Abstract
Pseudouridine (Ψ) is the most abundant of >150 nucleoside modifications in RNA. Although Ψ was discovered as the first modified nucleoside more than half a century ago, neither the enzymatic mechanism of its formation, nor the function of this modification are fully elucidated. We present the consistent picture of Ψ synthases, their substrates and their substrate positions in model organisms of all domains of life as it has emerged to date and point out the challenges that remain concerning higher eukaryotes and the elucidation of the enzymatic mechanism.
Keywords: enzymatic mechanism, modified nucleoside, pseudouridine, regulation, rRNA, RNA Modification, tRNA, snRNA
Abbreviations
- Pus
pseudouridine synthase
- Ψ
Psi, pseudouridine
- rRNA
ribosomal RNA
- snRNA
small nuclear RNA
- E. coli
Escherichia coli
- S. typhimurium
Salmonella typhimurium
- S. cerevisiae
Saccharomyces cerevisiae
- H. volcanii
Haloferax volcanii and/or Halobacterium volcanii.
Introduction
More than 150 nucleoside modifications fine-tune conformation, structure and function of RNA.1,2 In 1951 the first modified nucleoside was discovered in RNA hydrolysate3, shortly after termed the fifth nucleoside,4 identified as 5-ribosyl uracil5 and named pseudouridine (Ψ).6 Eventually the development of a tritium release assay for Ψ formation led to the identification of the first pseudouridine synthase gene,7 HisT, later renamed to TruA, which modifies tRNA in S. thyphimurium8 and E. coli.9 Although Ψ is the most abundant nucleoside modification,1 the actual advantage of pseudouridylation that warrants this abundance, remains hard to grasp and is usually described as stabilization by ‘additional hydrogen bonds’ and ‘improved base stacking’.10
The importance of Ψ is reflected and documented in the variety of existing reviews, be it general,11 centered on structural biology of either stand-alone protein Ψ synthases12,13 or H/ACA box ribonucleic particles (RNPs),14 or, even more recently, focused on H/ACA box RNPs and Ψ formation and function in snRNA and rRNA.15
In the last 2 decades a more consistent picture of Ψ synthesis and Ψ distribution in model organisms of all domains of life has emerged, of which the outlines will be presented here. Despite significant progress however, a clear catalytic role assignment to amino acids is still lacking, and hence the catalytic mechanism of Ψ formation remains elusive even now, almost 15 years after publication of the first cocrystal structure.16 We will outline why the elucidation of this mechanism remains a challenge, while research on Ψ is about to move to complex organisms and transcriptome wide analyses.
Physicochemical Properties of Ψ
Pseudouridine is a C-C glycosidic isomer of uridine (U), and the isomerization reaction, which incorporates the C5 into the glycosidic bond, is shown in Figure 1. Both nucleosides share a similar UV spectrum5 and identical molecular mass5, but differ in mass spectrometric dissociation.17,18 Early methods for the detection of Ψ were based on random alkaline hydrolysis followed by TLC detection of 32P-labeled nucleotides19, rendered semi-quantitative by biased, non-quantitative hydrolysis and incomplete labeling of Ψ.20 In a known sequence context, random hydrolysis can be substituted by site-specifically cleaving DNAzymes20 or RNase H21 or by making use of the decreased ligation efficiency of a complementary probing strand, thereby circumventing cleavage.22 The arguable most popular, albeit technically demanding technique for sequence specific Ψ detection includes specific derivatization of Ψ with CMCT followed by primer extension.23 This technique found recent application in genome-wide pseudouridine profiling by deep sequencing in yeast and human.24,25 Specific derivatization of Ψ with CMCT, acetonitrile, or methylvinylsulfone is also applied in sequence specific detection via LC-MS approaches, (reviewed in ref. 17). Incomplete reactions, side products, and unstable response factors prevent quantitative analysis by this approach. In consequence there is an increasing interest in derivatization-free MS/MS-based approaches that allow quantitative analysis,26,27 and which have recently included isotope labeling28,29
Figure 1.
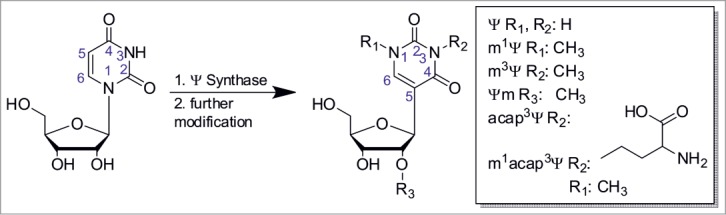
Isomerization of uridine into pseudouridine (Ψ). Post-isomerization several derivatives discovered to date1 can be formed by further modification at either position 1 (R1), 3 (R2) or 2’-O (R3), while several modifications at once are possible.
So called hypermodified Ψ derivatives are formed via further modification of Ψ. At present, they comprise Ψm, m1Ψ, (found in all domains of life, predominantly in tRNA1), m3Ψ (in rRNA of Eubacteria1), as well as 3-(3-amino-3-carboxypropyl)-Ψ (αχαπ3Ψ)30,31 and m1acap3Ψ1 in rRNA of eukaryotes (Fig. 1).
General Function/Structural Aspects of Ψ
Although the identical Watson-Crick faces of Ψ (Fig. 1) and U enable both to engage in classical Watson-Crick base pairing with adenosine (A), Ψ base pairs with any of the 4 major bases32,33 are more stable than their U equivalents. For Ψ-A base pairs, NMR revealed that NH1, which is situated in the major groove, was being protected from proton exchange with solvent water.10,34,35 This protective effect is probably caused by hydrogen bonding of ΨNH1 to the 5’-phosphate oxygen atoms via water, for which several lines of evidence lend support.11,33,36-40 Thus, conferred increased backbone rigidity may be the cause for a presumably secondary effect of Ψ formation: improved base stacking, which was concluded from a preference of the 3’-endo-conformation.10
The Presence of Ψ in Various RNAs
Ψ was first identified in rRNA,1 (recently reviewed by Ge and Yu15) and tRNA.1 Further occurrences of Ψ are known in small nuclear RNAs of various eukaryotes, as is reviewed in, e.g., refs 15,41. As an example, the Ψ in spliceosomal branch site of U2 snRNA will be discussed below in some detail. Further RNAs containing Ψ include snoRNAs U3 of rat and U8 of mouse1, tRNA-like domains of plant viruses,42,43 SRA RNA,44 and human telomerase RNA,25,45 long non-coding RNAs and mRNA.24,25 The following section will illustrate the distribution of Ψ in tRNA and rRNA, along with their respective enzymes, in all domains of life based on model organisms. In the subsequent sections, the functional and structural aspects of Ψ will be discussed in more detail.
Enzymatic Formation of Ψ residues
Enzyme families
Six families of pseudouridine synthases (Pus enzymes) have been identified, each named for a prominent representative: TruA, TruB, TruD, RsuA, RIuA, (reviewed in ref. 12) and Pus10p.46 They share the same overall fold and require an active site aspartate for catalysis,12,47 implying a common mechanism, to which we will turn our attention later. Different N- or C-terminal domains govern substrate specificity, as reviewed in ref.12 In contrast, few Ψ-hypermodification enzymes are known: E. coli m3Ψ methyltransferase RImH48,49 and 3 m1Ψ methyltransferases in Archaea50,51 and yeast.52
The most versatile enzyme family may be the ribonucleic particles (RNPs) depicted in Figure 2. These particles contain a subgroup of small nucleolar RNAs (snoRNAs), called H/ACA RNAs, and were proven to catalyze Ψ formation, at first in eukaryotes,53,54 later in Archaea.55 The snoRNA (called sRNA in Archaea) acts as guide for the protein components with Nop10 and the Ψ synthase NAP57 (higher eukaryotes) or Cbf5 (yeast, Archaea) as minimal requirements.56,57 Non-essential components Gar1 and L7Ae (or Nhp2 in Eukarya) are involved in catalysis and product release58 or in substrate binding by interaction with Nop1059-61, respectively. Cbf5 is also capable of guide RNA free catalysis, the activity of which is increased by Gar1 and Nop10.62 Investigation of guide RNA specificity63 enabled artificial guide RNAs to target specific uridines for Ψ formation.64-66
Figure 2.
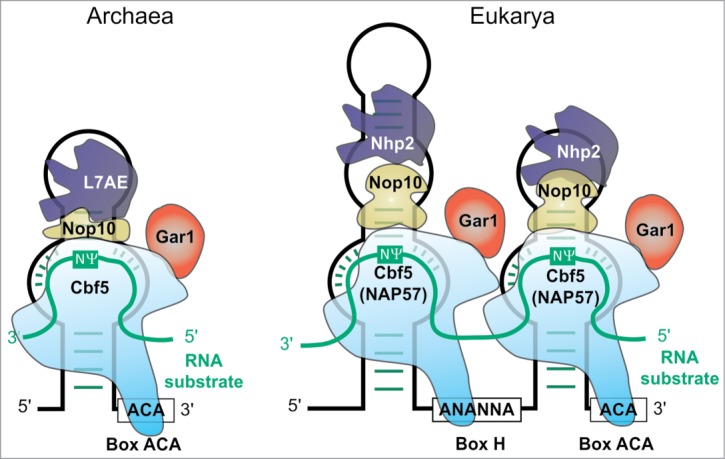
Structure of the archaeal ACA RNP198 (left) and the eukaryotic H/ACA RNP199 (right). Guide RNA in black, substrate RNA turquoise. Catalytically active component is light blue Cbf5 (NAP57)
From biochemical data67 and crystal structures68 a specific degradation pathway for Ψ in Eubacteria is evident: Ψ is first phosphorylated by a dedicated kinase and subsequently converted to uracil and ribose-5’-phosphate. The remarkable cleavage of a C-C glycosidic bond was reported to be reversible67 and to proceeds via a ribose ring opening mechanism.68 Mammals do not degrade Ψ, but urinary excrete the intact nucleoside.69,70 Recently, a pseudouridine-5’-phosphatase that dephosphorylates Ψ in human was described.71 As assays performed in cell extracts indicated conversion of pseudouridine-5’-phosphate into triphosphate,72 dephosphorylation might prevent accidental incorporation of pseudouridine into RNA transcripts. 71
Occurrence and formation of Ψ in model organisms of all domains of life
As indicated above, pseudouridine formation in cellular RNAs is ensured either by stand-alone protein enzymes or by H/ACA sno(s)RNA-dependent RNP particles or by both. In Eubacteria, E. coli is taken as a model (Fig. 3), Ψ synthases acting on RNA belong to 5 distinct families, the Pus10-related family was not detected. Altogether 11 enzymes ensure complete modification of tRNAs and rRNAs in Eubacteria. Pseudouridine modification of other eubacterial RNAs have never been reported in the literature. Although no knockout of a single Ψ synthase has proven to be lethal in Eubacteria, certain single-knockouts suffer from disadvantages compared to their unaffected counterparts.73-75
Figure 3.
