been linked to scleroderma fibrosis [70], aortic stenosis [60], atrial fibrosis [1], renal fibrosis [9], and pulmonary fibrosis [39]. In addition, redox imbalance impairs the myofibroblast-mediated fibrosis resolution in a mouse lung fibrosis model [22], and the lipid metabolism pathway is deregulated in miR-21 knockout mice with injury-induced kidney fibrosis [9]. Interestingly, aldo/keto reductases and lipid synthesis are both down-regulated in our miR-21 knockout HeLa cells (Table 1). Thus, this study reveals a set of miR-21 targets that may play an important role in miR-21 mediated regulation of TGFβ and fibrosis. Our work further suggests that modulation of the ECM may be an important mechanism by which miR-21 contributes to pathogenesis. In conclusion, the miR-21 null cells isolated and the genes/pathways identified in this study provide important first steps in elucidating the regulatory mechanism involving miR-21 in malignant and fibrotic diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Harry Gao, Jinhui Wang, and Hanjun Qin for performing Solexa RNA sequencing; Sofia Loera for performing tumor pathology; and our City of Hope colleagues Emily Wang, Mei Kong, Rama Natarajan, Mark Boldin, Adam Bailis, Ruby Chen, Joseph Chao, and Lindsey Skrdlant for suggestions and helpful discussion. BC and XC were supported in part by the P.R.C. National and Fujian Provincial Key Clinical Specialty Discipline Construction Programs and from the Natural Science Foundation of Fujian Province of China (Grant No. 2014J01328). YJW was supported by P.R.C. National and Zhejiang Provincial Natural Science Foundations, Zhejiang Provincial Major Science and Technology Project, and China Scholarship Council. JKY is supported by grant RB3-02161 from California Institute of Regenerative Medicine. This work was supported by Nesvig Lymphoma Research Foundation, Beckman Research Institute Excellence Awards, and the Ramesh Kesanupalli family to RJL, as well as by NIH P30-CA033572 for shared research resources at City of Hope. The animal experiments were done according to protocol #12026 as approved by the Institutional Animal Care and Use Committee.
Abbreviations
- TALEN
Transcription activator-like effectors nuclease
- miR-21
microRNA-21
- TGFβ
transforming growth factor beta
- RNA-Seq
Next-generation shotgun RNA sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
BC, XC, JH, JY, and RL conceived and designed the experiments; BC, XC, and JW performed the experiments; BC, XC, XW, TL, JK, JW, SV, JY, and RL analyzed the data; XW, YW, HH contributed with reagents and ideas; and BC, XC, and RL wrote the paper.
Conflict of Interest
The authors declare no competing financial interests.
REFERENCES
- 1.Adam O, Lohfelm B, Thum T, Gupta SK, Puhl SL, Schafers HJ, Bohm M, Laufs U. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic research in cardiology. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 2.Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–211. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 4.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chinese journal of cancer. 2011;30:371–380. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, MacKenna DA, Orkin SH, Duffield JS. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Science translational medicine. 2012;4:121ra118. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Tsai YH, Fang Y, Tseng SH. Micro-RNA-21 regulates the sensitivity to cisplatin in human neuroblastoma cells. Journal of pediatric surgery. 2012;47:1797–1805. doi: 10.1016/j.jpedsurg.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. Journal of cardiovascular translational research. 2010;3:251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cher ML, Biliran HR, Jr., Bhagat S, Meng Y, Che M, Lockett J, Abrams J, Fridman R, Zachareas M, Sheng S. Maspin expression inhibits osteolysis, tumor growth, and angiogenesis in a model of prostate cancer bone metastasis. Proc Natl Acad Sci U S A. 2003;100:7847–7852. doi: 10.1073/pnas.1331360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 16.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaj T, Gersbach CA, Barbas CF, 3rd, ZFN TALEN. CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013 doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greer BE, Koh WJ, Abu-Rustum NR, Apte SM, Campos SM, Chan J, Cho KR, Copeland L, Crispens MA, Dupont N, Eifel PJ, Gaffney DK, Huh WK, Kapp DS, Lurain JR, 3rd, Martin L, Morgan MA, Morgan RJ, Jr., Mutch D, Remmenga SW, Reynolds RK, Small W, Jr., Teng N, Valea FA. N. National Comprehensive Cancer, Cervical cancer. Journal of the National Comprehensive Cancer Network : JNCCN. 2010;8:1388–1416. doi: 10.6004/jnccn.2010.0104. [DOI] [PubMed] [Google Scholar]
- 20.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 21.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, Meldrum E, Sanders YY, Thannickal VJ. Reversal of persistent fibrosis in aging by targeting nox4-nrf2 redox imbalance. Science translational medicine. 2014;6:231ra247. doi: 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiemer SE, Szymaniak AD, Varelas X. The transcriptional regulators TAZ and YAP direct transforming growth factor beta-induced tumorigenic phenotypes in breast cancer cells. J Biol Chem. 2014;289:13461–13474. doi: 10.1074/jbc.M113.529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nature biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu R, Wallace J, Dahlem TJ, Grunwald DJ, O'Connell RM. Targeting human microRNA genes using engineered Tal-effector nucleases (TALENs) PloS one. 2013;8:e63074. doi: 10.1371/journal.pone.0063074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 28.Ishida M, Selaru FM. miRNA-Based Therapeutic Strategies. Current anesthesiology reports. 2013;1:63–70. doi: 10.1007/s40139-012-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim H, Hwang JS, Lee B, Hong J, Lee S. Newly Identified Cancer-Associated Role of Human Neuronal Growth Regulator 1 (NEGR1) Journal of Cancer. 2014;5:598–608. doi: 10.7150/jca.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YK, Wee G, Park J, Kim J, Baek D, Kim JS, Kim VN. TALEN-based knockout library for human microRNAs. Nature structural & molecular biology. 2013;20:1458–1464. doi: 10.1038/nsmb.2701. [DOI] [PubMed] [Google Scholar]