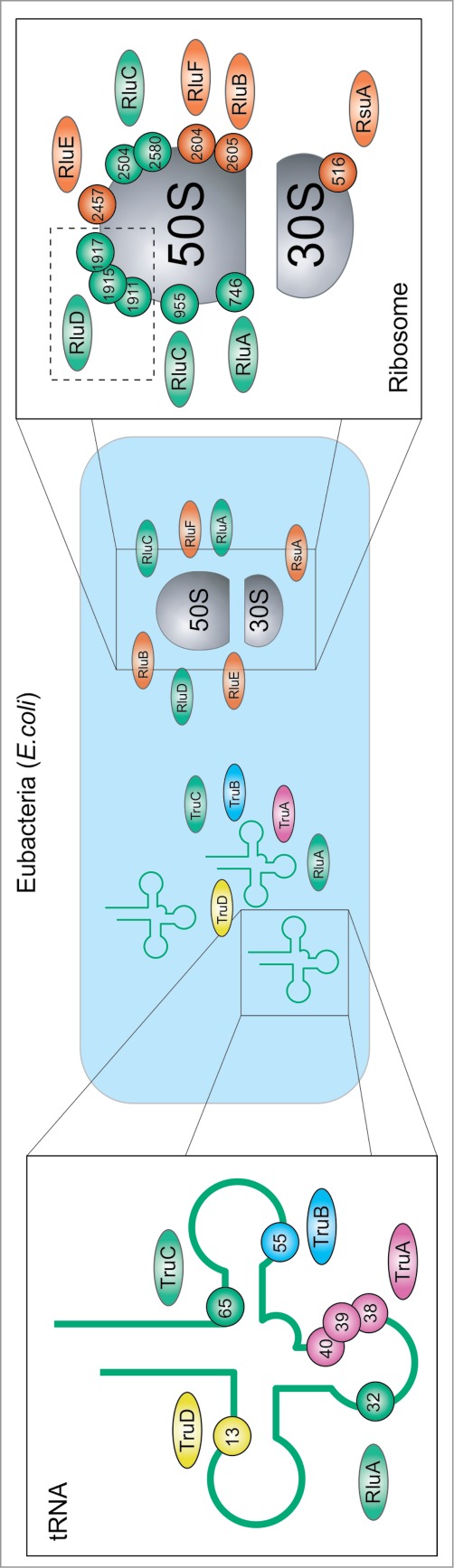
Distribution of Ψ and Ψ synthases in E. coli: Enzymes and their substrates positions color-coded: TruA in purple, RIuA and family members green, RsuA family members orange, TruB blue, all reviewed in,200 and TruD124 yellow. Substrate residues of RIuD are shown in a dashed box to indicate model helix H69.
In this light it may seem surprising that the total number of pseudouridine modification sites is much higher, and thus many enzymes demonstrate so-called region-specificity (like TruA or RluD, see Fig. 3) or even multisite-specificity (RluA and RluC).73,74 This balance between substrate specificity and promiscuity typical for Ψ synthases is evident in, e.g., E. coli TruA, the only dimeric Ψ synthase, which uses the intrinsic flexibility of its substrate tRNA to access either positions 38, 39 or 40.76 In contrast the specificity of RIuF and RIuB for adjacent sites in the ribosome, is achieved by substrate binding in different conformations.77,78 This specificity is compromised by a weak activity of RIuF for the substrate position of RIuB.79 TruB recognizes the shape of the T-stem loop and therewith its substrate position in its single substrate tRNA.80 Strikingly, the preference for structured 50S subunits over free 23S rRNA of RIuD81 coincides with few sequence requirements, in contrast to the associated m3Ψ methyltransferase RImH.82
The modification pattern of archaeal RNAs (including pseudouridine residues) was only studied for a limited number of species, among which the halophilic Archaea H. volcanii is the best studied organism (see Fig. 4 for modification positions and responsible enzymes). Direct RNA sequencing of isolated tRNA species83,84 pointed out a modification profile similar to the one observed in bacteria, but Ψ32 was absent and some additional sites were detected in D-and TΨ-loops. Genomic studies and direct analysis of Ψ synthase activities85,86 confirmed the absence of RIuA-related activities in Archaea, while instead, an additional family of Pus10-related proteins was found. One of the best studied members of this family, Pus10p from H. volcanii,87 fills out the role of the TruB enzymes by acting as Ψ synthase on positions 54 and 55 in archaeal tRNA,88 using a different recognition mechanism for each position.89 Recognition by Pus10 proteins probably involves the characteristic N-terminal THUMP domain,47 that binds to the tRNA acceptor stem in a docking model of the human Pus10 homolog.90 This binding mode is supported by a recent cocrystal structure of a THUMP domain-containing enzyme, 4-thiouridine synthetase.91 Generation of Ψ55 is undisturbed by deletion of Cbf5,87 which can also generate Ψ55 in vitro.92 Whether Cbf5 can substitute Pus10p in generating Ψ55 in vivo cannot be tested since a Pus10p knockout is lethal.87 In contrast to Eubacteria, the pseudouridine formation in archaeal rRNA is insured by H/ACA sRNA RNPs,86,87 and thus the RsuA related family is also missing. Several rare sites of pseudouridine modification in archaeal tRNAs still have not been assigned to a particular enzyme,86,87 but highly promiscuous enzymes like TruD (Pus7)93 may be responsible for Ψ formation at these locations.
Figure 4.
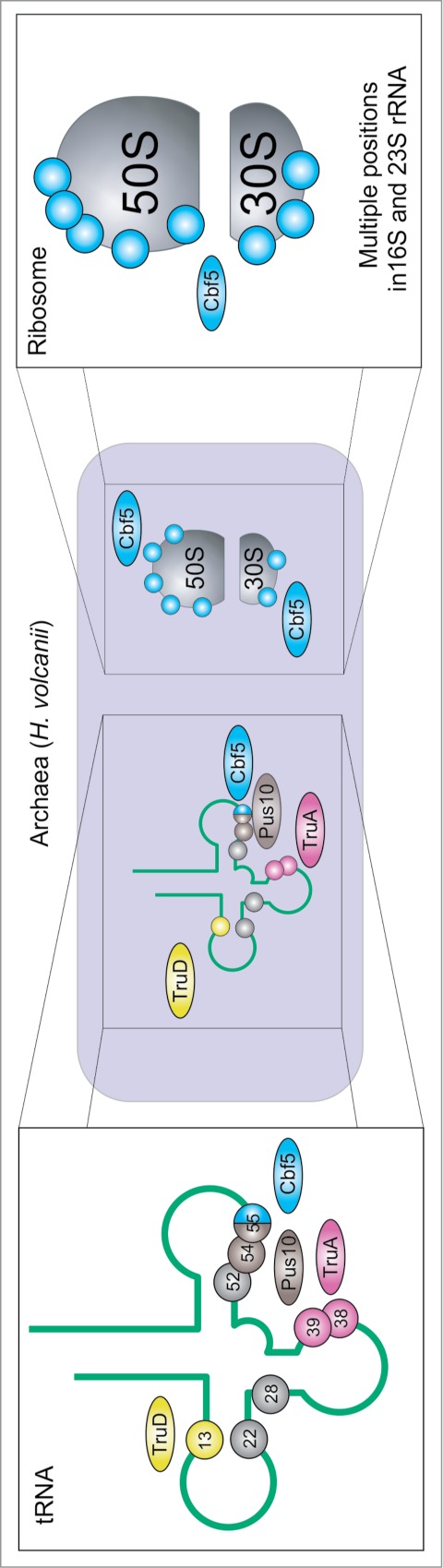
Distribution of Ψ and Ψ synthases in H. volcanii: TruA purple87, TruD86 yellow, Pus10p87 brown, Cbf586,87 blue, positions with yet unknown enzyme86 in gray. Note that position 52 is only partially modified83 and that ribosomal Ψs are only available for 16S and not for 23S and 5S rRNA. 86
The best studied lower eukaryote, S. cerevisiae, displays 4 common Ψ synthase families (see Fig. 5, RsuA- and Pus10-related families are missing). In Eukarya, not only tRNAs and rRNA are modified to pseudouridine, but also snRNAs. Recent genome-wide pseudouridine profiling even revealed hundreds of Ψs in mRNA and provided further evidence on Ψs in snoRNAs.24,25 Additional complexity comes from distinct cellular compartments (and their respective specific RNA species) coexisting in eukaryotic cells. Thus, nuclear (cytoplasmic) tRNAs and rRNA are not necessarily modified by the same machinery as their mitochondrial counterparts. This duality clearly exists for Pus1/Pus294,95 and Pus8/Pus996 pairs for tRNA modification and for Cbf5/Pus515,97 for rRNA pseudouridine formation. However, some enzymes like Pus398, Pus499 and Pus6100 have dual functions and are partially imported to mitochondrial compartment. As for bacterial Ψ synthases, many yeast enzymes demonstrate both region-specificity and multisite-specificity to account for the large number of modification in all types of cellular RNAs. Formation of some pseudouridine residues in yeast RNAs, notably U2 snRNA and U6 snRNA (dashed circles in Fig. 5, see also below) and mRNA is stress-regulated.24,66,101 Upon heat shock the localization of Pus7p changes from nuclear to in part cytosolic.25 TruA family member Pus1p, as well as yeast TruD homolog Pus7p,102 in contrast to their bacterial counterparts, modify various positions in a large variety of substrates, including U2 snRNA66,103,104, various positions in various tRNAs95,105,106, 5S rRNA107 and mRNA.24,25 The loose specificity of eukaryotic Pus1p may be related to its additional C-terminal domain, which, in contrast to E. coli TruA, causes it to act as monomer.108 This difference in structure results in substrate specificity for a minimal substrate defined solely by shape and not by sequence.109 In contrast, Pus7p acts on a specific recognition sequence.102 This striking difference in substrate recognition could be confirmed in pseudouridine profiling of mRNA.24,25
Figure 5.
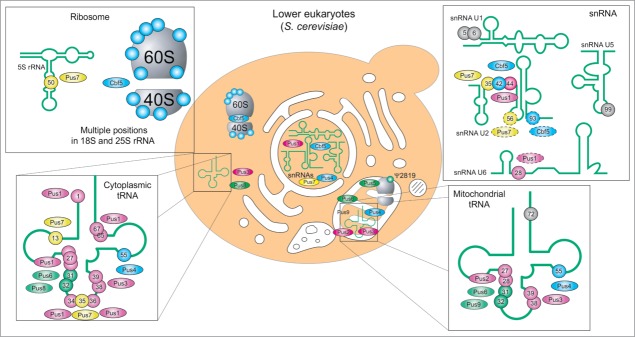
Distribution of pseudouridine and pseudouridine synthases in yeast: Cellular location of enzyme and substrates as well as substrate position are given for TruA family members Pus1p95,103,106, Pus2p94 and Pus3p98 (purple), RIuA family members Pus5p,97 Pus6p,100 Pus8p96 and Pus9p96 (green), TruD homolog Pus7p104,105,107 (yellow) and stand-alone TruB homolog Pus4p99 (blue), as well as the RNA-guided TruB homolog Cbf5 (blue) for U2 RNA201 und U5 snRNA25. Modification sites without attributed enzymatic activity are indicated in gray. Mitochondrial LSU rRNA contains only one Ψ residue at position 2819 generated by Pus5.97 Note that for clarity the at least 44 ribosomal Ψs formed by Cbf515 are only suggested and that U2 snRNA positions 56 and 93 and U6 snRNA at position 28 have a dashed outline due to their inducibility.66,100 Pus7p is shown in the cytoplasm with dashed outline, since the enzymes changes its localization from nuclear to cytoplasmic upon heat shock.25
Occurrence and formation of Ψ in human
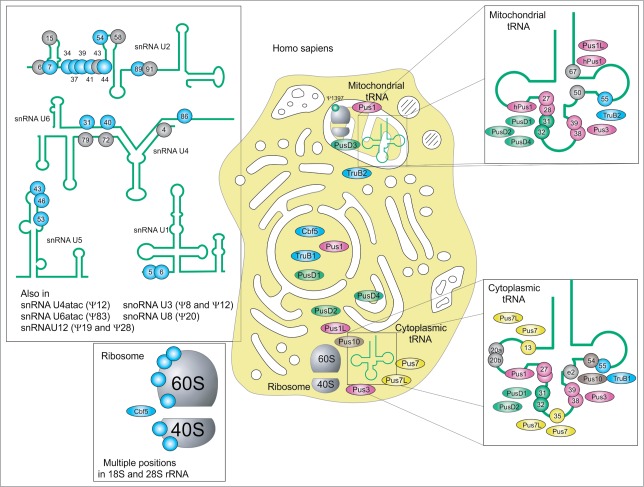
The precise pseudouridylation pattern of human RNAs remains only partially uncovered (see Fig. 6). Despite hard efforts in direct RNA sequencing of cytoplasmic and mitochondrial tRNAs, only some species have been analyzed in detail,1 and some existing pseudouridine sites still escape identification. However, the overall profile of human tRNA modification is similar to the one from S. cerevisiae, even if some minor sites have not (yet) been detected in human. For instance, Ψ32, very common in Eubacteria and in yeast, has been mapped in only one cytoplasmic human tRNA so far, tRNAHis.1 Known human Ψ synthases belong to 5 families, only the RsuA-related family is missing (like it is also the case for S. cerevisiae). One can also notice duplication of some Ψ synthase genes, as Pus1/Pus1L, Pus7/Pus7L, TruB1/TruB2.110 All stand-alone human Ψ synthases are supposed to modify mostly tRNAs, since the great majority of known sites in rRNA and snRNA are attributed to specific H/ACA-snoRNA-guided machinery. However, the implication of stand-alone enzymes (like highly promiscuous Pus1 or/and Pus7) in modification of these species cannot be formally excluded. Only a few predicted human Ψ synthases have been studied up to date, only the specificity of hPus1 was experimentally confirmed,109,111 assignment of the other proteins is mostly based on the sequence homology and the properties of the human and archaeal counterparts and thus remains only tentative. Some of human Ψ synthases are predicted to have preferential mitochondrial localization and are thus supposed to modify tRNAs in this compartment. Like in S. cerevisiae, Ψ13 and Ψ35 are missing in mitochondrial tRNA, while other sites are quite well conserved.
Figure 6.
Distribution of Ψ and Ψ synthases in Homo sapiens: Cellular location of substrates and substrate position are given for TruA family members Pus1,111 Pus1L, Pus3 (UniProt Acc. number Q9BZE2) (purple), TruB family members TruB1110, TruB2110 and Cbf5202 (blue), TruD family members Pus7 and Pus7L (UniProt Acc. number Q9H0K6), RIuA family members PusD1 (UniProt Acc. number Q9UJJ7.1), PusD3 (UniProt Acc. number Q6P087.3) and PusD4 (UniProt Acc. number Q96CM3.1) (green) and Pus1090 (brown). In addition to tRNA and rRNA snRNA and snoRNA are modified. Note that, to current knowledge, Ψ-positions in snRNAs U2, U4 and U6, exclusively formed by H/ACA Box RNPs.203 Positions with known or putative guide RNAs are depicted in blue, while gray positions await guide RNA identification.203 Not shown are Ψ-containing SRA RNA204 and human telomerase RNA.45
Regulation of and via Ψ
Levels of Ψ differ from tissue to tissue112 and may be cell cycle dependent.113 This implies that Ψ levels are regulated and, in turn, that there is a biological benefit to this regulation. Consistent with this picture, additional Ψs can be induced in yeast U2 snRNA and U6 snRNA in site-specific and stimulus specific manner66,101 and the mTOR pathway induces a higher Ψ content in 28S rRNA of CHO cell cultures.114 In mouse, Ψ is directly involved in activation of nuclear receptors via pseudouridylation of steroid receptor RNA activator (SRA).115 Such regulatory function in transcription is related to the concept of a regulatory role of Ψ in translation. Interestingly, Ψ can suppress non-sense codons in vitro and in vivo, if it is artificially and site-specifically introduced into mRNA.65 This led to a detailed study on possible effects pseudouridine modified nonsense and sense codons.116 Nonsense suppression may be caused via a Ψ-A base pair, which is thought to stabilize the 2 non-canonical base pairs completing the codon-anticodon interaction.117 Indeed 2 recent studies reported various inducible Ψs in yeast mRNA.24,25 Further investigation identified the enzyme Pus7p to be mainly responsible for heat shock induced pseudouridylation in yeast: A change in localization of the enzyme from mainly nuclear to also cytosolic seems to allow mRNA pseudouridylation that presumably contributes to mRNA stability.25
The mechanism of Ψ formation
Kinetics
Kinetic studies on Ψ synthases depict them as slow in catalysis under multiple turnover conditions (see Table 1) with changes in RNA conformation118, catalysis119,120 and catalysis or product release119 as rate limiting steps.
Table 1.