- 31.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. Journal of cellular and molecular medicine. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 33.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA biology. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 35.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stutz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, Gagneur J, Korbel JO, Huber W, Steinmetz LM. The genomic and transcriptomic landscape of a HeLa cell line. G3. 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HY, Doudna JA. TRBP alters human precursor microRNA processing in vitro. RNA. 2012;18:2012–2019. doi: 10.1261/rna.035501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nature reviews. Drug discovery. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 39.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. The Journal of experimental medicine. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, Liu MF, Li Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nature reviews. Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- 42.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 43.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nature biotechnology. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 45.Murata K, Ota S, Niki T, Goto A, Li CP, Ruriko UM, Ishikawa S, Aburatani H, Kuriyama T, Fukayama M. p63 - Key molecule in the early phase of epithelial abnormality in idiopathic pulmonary fibrosis. Experimental and molecular pathology. 2007;83:367–376. doi: 10.1016/j.yexmp.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Nakken KE, Nygard S, Haaland T, Berge KE, Arnkvaern K, Odegaard A, Labori KJ, Raeder MG. Multiple inflammatory-, tissue remodelling- and fibrosis genes are differentially transcribed in the livers of Abcb4 (−/ − ) mice harbouring chronic cholangitis. Scandinavian journal of gastroenterology. 2007;42:1245–1255. doi: 10.1080/00365520701320521. [DOI] [PubMed] [Google Scholar]
- 47.Pajares MJ, Agorreta J, Salvo E, Behrens C, Wistuba II, Montuenga LM, Pio R, Rouzaut A. TGFBI expression is an independent predictor of survival in adjuvant-treated lung squamous cell carcinoma patients. British journal of cancer. 2014;110:1545–1551. doi: 10.1038/bjc.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. The Journal of clinical investigation. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pauwels K, Podevin N, Breyer D, Carroll D, Herman P. Engineering nucleases for gene targeting: safety and regulatory considerations. New biotechnology. 2013 doi: 10.1016/j.nbt.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG, Inflammation LSCRP. Host Response to Injury, Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takita J, Chen Y, Okubo J, Sanada M, Adachi M, Ohki K, Nishimura R, Hanada R, Igarashi T, Hayashi Y, Ogawa S. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer science. 2011;102:1645–1650. doi: 10.1111/j.1349-7006.2011.01995.x. [DOI] [PubMed] [Google Scholar]
- 54.Taniguchi K, Kajiyama T, Kambara H. Quantitative analysis of gene expression in a single cell by qPCR. Nature methods. 2009;6:503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- 55.Tay Y, Song SJ, Pandolfi PP. The Lilliputians and the Giant: An Emerging Oncogenic microRNA Network that Suppresses the PTEN Tumor Suppressor In Vivo. MicroRNA. 2013;2:127–136. doi: 10.2174/22115366113029990017. [DOI] [PubMed] [Google Scholar]
- 56.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 57.Uhde-Stone C, Sarkar N, Antes T, Otoc N, Kim Y, Jiang YJ, Lu B. A TALEN-based strategy for efficient bi-allelic miRNA ablation in human cells. RNA. 2014 doi: 10.1261/rna.042010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 59.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nature methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Villar AV, Garcia R, Merino D, Llano M, Cobo M, Montalvo C, Martin-Duran R, Hurle MA, Nistal JF. Myocardial and circulating levels of microRNA-21 reflect left ventricular fibrosis in aortic stenosis patients. International journal of cardiology. 2013;167:2875–2881. doi: 10.1016/j.ijcard.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 61.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr., Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer research. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang SE, Lin R-J. MicroRNA and HER2-overexpressing Cancer. MicroRNA. 2013;2:137–147. doi: 10.2174/22115366113029990011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Wang Y, Huang H, Chen B, Chen X, Hu J, Chang T, Lin RJ, Yee JK. Precise Gene Modification Mediated by TALEN and Single-Stranded Oligodeoxynucleotides in Human Cells. PloS one. 2014;9:e93575. doi: 10.1371/journal.pone.0093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright DA, Li T, Yang B, Spalding MH. TALEN-mediated genome editing: prospects and perspectives. The Biochemical journal. 2014;462:15–24. doi: 10.1042/BJ20140295. [DOI] [PubMed] [Google Scholar]
- 65.Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–168. doi: 10.1016/j.tox.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 66.Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–542. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 67.Yu H, Konigshoff M, Jayachandran A, Handley D, Seeger W, Kaminski N, Eickelberg O. Transgelin is a direct target of TGF-beta/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:1778–1789. doi: 10.1096/fj.07-083857. [DOI] [PubMed] [Google Scholar]
- 68.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou P, Xu W, Peng X, Luo Z, Xing Q, Chen X, Hou C, Liang W, Zhou J, Wu X, Songyang Z, Jiang S. Large-scale screens of miRNA-mRNA interactions unveiled that the 3′UTR of a gene is targeted by multiple miRNAs. PloS one. 2013;8:e68204. doi: 10.1371/journal.pone.0068204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, Xiao X, You Y, Zuo X. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. Journal of clinical immunology. 2013;33:1100–1109. doi: 10.1007/s10875-013-9896-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