Overview on Km and kcat of Ψ synthases
| Enzyme | Organism | Family | KM / nM | kcat / s−1 |
|---|---|---|---|---|
| RIuD205 | E. coli | RIuD | 980 ± 180 | ∼0.033 |
| TruB116,120, 123,135 | E. coli | TruB | 146-780 | 0.12-0.7 |
| TruA119,131 | E. coli | TruA | 940 | 0.18-0.7 |
| RIuA138, 206,207 | E. coli | RIuA | 108-308 | 0.1 |
| TruD208 | E. coli | TruD | 380 | 0.001 |
| Pus1p111 | H. sapiens | TruA | 32 | — |
| Pus1p118 | S. cerevisiae | TruA | 420-740 | ∼0.006 |
| Pus10p47 | P. furiosus | Pus10p | 400 | 0.9 |
Judging from the apo-enzyme121, cocrystal structure16 and kinetic studies119,120 E. coli TruB, serving as a general role model for Ψ synthases, acts via an induced fit mechanism that consists of at least 4 steps: (i) initial RNA binding (ii) induced fit (iii) catalysis (iv) product release. The process of base-flipping involves a non-essential12,122 histidine5 for TruB family members or an arginine in other Ψ synthases.12,47,77,78 The most obvious explanation for Ψ formation being slow is that the chemistry of the reaction is rate limiting and may not allow faster catalysis.119 Several Ψ synthases were found to act more efficiently on weakly structured RNAs and avoid modification of stable RNAs.76,80,102,123,124 Stabilizing RNA modifications are of cooperative and/or pleiotropic nature73,125,126 and single modifications were often found to be non-quantitative.83,127
Mechanistic studies on Ψ synthases using 5-fluorouridine
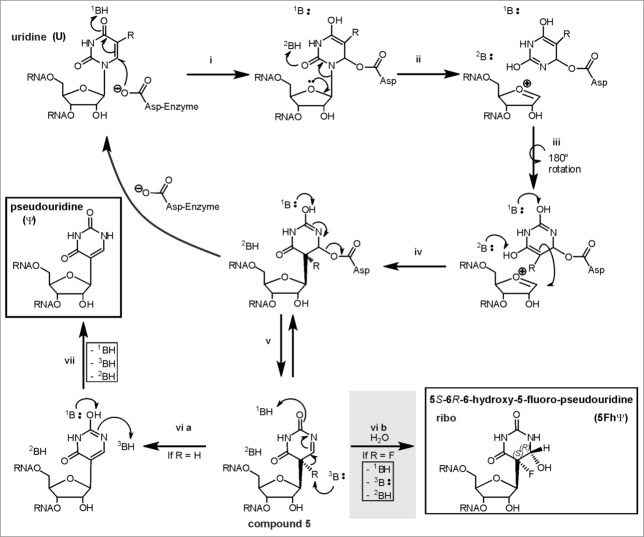
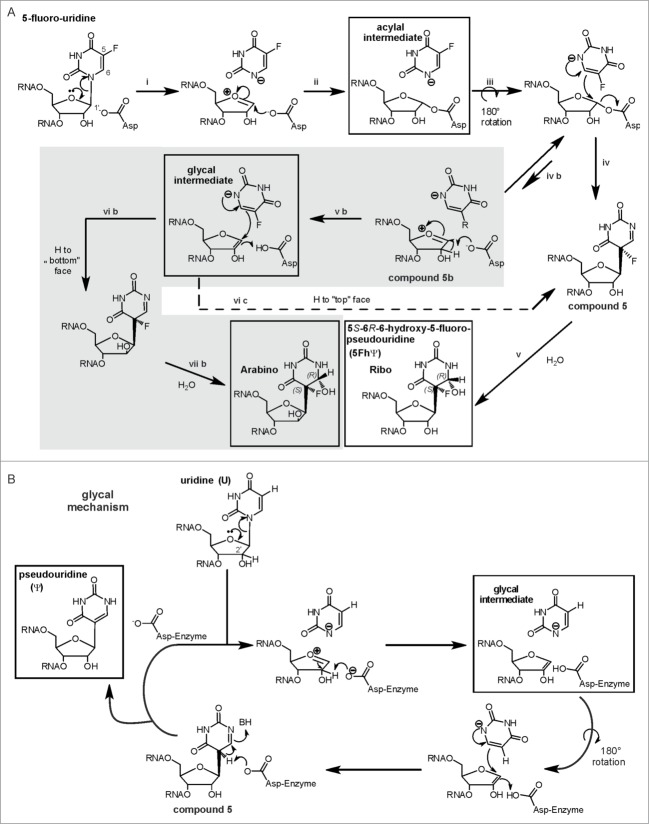
Inhibition of Ψ synthases by the anti-cancer drug 5-fluorouracil128 (5FU) was investigated in several organisms.129-132 While the original target of 5FU is thymidylate synthase, it may also inhibit formation of ribothymidine if incorporated into RNA, where it is also commonly regarded as inhibitor of Ψ formation. SDS-PAGE stable, but heat disruptable 5FU-RNA-Ψ synthase complexes,77,131-134 requiring the catalytic aspartate, were reported for several Pus enzymes, leading to the proposal of a Michael addition like mechanism of Ψ formation.131,133 In this mechanism, the catalytic aspartate would attack the Michael acceptor C6 of the base (see Fig. 7), while the alternative, so called “acylal mechanism” would involve an aspartate attack on the C1’ of the ribose (see non-gray reaction path in Fig. 8a).133
Figure 7.
The “Michael” addition-like mechanism of Ψ formation modified from Czudnochowski and coworkers.78 The substrate is either 5-fluorouridine (R = F) or uridine (R = H ). To account for the “generally accepted covalent adduct” of the substrate base’ C6 to the catalytic aspartate of the enzyme (if the substrate is 5FU), the aspartate would have to attack in an Michael addition-like manner. The protonation- and deprotonation steps proposed by Czudnochowski et al. would be carried out by yet unidentified bases (1B, 2B, 3B). Please note that turnover of U and 5FU both result in compound 5. This final intermediate is either deprotonated to eventually result in pseudouridine or hydrated in case of 5FU (gray shaded reaction step) to generate 5S-6R-6-hydroxy-5-fluoro-pseudouridine.
Figure 8.
(See previous page). The acylal mechanism and the glycal mechanism for Ψ formation in a version modified from ref. 139. (A) In case of 5FU the acylal intermediate can result in compound 5 to eventually yield the 5S-6R-6-hydroxy-5-fluoro-pseudouridine found in the crystal structures. However, an equilibrium of the 5FU-acylal intermediate with an oxocarbonium intermediate (compound 5b) might open an additional gray shaded reaction manifold exclusively to 5FU. This would account for the arabino-isomer as minor product of E. coli TruB action on 5FU RNA that was discovered by Miracco and Mueller.139 Pseuoduridine could be formed by the not-shaded acylal mechanism, the only difference would be the last step: The ‘F’ would be a proton that is abstracted to generate the product. (B) Miracco and Mueller proposed that pseudouridine could also be formed by a third glycal mechanism. This mechanism resembles the gray reaction manifold in a) but yield only one product in ribo conformation.
Cocrystal structures of active, e.g. refs.16,77,78,121,134-136, but not of inactive137 Ψ synthases with 5FU RNA contain a hydrated and rearranged 5FU, 5S-6R-6-hydroxy-5-fluoro-pseudouridine. Evidence that the hydration is caused by attack of water132,138,139 does not favor one mechanism over the other, but is strengthened by a fortuitous adduct of RNA with Ψ synthase RIuB, where a conserved, but not catalytically essential135 Tyrosine78 substitutes water. In one case the SDS PAGE stable adduct proved sensitive to X-ray exposure136, implying that the covalent adduct cannot be visualized in crystals because it was destroyed during measurement.
One Ψ synthase, E. coli TruB, failed to form a SDS PAGE stable complex with 5FU-containing RNA and failed to be inhibited in kinetic studies,132 which is consistent with turnover of 5FU to the same rearranged, hydrated product by several E. coli enzymes.139 In depth NMR analysis of E. coli TruB-5FU-RNA products, revealed a second, minor product in the arabino conformation, specifically resulting from turnover of 5FU-RNA.140 To account for the lack of arabino product in U turnover compared to 5FU turnover, Miracco and Mueller suggested that U and 5FU might be turned over by different mechanism. Pseudouridine could either be formed by the acylal mechanism, which is shown in the non-gray reaction path of Figure 8a, or by a third ‘glycal mechanism’ shown in Figure 8b. Miracco and Mueller hypothesize that 5FU turnover by the acylal mechanism (Fig. 8a) might open an additional, reaction manifold, shaded gray in Figure 8A, which is unavailable to uridine.140 They suggested that step ‘iv b’ and the following reaction path leading to the arabino product are restricted to 5FU due to lower reactivity: The electron-withdrawing fluorine substituent might stabilize the free anion of the fluorinated pyrimidine, thereby decreasing its nucleophilicity.140 Please note that the glycal intermediate in Figure 8a can, in contrast to its counterpart in Figure 8b, be converted to either the ribo product (“H to top face”) or to the arabino product (“H to bottom face”), again due to the assumed long lifetime of the intermediate. E. coli RIuA might also form an arabino product, as 2 products detectable in preliminary NMR data imply.139 Undoubtedly, this analysis is the most sophisticated and most reliable analysis of 5FU-turnover by a Ψ synthase reported until now. In this respect it is particularly surprising that a minor arabino product was not reported in any of the available cocrystal structures of 5FU-RNA and Ψ synthases. We checked the B-factors of the respective O2’ in cocrystal structures of 3 different enzyme families for irregularities: Indeed we found them to be mostly unremarkable.16,76,77,120,133-135 This indicates a confidence of the ribo conformation compared to an arabino conformation that is similar to the accuracy of the whole structure. Seemingly the arabino product is either not contained in the crystals or not detectable for yet unknown reasons.
Of note, related modification enzymes use both, the attack on C1’68,141 and the C6 Michael addition mechanism, respectively.142-147 The most instructive hint in this case might be, that related transglycosylases actually proceed by a C1’ attack as reviewed in ref. 148.
Functions of Ψ residues in RNAs
Structural effects - tRNA
The most conserved Ψ modifications stabilize the tertiary structure of tRNA, be it at position 32,149 39150,151 or 55.152 Conformational effects caused by Ψ39 influence anticodon recognition153,154, missreading and frame shifting in yeast (together with Ψ38)155, and interaction with HIV RNA.156
Several eukaryotic cytoplasmic tRNAs carry Ψ at the anticodon positions 34, 35 and 36, where the modification is introduced in intron-dependent manner, as reviewed in ref. 157. Ψ35, the only modification tolerated at that position158, is especially conserved in tRNATyr of a large variety of eukaryotes1, including, e.g., the amobea Tetrahymena thermophila159 and Xenopus.160 Presumably, Ψ35 confers superior stabilization to the anticodon by replacing a (U33)O2’-HC5(U35) hydrogen bond by the stronger (U33)O2’-H-N1(Ψ35).158 Until now there is no mechanistic basis for other anticodon Ψs, namely at positions 34 and 36, that can occur single1 or as pair.161
The function of Ψ in mitochondrial tRNAs is less characterized. In case of human Ψ occurs at positions 27, 28, 41, 42, 49, 40, 50 and 67, and occasionally at 55.162 A well understood, but special case demonstrating a possible role of nucleoside modifications is human mitochondrial tRNALys. The conformational equilibrium of this tRNA is influenced by nucleoside modifications, including 2 Ψs. These Ψs, located at positions 27 and 28, have, in contrast to the usual role of Ψ, a slight destabilizing effect on the canonical cloverleaf structure.163
Role of Ψ in the helix 69 of the ribosome
The role of Ψ in ribosomes was reviewed recently,15 a deeply investigated motif conserved over all domains is helix 69 (H69). The three Ψs in the isolated H69 of E. coli (indicated by the dashed box in Fig. 3) show complicated pleiotropic effects,164 potentially involving increased base stacking and N1H hydrogen bonding165,166 and influence of a m3Ψ modification167 and pH.168 These effects are equally present in human H69169,170 and in whole ribosomes171, and influence ribosomal subunit association.172 These conformational effects still await full clarification.
Ψ in spliceosomal branch-site architecture
Ψ in small nuclear RNAs was thoroughly reviewed recently, e.g. in refs. 15,41 A prominent example is a Ψ residue in eukaryotic U2 RNA that stabilizes and fine-tunes spliceosomal branch-site interaction39,173, involving a water-ΨNH1 hydrogen bond.174,175
Functional importance for RNA
Ψ in artificial mRNAs
Synthetic replacement of all uridines by Ψ renders mRNAs non-immunogenic176, increases biological stability176-178 and enhances translation in vivo176,179,180, while reducing PKR activation.181 In contrast, studies with in vitro assays suggested that mRNAs where all Us were changed to Ψs inhibit translation at the initiation and elongation levels.182
Ψ in eukaryotic mRNAs
Recently at least 260 Ψs in 238 mRNAs of Saccharomyces cerevisiae could be identified with most frequent occurrences in the GUA valine codon and an initial screen of highly expressed genes identified 96 Ψs in 89 human mRNAs.24 A second study could link 41 Ψs in 41 mRNAs to specific Ψ synthases in yeast and 136 mRNA sites in human to specific Ψ synthases.25 Although the majority of modifications could be induced by starvation24 or heat shock25, their actual functional relevance remains to be proven. In case of yeast most pseudouridines are introduced not by H/ACA box RNPs but by 4 out of 9 stand-alone protein Ψ synthases: Mainly by Pus1p and Pus7p, but also by Pus2p and Pus4p.24,25 Occurrences of Ψ in mRNA are widely distributed over coding, as well as non-coding 5’ and 3’ sequences. It is therefore possible that a portion of modification sites mimic Pus substrates rather by coincidence than due to an actual advantage gained from pseudouridylation.24
RluD/ribosomal assembly
Knockout of RIuD, the enzyme generating the 3 Ψs H69 of the E. coli ribosome (see dashed box in Fig. 3), interferes with ribosome assembly,183 implying requirement for normal growth in E. coli K12,184 in contrast to wild type E. coli.185 A mutated release factor 2 rescues ΔRIuD E. coli K12,186 which is consistent with increased affinity of native release factor 2 to pseudouridylated H69.187 In yeast the loss of Ψs in Helix69 impairs growth and influences ribosome synthesis188 and function synergistically,189 but also with pleiotropic effects.188,190
Implications in human pathologies
Pseudouridine related enzymes have been implicated in various human diseases, e. g. in Crohn's disease and Celiac disease191 and X-linked ichthyosis.71 The involvement of NEP1, a N1-Ψ specific methyltransferase, in the Bowen-Conradi syndrome192 and dyskerin in X-linked dyskeratosis congenita193 may not be directly related to Ψ, but rather caused by involvement of the proteins in ribosomal assembly194 and telomere maintenance, respectively.195,196 A recent study detected a slightly lower pseudouridylation level in dyskeratosis congentia patients compared to healthy individuals and verified Ψs in the telomerase RNA component that may be involved in the disease.25
A mutation in the human PUS1 gene leads to hypomodification in mitochondrial tRNAs by preventing hPus1p activity, resulting in mitochondrial myopathy and sideroblastic anemia.197 Due to the wide substrate specificity of Pus1p discussed above, hypomodification of RNAs other than tRNA might contribute to the disease. 162
Conclusions and outlook
To date enzymes and substrate positions for Ψ formation are quite well understood in the major model organisms E. coli, H. volcanii and S. cerevisiae. In contrast the chemical mechanism of Ψ formation is as elusive as ever. Possible are either an acylal mechanism139, where the catalytic Asp acts as general base as inferred from the pH dependency of the TruB reaction122 or a Michael addition mechanism that would not account for a (still not directly characterized) covalent adduct of the enzyme to C6 of the target base in RNA.78 These mechanistic studies suffer from ambiguous mutagenesis approaches, which were unable to identify the major basic and acidic residues required for either mechanisms (abbreviated as ‘B’ in Fig. 7 and 8).
The next task on hand is undoubtedly the functional characterization of Ψ in mRNA and elucidating the modifications regulatory properties. Such properties should intensify the interest in human Ψ synthases, of which only hPus1p is characterized109,111 and all others lack evidence on protein level (Fig. 6).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kellner S, Neumann J, Rosenkranz D, Lebedeva S, Ketting RF, Zischler H, Schneider D, Helm M. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem Commun 2014; 50:3516-8; PMID:24567952; http://dx.doi.org/ 10.1039/c3cc49114e [DOI] [PubMed] [Google Scholar]
- 3. Cohn WE, Volkin E. Nucleoside-5′-hosphates from ribonucleic acid. Nature 1951; 167:483-4; http://dx.doi.org/ 10.1038/167483a0 [DOI] [Google Scholar]
- 4. Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Bio Chem 1957; 227:907-15; PMID:13463012 [PubMed] [Google Scholar]
- 5. Yu CT, Allen FW. Studies on an isomer of uridine isolated from ribonucleic acids. Biochim Biophys Acta 1959; 32:393-406; PMID:13846687; http://dx.doi.org/ 10.1016/0006-3002(59)90612-2 [DOI] [PubMed] [Google Scholar]
- 6. Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta 1959; 32:569-71; PMID:13846687 [DOI] [PubMed] [Google Scholar]
- 7. Cortese R, Kammen HO, Spengler SJ, Ames BN. Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem 1974; 249:1103-8; PMID:4592259 [PubMed] [Google Scholar]
- 8. Ciampi MS, Arena F, Cortese R. Biosynthesis of pseudouridine in the in vitro transcribed tRNATyr precursor. FEBS Lett 1977; 77:75-82; PMID:323061; http://dx.doi.org/ 10.1016/0014-5793(77)80196-8 [DOI] [PubMed] [Google Scholar]
- 9. Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J Bio Chem 1988; 263:2255-63; PMID:3276686 [PubMed] [Google Scholar]
- 10. Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 1995; 23:5020-6; PMID:8559660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB life 2000; 49:341-51; PMID:10902565; http://dx.doi.org/ 10.1080/152165400410182 [DOI] [PubMed] [Google Scholar]
- 12. Hamma T, Ferré-D'Amaré AR. Pseudouridine synthases. Chem Bio 2006; 13:1125-35; PMID:17113994; http://dx.doi.org/ 10.1016/j.chembiol.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 13. Hur S, Stroud RM, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J Biol Chem 2006; 281:38969-73; PMID:17085441; http://dx.doi.org/ 10.1074/jbc.R600034200] [DOI] [PubMed] [Google Scholar]
- 14. Hamma T, Ferré-D'Amaré AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem 2010; 285:805-9; PMID:19917616; http://dx.doi.org/ 10.1074/jbc.R109.076893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ge J, Yu Y-T. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 2013; 38:210-8; PMID:23391857; http://dx.doi.org/ 10.1016/j.tibs.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoang C, Ferré-D'Amaré AR. Cocrystal structure of a tRNA $$55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 2001; 107:929-39; PMID:11779468; http://dx.doi.org/ 10.1016/S0092-8674(01)00618-3 [DOI] [PubMed] [Google Scholar]
- 17. Durairaj A, Limbach PA. Mass spectrometry of the fifth nucleoside: a review of the identification of pseudouridine in nucleic acids. Anal Chim Acta 2008; 623:117-25; PMID:18620915; http://dx.doi.org/ 10.1016/j.aca.2008.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pomerantz SC, McCloskey JA. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal Chem 2005; 77:4687-97; PMID:16053277; http://dx.doi.org/ 10.1021/ac058023p [DOI] [PubMed] [Google Scholar]
- 19. Stanley J, Vassilenko S. A different approach to RNA sequencing. Nature 1978; 274:87-9; PMID:662002; http://dx.doi.org/ 10.1038/274087a0] [DOI] [PubMed] [Google Scholar]
- 20. Hengesbach M, Meusburger M, Lyko F, Helm M. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA 2008; 14:180-7; PMID:17998290; http://dx.doi.org/ 10.1261/rna.742708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao X, Yu YT. Detection and quantitation of RNA base modifications. RNA 2004; 10:996-1002; PMID:15146083; http://dx.doi.org/ 10.1261/rna.7110804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dai Q, Fong R, Saikia M, Stephenson D, Yu YT, Pan T, Piccirilli JA. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res 2007; 35:6322-9; PMID:17881375; http://dx.doi.org/ 10.1093/nar/gkm657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ofengand J, Del Campo M, Kaya Y. Mapping pseudouridines in RNA molecules. Methods 2001; 25:365-73; PMID:11860291; http://dx.doi.org/ 10.1006/meth.2001.1249 [DOI] [PubMed] [Google Scholar]
- 24. Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; PMID:25192136; http://dx.doi.org/ 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, et al. Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://dx.doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Addepalli B, Limbach PA. Mass spectrometry-based quantification of pseudouridine in RNA. J Am Soc Mass Spectrom 2011; 22:1363-72; PMID:21953190[http://dx.doi.org/ 10.1007/s13361-011-0137-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taucher M, Ganisl B, Breuker K. Identification, localization, and relative quantitation of pseudouridine in RNA by tandem mass spectrometry of hydrolysis products. Int J Mass Spectrom 2011; 304:91-7; PMID:21960742; http://dx.doi.org/ 10.1016/j.ijms.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meng Z, Limbach PA. Quantitation of ribonucleic acids using 18O labeling and mass spectrometry. Anal Chem 2005; 77:1891-5; PMID:15762601; http://dx.doi.org/ 10.1021/ac048801y [DOI] [PubMed] [Google Scholar]
- 29. Popova AM, Williamson JR. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J Am Chem Soc 2014; 136:2058-69; PMID:24422502; http://dx.doi.org/ 10.1021/ja412084b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saponara AG, Enger MD. The isolation from ribonucleic acid of substituted uridines containing alpha-aminobutyrate moieties derived from methionine. Biochim Biophys Acta 1974; 349:61-77; PMID:11400439; http://dx.doi.org/ 10.1016/0005-2787(74)90009-4 [DOI] [PubMed] [Google Scholar]
- 31. Brand RC, Klootwijk J, Planta RJ, Maden BE. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J 1978; 169:71-7; PMID:629754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hudson GA, Bloomingdale RJ, Znosko BM. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 2013; 19:1474-82; PMID:24062573; http://dx.doi.org/ 10.1261/rna.039610.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic acids research 2013; 42:3492-501; PMID:24369424; http://dx.doi.org/ 10.1093/nar/gkt1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hall KB, McLaughlin LW. Properties of pseudouridine N1 imino protons located in the major groove of an A-form RNA duplex. Nucleic Acids Res 1992; 20:1883-9; PMID:1579489; http://dx.doi.org/ 10.1093/nar/20.8.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall KB, McLaughlin LW. Properties of a U1/mRNA 5' splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry 1991; 30:1795-801; PMID:1993194; http://dx.doi.org/ 10.1021/bi00221a010 [DOI] [PubMed] [Google Scholar]
- 36. Davis DR, Poulter CD. 1H-15N NMR studies of Escherichia coli tRNA(Phe) from hisT mutants: a structural role for pseudouridine. Biochemistry 1991; 30:4223-31; PMID:2021615 [DOI] [PubMed] [Google Scholar]
- 37. Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 1994; 33:7560-7; PMID:8011621; http://dx.doi.org/ 10.1021/bi00190a008 [DOI] [PubMed] [Google Scholar]
- 38. Auffinger P WEE. Effects of pseudouridylation on tRNA hydration and dynamics. In: Grosjean H. BR, ed. Modification and Editing of RNA Washington: ASM Press, 1998:103-12. [Google Scholar]
- 39. Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struc Biol 2002; 9:958-65; PMID:12426583; http://dx.doi.org/ 10.1038/nsb873 [DOI] [PubMed] [Google Scholar]
- 40. Hayrapetyan A S-LS, Helm M. Function of modified nucleosides in RNA stabilization. In: G H., ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function, and Evolution. Austin, TX: Landes Bioscience, 2009:550-63. [Google Scholar]
- 41. Wu G, Yu AT, Kantartzis A, Yu YT. Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation. Wiley Interdiscip Rev RNA 2011; 2:571-81; PMID:21957045; http://dx.doi.org/ 10.1002/wrna.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Becker HF, Motorin Y, Florentz C, Giege R, Grosjean H. Pseudouridine and ribothymidine formation in the tRNA-like domain of turnip yellow mosaic virus RNA. Nucleic Acids Res 1998; 26:3991-7; PMID:9705510; http://dx.doi.org/ 10.1093/nar/26.17.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baumstark T, Ahlquist P. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA 2001; 7:1652-70; PMID:11720293 [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p-and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol Endocrinol 2007; 21:686-99; PMID:17170069; http://dx.doi.org/ 10.1210/me.2006-0414 [DOI] [PubMed] [Google Scholar]
- 45. Kim N-K, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6. 1 hairpin in human telomerase RNA. Nucleic Acids Res 2010; 38:6746-56; PMID:20554853; http://dx.doi.org/ 10.1093/nar/gkq525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe Y, Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 2000; 28:2342-52; PMID:10871366; http://dx.doi.org/ 10.1093/nar/28.12.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kamalampeta R, Keffer-Wilkes LC, Kothe U. tRNA binding, positioning and modification by the pseudouridine synthase Pus10. J Mol Biol 2013; 425:3863-74; PMID:23743107; http://dx.doi.org/ 10.1016/j.jmb.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 48. Ero R, Peil L, Liiv A, Remme J. Identification of pseudouridine methyltransferase in Escherichia coli. RNA 2008; 14:2223-33; PMID:18755836; http://dx.doi.org/ 10.1261/rna.1186608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Douthwaite S. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA 2008; 14:2234-44; PMID:18755835; http://dx.doi.org/ 10.1261/rna.1198108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res 2010; 38:2387-98; PMID:20047967; http://dx.doi.org/ 10.1093/nar/gkp1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wurm JP, Griese M, Bahr U, Held M, Heckel A, Karas M, Soppa J, Wöhnert J. Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. RNA 2012; 18:412-20; PMID:22274954; http://dx.doi.org/ 10.1261/rna.028498.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res 2011; 39:1526-37; PMID:20972225; http://dx.doi.org/ 10.1093/nar/gkq931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; [http://dx.doi.org/ 10.1016/S0092-8674(00)80263-9 [DOI] [PubMed] [Google Scholar]
- 54. Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997; 89:565-73; PMID:9160748; http://dx.doi.org/ 10.1016/S0092-8674(00)80238-X [DOI] [PubMed] [Google Scholar]
- 55. Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A 2002; 99:7536-41; PMID:12032318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 2005; 33:3133-44; PMID:15933208; http://dx.doi.org/ 10.1093/nar/gki630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev 2005; 19:1238-48; PMID:15870259; http://dx.doi.org/ 10.1101/gad.1309605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang X, Duan J, Li S, Wang P, Ma S, Ye K, Zhao XS. Kinetic and thermodynamic characterization of the reaction pathway of box H/ACA RNA-guided pseudouridine formation. Nucleic Acids Res 2012; 40:10925-36; PMID:23012266; http://dx.doi.org/ 10.1093/nar/gks882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fourmann J-B, Tillault A-S, Blaud M, Leclerc F, Branlant C, Charpentier B. Comparative study of two bBox H/ACA ribonucleoprotein pseudouridine-synthases: relation between conformational dynamics of the Guide RNA, enzyme assembly and activity. PloS One 2013; 8:e70313; PMID:23922977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liang B, Xue S, Terns RM, Terns MP, Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struc Mol Biol 2007; 14:1189-95; PMID:18059286; http://dx.doi.org/ 10.1038/nsmb1336 [DOI] [PubMed] [Google Scholar]
- 61. Liang B, Kahen EJ, Calvin K, Zhou J, Blanco M, Li H. Long-distance placement of substrate RNA by H/ACA proteins. RNA 2008; 14:2086-94; PMID:18755842; http://dx.doi.org/ 10.1261/rna.1109808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamalampeta R, Kothe U. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep 2012; 2:663; PMID:22993689; http://dx.doi.org/ 10.1038/srep00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xiao M, Yang C, Schattner P, Yu YT. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 2009; 15:176-86; PMID:19033376; http://dx.doi.org/ 10.1261/rna.1361509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen C, Zhao X, Kierzek R, Yu YT. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Mol Cell Biol 2010; 30:4108-19; PMID:20606010; http://dx.doi.org/ 10.1128/MCB.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karijolich J, Yu Y-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011; 474:395-8; PMID:21677757; http://dx.doi.org/ 10.1038/nature10165] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 2011; 30:79-89; PMID:21131909; http://dx.doi.org/ 10.1038/emboj.2010.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Preumont A, Snoussi K, Stroobant V, Collet JF, Van Schaftingen E. Molecular identification of pseudouridine-metabolizing enzymes. JBio Chem 2008; 283:25238-46; PMID:18591240; http://dx.doi.org/ 10.1074/jbc.M804122200 [DOI] [PubMed] [Google Scholar]
- 68. Huang S, Mahanta N, Begley TP, Ealick SE. Pseudouridine monophosphate glycosidase: a new glycosidase mechanism. Biochemistry 2012; 51:9245-55; PMID:23066817; http://dx.doi.org/ 10.1021/bi3006829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drahovsky D, Winkler A, Skoda J. Increased urinary pseudouridine excretion in rats following irradiation. Nature 1964; 201:411-2; PMID:14110019[http://dx.doi.org/ 10.1038/201411a0 [DOI] [PubMed] [Google Scholar]
- 70. Rasmuson T, Bjork GR. Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol 1995; 34:61-7; PMID:7865238; http://dx.doi.org/ 10.3109/02841869509093640 [DOI] [PubMed] [Google Scholar]
- 71. Preumont A, Rzem R, Vertommen D, Van Schaftingen E. HDHD1, which is often deleted in X-linked ichthyosis, encodes a pseudouridine-5′-phosphatase. Biochem J 2010; 431:237-44; PMID:20722631; http://dx.doi.org/ 10.1042/BJ20100174 [DOI] [PubMed] [Google Scholar]
- 72. Goldberg IH, Rabinowitz M. The incorporation of 5-ribosyluracil triphosphate into RNA in nuclear extracts of mammalian cells. Biochem Biophys Res Commun 1961; 6:394-8; PMID:13899698; http://dx.doi.org/ 10.1016/0006-291X(61)90152-8 [DOI] [PubMed] [Google Scholar]
- 73. Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res 2011; 39:2304-18; PMID:21097467; http://dx.doi.org/ 10.1093/nar/gkq1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kinghorn SM, O’Byrne CP, Booth IR, Stansfield I. Physiological analysis of the role of truB in Escherichia coli: a role for tRNA modification in extreme temperature resistance. Microbiology 2002; 148:3511-20; PMID:12427942 [DOI] [PubMed] [Google Scholar]
- 75. Urbonavicius J, Durand JM, Bjork GR. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol 2002; 184:5348-57; PMID:12218021; http://dx.doi.org/ 10.1128/JB.184.19.5348-5357.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell 2007; 26:189-203; PMID:17466622; http://dx.doi.org/ 10.1016/j.molcel.2007.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alian A, DeGiovanni A, Griner SL, Finer-Moore JS, Stroud RM. Crystal structure of an RluF–RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome. J Mol Biol 2009; 388:785-800; PMID:19298824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Czudnochowski N, Ashley GW, Santi DV, Alian A, Finer-Moore J, Stroud RM. The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs. Nucleic Acids Res 2013; 42:2037-48; PMID:24214967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 2001; 7:1603-15; PMID:11720289 [PMC free article] [PubMed] [Google Scholar]
- 80. Gu X, Yu M, Ivanetich KM, Santi DV. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 1998; 37:339-43; PMID:9425055; http://dx.doi.org/ 10.1021/bi971590p [DOI] [PubMed] [Google Scholar]
- 81. Vaidyanathan PP, Deutscher MP, Malhotra A. RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA 2007; 13:1868-76; PMID:17872507; http://dx.doi.org/ 10.1261/rna.711207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Leppik M, Ero R, Liiv A, Kipper K, Remme J. Different sensitivity of H69 modification enzymes RluD and RlmH to mutations in Escherichia coli 23S rRNA. Biochimie 2012; 94:1080-9; PMID:22586702; http://dx.doi.org/ 10.1016/j.biochi.2012.02.023 [DOI] [PubMed] [Google Scholar]
- 83. Gupta R. Halobacterium-volcanii Transfer-Rnas - identification of 41 Transfer-Rnas covering all Amino-Acids, and the sSequences of 33 class-I Transfer-Rnas. JBiol Chem 1984; 259:9461-71; PMID:6746655 [PubMed] [Google Scholar]
- 84. Gupta R. Transfer-Rnas of halobacterium-volcanii - sequences of 5 leucine and 3 serine Transfer-Rnas. Syst Appl Microbiol 1986; 7:102-5. [Google Scholar]
- 85. Constantinesco F, Motorin Y, Grosjean H. Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res 1999; 27:1308-15; PMID:9973619; http://dx.doi.org/ 10.1093/nar/27.5.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Grosjean H, Gaspin C, Marck C, Decatur WA, de Crecy-Lagard V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC genomics 2008; 9:470; PMID:18844986; http://dx.doi.org/ 10.1186/1471-2164-9-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crécy-Lagard V, Gupta R. Pseudouridine formation in archaeal RNAs: the case of Haloferax volcanii. RNA 2011; 17:1367-80; PMID:21628430; http://dx.doi.org/ 10.1261/rna.2712811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gurha P, Gupta R. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 2008; 14:2521-7; PMID:18952823; http://dx.doi.org/ 10.1261/rna.1276508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Joardar A, Jana S, Fitzek E, Gurha P, Majumder M, Chatterjee K, Geisler M, Gupta R. Role of forefinger and thumb loops in production of $$54 and $$55 in tRNAs by archaeal Pus10. RNA 2013; 19:1279-94; PMID:23898217; http://dx.doi.org/ 10.1261/rna.039230.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 2007; 373:1243-54; PMID:17900615; http://dx.doi.org/ 10.1016/j.jmb.2007.08.053 [DOI] [PubMed] [Google Scholar]
- 91. Neumann P, Lakomek K, Naumann PT, Erwin WM, Lauhon CT, Ficner R. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res 2014; 42:6673-85; PMID:24705700[http://dx.doi.org/ 10.1093/nar/gku249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 2006; 34:4293-301; PMID:16920741; http://dx.doi.org/ 10.1093/nar/gkl530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Muller S, Urban A, Hecker A, Leclerc F, Branlant C, Motorin Y. Deficiency of the tRNATyr: Y35-synthase aPus7 in Archaea of the Sulfolobales order might be rescued by the H/ACA sRNA-guided machinery. Nucleic Acids Res 2009; 37:1308-22; PMID:19139072; http://dx.doi.org/ 10.1093/nar/gkn1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Behm-Ansmant I, Branlant C, Motorin Y. The saccharomyces cerevisiae Pus2 protein encoded by YGL063w ORF is a mitochondrial tRNA: Psi27/28-synthase. RNA 2007; 13:1641-7; PMID:17684231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 1998; 4:856-69; PMID:9671058; http://dx.doi.org/ 10.1017/S1355838298980396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Behm-Ansmant I, Grosjean H, Massenet S, Motorin Y, Branlant C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J Biol Chem 2004; 279:52998-3006; PMID:15466869; http://dx.doi.org/ 10.1074/jbc.M409581200 [DOI] [PubMed] [Google Scholar]
- 97. Ansmant I, Massenet S, Grosjean H, Motorin Y, Branlant C. Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of psi(2819) in 21S mitochondrial ribosomal RNA. Nucleic Acids Res 2000; 28:1941-6; PMID:10756195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J Biol Chem 1998; 273:1316-23; PMID:9430663; http://dx.doi.org/ 10.1074/jbc.273.3.1316 [DOI] [PubMed] [Google Scholar]
- 99. Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 1997; 25:4493-9; PMID:9358157; http://dx.doi.org/ 10.1093/nar/25.22.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ansmant I, Motorin Y, Massenet S, Grosjean H, Branlant C. Identification and characterization of the tRNA:Psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. The Journal of biological chemistry 2001; 276:34934-40; PMID:11406626; http://dx.doi.org/ 10.1074/jbc.M103131200 [DOI] [PubMed] [Google Scholar]
- 101. Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 2014; 8:966-73; PMID:25127136; http://dx.doi.org/ 10.1016/j.celrep.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Urban A, Behm-Ansmant I, Branlant C, Motorin Y. RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J Biol Chem 2009; 284:5845-58; PMID:19114708; http://dx.doi.org/ 10.1074/jbc.M807986200 [DOI] [PubMed] [Google Scholar]
- 103. Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 1999; 19:2142-54; PMID:10022901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 2003; 22:1889-97; PMID:12682021; http://dx.doi.org/ 10.1093/emboj/cdg191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 2003; 9:1371-82; PMID:14561887; http://dx.doi.org/ 10.1261/rna.5520403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. A previously unidentified activity of yeast and mouse RNA: pseudouridine synthases 1 (Pus1p) on tRNAs. RNA 2006; 12:1583-93; PMID:16804160; http://dx.doi.org/ 10.1261/rna.100806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol Cell Biol 2008; 28:3089-100; PMID:18332121; http://dx.doi.org/ 10.1128/MCB.01574-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Czudnochowski N, Wang AL, Finer-Moore J, Stroud RM. In human pseudouridine synthase 1 (hPus1) a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J Mol Biol 2013; 425:3875-87; PMID:23707380; http://dx.doi.org/ 10.1016/j.jmb.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sibert BS, Patton JR. Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements. Nucleic Acids Res 2012; 40:2107-18; PMID:22102571; http://dx.doi.org/ 10.1093/nar/gkr1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zucchini C, Strippoli P, Biolchi A, Solmi R, Lenzi L, D'Addabbo P, Carinci P, Valvassori L. The human TruB family of pseudouridine synthase genes, including the Dyskeratosis Congenita 1 gene and the novel member TRUB1. Int J Mol Med 2003; 11:697-704; PMID:12736709 [PubMed] [Google Scholar]
- 111. Sibert BS, Fischel-Ghodsian N, Patton JR. Partial activity is seen with many substitutions of highly conserved active site residues in human Pseudouridine synthase 1. RNA 2008; 14:1895-906; PMID:18648068; http://dx.doi.org/ 10.1261/rna.984508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Brandmayr C, Wagner M, Bruckl T, Globisch D, Pearson D, Kneuttinger AC, Reiter V, Hienzsch A, Koch S, Thoma I. Isotope-based analysis of modified tRNA nucleosides correlates modification density with translational efficiency. Angew Chem 2012; 51:11162-5; PMID:23037940; http://dx.doi.org/ 10.1002/anie.201203769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CT, Dedon PC, Begley TJ. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle 2012; 11:3656-65; PMID:22935709; http://dx.doi.org/ 10.4161/cc.21919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Courtes FC, Gu C, Wong NS, Dedon PC, Yap MG, Lee DY. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. J Biotech 2014; 174:16-21; PMID:24480570; http://dx.doi.org/ 10.1016/j.jbiotec.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 2004; 15:549-58; PMID:15327771; http://dx.doi.org/ 10.1016/j.molcel.2004.06.044 [DOI] [PubMed] [Google Scholar]
- 116. Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA 2012; 18:355-67; PMID:22282339; http://dx.doi.org/ 10.1261/rna.031351.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fernandez IS, Ng CL, Kelley AC, Wu G, Yu Y-T, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 2013; 500:107-10; PMID:23812587; http://dx.doi.org/ 10.1038/nature12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Arluison V, Buckle M, Grosjean H. Pseudouridine synthetase Pus1 of Saccharomyces cerevisiae: kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA. J Mol Biol 1999; 289:491-502; PMID:10356324; http://dx.doi.org/ 10.1006/jmbi.1999.2789 [DOI] [PubMed] [Google Scholar]
- 119. Wright JR, Keffer-Wilkes LC, Dobing SR, Kothe U. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA 2011; 17:2074-84; PMID:21998096[http://dx.doi.org/ 10.1261/rna.2905811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Friedt J, Leavens FM, Mercier E, Wieden H-J, Kothe U. An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. Nucleic Acids Res 2014; 42:3857-70; PMID:24371284; http://dx.doi.org/ 10.1093/nar/gkt1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Nat Acad Sci U S A 2003; 100:12648-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hamilton CS, Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. Arch Biochem Biophys 2005; 433:322-34; PMID:15581587; http://dx.doi.org/ 10.1016/j.abb.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 123. Watanabe M, Nameki N, Matsuo-Takasaki M, Nishimura S, Okada N. tRNA recognition of tRNA-guanine transglycosylase from a hyperthermophilic archaeon, Pyrococcus horikoshii. J Biol Chem 2001; 276:2387-94; PMID:11060284; http://dx.doi.org/ 10.1074/jbc.M005043200 [DOI] [PubMed] [Google Scholar]
- 124. Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 2003; 9:711-21; PMID:12756329; http://dx.doi.org/ 10.1261/rna.5230603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry 2010; 49:4934-44; PMID:20459084; http://dx.doi.org/ 10.1021/bi100408z [DOI] [PubMed] [Google Scholar]
- 126. Helm M, Alfonzo JD. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 2014; 21:174-85; PMID:24315934; http://dx.doi.org/ 10.1016/j.chembiol.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Buchhaupt M, Sharma S, Kellner S, Oswald S, Paetzold M, Peifer C, Watzinger P, Schrader J, Helm M, Entian KD. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PloS One 2014; 9:e89640; PMID:24586927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003; 3:330-8; PMID:12724731; http://dx.doi.org/ 10.1038/nrc1074 [DOI] [PubMed] [Google Scholar]
- 129. Samuelsson T. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res 1991; 19:6139-44; PMID:1956773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Patton JR. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry 1993; 32:8939-44; PMID:8364039 [DOI] [PubMed] [Google Scholar]
- 131. Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry 1998; 37:344-51; PMID:9425056; http://dx.doi.org/ 10.1021/bi971874+ [DOI] [PubMed] [Google Scholar]
- 132. Spedaliere CJ, Mueller EG. Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine. RNA 2004; 10:192-9; PMID:14730018; http://dx.doi.org/ 10.1261/rna.5100104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc Nat Acad Sci U S A 1999; 96:14270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Phannachet K, Huang RH. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res 2004; 32:1422-9; PMID:14990747; http://dx.doi.org/ 10.1093/nar/gkh287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Phannachet K, Elias Y, Huang RH. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry 2005; 44:15488-94; PMID:16300397; http://dx.doi.org/ 10.1021/bi050961w [DOI] [PubMed] [Google Scholar]
- 136. Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferré-D'Amaré AR. Crystal structure of pseudouridine synthase RluA: Indirect sequence readout through protein-induced RNA structure. Mol Cell 2006; 24:535-45; PMID:17188032 [DOI] [PubMed] [Google Scholar]
- 137. Hoang C, Hamilton CS, G Mueller E, Ferré-D'Amaré AR. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci 2005; 14:2201-6; PMID:15987897; http://dx.doi.org/ 10.1110/ps.051493605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hamilton CS, Greco TM, Vizthum CA, Ginter JM, Johnston MV, Mueller EG. Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine. Biochemistry 2006; 45:12029-38; PMID:17002302; http://dx.doi.org/ 10.1021/bi061293x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. McDonald MK, Miracco EJ, Chen J, Xie Y, Mueller EG. The handling of the mechanistic probe 5-fluorouridine by the pseudouridine synthase TruA and its consistency with the handling of the same probe by the pseudouridine synthases TruB and RluA. Biochemistry 2010; 50:426-36; PMID:21142053; http://dx.doi.org/ 10.1021/bi101737z] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Miracco EJ, Mueller EG. The products of 5-fluorouridine by the action of the pseudouridine synthase TruB disfavor one mechanism and suggest another. J Am Chem Soc 2011; 133:11826-9; PMID:21744792; http://dx.doi.org/ 10.1021/ja201179f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Silva RG, Schramm VL. Uridine phosphorylase from Trypanosoma cruzi: kinetic and chemical mechanisms. Biochemistry 2011; 50:9158-66; PMID:21932786; http://dx.doi.org/ 10.1021/bi2013382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Oja T, Niiranen L, Sandalova T, Klika KD, Niemi J, Mantsala P, Schneider G, Metsä-Ketelä M. Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway. Proc Nat Acad Sci U S A 2013; 110:1291-6; PMID:23297194; http://dx.doi.org/ 10.1073/pnas.1207407110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Graves KL, Butler MM, Hardy LW. Roles of Cys148 and Asp179 in catalysis by deoxycytidylate hydroxymethylase from bacteriophage T4 examined by site-directed mutagenesis. Biochemistry 1992; 31:10315-21; PMID:1420151; http://dx.doi.org/ 10.1021/bi00157a020 [DOI] [PubMed] [Google Scholar]
- 144. Kunitani MG, Santi DV. On the mechanism of 2′-deoxyuridylate hydroxymethylase. Biochemistry 1980; 19:1271-5; PMID:6770896; http://dx.doi.org/ 10.1021/bi00548a001 [DOI] [PubMed] [Google Scholar]
- 145. Wu JC, Santi DV. Kinetic and Catalytic Mechanism of Hhai Methyltransferase. J Biol Chem 1987; 262:4778-86; PMID:3558369 [PubMed] [Google Scholar]
- 146. Motorin Y, Helm M. RNA nucleotide methylation. Wiley interdisciplinary reviews RNA 2011; 2:611-31; PMID:21823225; http://dx.doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 147. Santi DV, McHenry CS, Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry 1974; 13:471-81; PMID:4203910; http://dx.doi.org/ 10.1021/bi00700a012 [DOI] [PubMed] [Google Scholar]
- 148. Stengl B, Reuter K, Klebe G. Mechanism and substrate specificity of tRNA-guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism. Chembiochem 2005; 6:1926-39; PMID:16206323; http://dx.doi.org/ 10.1002/cbic.200500063 [DOI] [PubMed] [Google Scholar]
- 149. Cabello-Villegas J, Nikonowicz EP. Solution structure of $$32-modified anticodon stem–loop of Escherichia coli tRNAPhe. Nucleic Acids Res 2005; 33:6961-71; PMID:16377777; http://dx.doi.org/ 10.1093/nar/gki1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Denmon AP, Wang J, Nikonowicz EP. Conformation Effects of Base Modification on the Anticodon Stem–Loop of Bacillus subtilis tRNATyr. J Mol Biol 2011; 412:285-303; PMID:21782828; http://dx.doi.org/ 10.1016/j.jmb.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Vendeix FA, Murphy IV FV, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Sproat B, Malkiewicz A, Agris PF. Human tRNALys3UUU is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-EnolTautomerism. J Mol Biol 2012; 416:467; PMID:22227389; http://dx.doi.org/ 10.1016/j.jmb.2011.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res 2002; 30:4751-60; PMID:12409466; http://dx.doi.org/ 10.1093/nar/gkf595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of psi at tRNA's position 39. Nucleic Acids Res 1999; 27:3543-9; PMID:10446245; http://dx.doi.org/ 10.1093/nar/27.17.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 1999; 285:115-31; PMID:9878393; http://dx.doi.org/ 10.1006/jmbi.1998.2297 [DOI] [PubMed] [Google Scholar]
- 155. Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Bio Che 2002; 277:30445-53; PMID:12058040; http://dx.doi.org/ 10.1074/jbc.M203456200 [DOI] [PubMed] [Google Scholar]
- 156. Bilbille Y, Vendeix FA, Guenther R, Malkiewicz A, Ariza X, Vilarrasa J, Agris PF. The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Res 2009; 37:3342-53; PMID:19324888; http://dx.doi.org/ 10.1093/nar/gkp187] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Grosjean H, Szweykowska-Kulinska Z, Motorin Y, Fasiolo F, Simos G. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie 1997; 79:293-302; PMID:9258438; http://dx.doi.org/ 10.1016/S0300-9084(97)83517-1 [DOI] [PubMed] [Google Scholar]
- 158. Auffinger P, Westhof E. An extended structural signature for the tRNA anticodon loop. RNA 2001; 7:334-41; PMID:11333014; http://dx.doi.org/ 10.1017/S1355838201002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Junker V, Teichmann T, Hekele A, Fingerhut C, Beier H. The tRNATyr-isoacceptors and their genes in the ciliate Tetrahymena thermophila: cytoplasmic tRNATyr has a QPsiA anticodon and is coded by multiple intron-containing genes. Nucleic Acids Res 1997; 25:4194-200; PMID:9336446; http://dx.doi.org/ 10.1093/nar/25.21.4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Nishikura K, De Robertis EM. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 1981; 145:405-20; PMID:7196457; http://dx.doi.org/ 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- 161. Szweykowska-Kulinska Z, Senger B, Keith G, Fasiolo F, Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile). EMBO J 1994; 13:4636-44; PMID:7925304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annual Rev Genet 2011; 45:299-329; PMID:21910628; http://dx.doi.org/ 10.1146/annurev-genet-110410-132531 [DOI] [PubMed] [Google Scholar]
- 163. Kobitski AY, Hengesbach M, Seidu-Larry S, Dammertz K, Chow CS, Van Aerschot A, Nienhaus GU, Helm M. Single-molecule FRET reveals a cooperative effect of two methyl froup modifications in the folding of human mitochondrial tRNALys. Chem Biol 2011; 18:928-36; PMID:21802013; http://dx.doi.org/ 10.1016/j.chembiol.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 164. Meroueh M, Grohar PJ, Qiu J, SantaLucia J, Jr., Scaringe SA, Chow CS. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucleic Acids Res 2000; 28:2075-83; PMID:10773075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Desaulniers JP, Chang YC, Aduri R, Abeysirigunawardena SC, SantaLucia J, Jr., Chow CS. Pseudouridines in rRNA helix 69 play a role in loop stacking interactions. Org biomol Chem 2008; 6:3892-5; PMID:18931791; http://dx.doi.org/ 10.1039/b812731j [DOI] [PubMed] [Google Scholar]
- 166. Jiang J, Aduri R, Chow CS, SantaLucia J. Structure modulation of helix 69 from Escherichia coli 23S ribosomal RNA by pseudouridylations. Nucleic Acids Res 2013: 42:3971-81; PMID:24371282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Chui HM, Desaulniers JP, Scaringe SA, Chow CS. Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m(3)Psi and Psi. J Org Chem 2002; 67:8847-54; PMID:12467398; http://dx.doi.org/ 10.1021/jo026364m [DOI] [PubMed] [Google Scholar]
- 168. Abeysirigunawardena SC, Chow CS. pH-dependent structural changes of helix 69 from Escherichia coli 23S ribosomal RNA. RNA 2008; 14:782-92; PMID:18268024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Sakakibara Y, Abeysirigunawardena SC, Duc A-Ce E, Dremann DN, Chow CS. Ligand-and pH-Induced conformational changes of RNA domain helix 69 revealed by 2-aminopurine fluorescence. Angew Chem Int Ed 2012; 51:12095-8; PMID:23097282; http://dx.doi.org/ 10.1002/anie.201206000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sumita M, Jiang J, SantaLucia J, Chow CS. Comparison of solution conformations and stabilities of modified helix 69 rRNA analogs from bacteria and human. Biopolymers 2012; 97:94-106; PMID:21858779; http://dx.doi.org/ 10.1002/bip.21706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Sakakibara Y, Chow CS. Probing conformational states of modified helix 69 in 50S ribosomes. J Am Chem Soc 2011; 133:8396-9; PMID:21557607; http://dx.doi.org/ 10.1021/ja2005658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Sakakibara Y, Chow CS. Role of pseudouridine in structural rearrangements of helix 69 during bacterial ribosome assembly. ACS Chem Biol 2012; 7:871-8; PMID:22324880; http://dx.doi.org/ 10.1021/cb200497q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 2001; 7:833-45; PMID:11424937; http://dx.doi.org/ 10.1017/S1355838201002308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Nat Acad Sci U S A 2002; 99:12697-702; PMID:1224234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Lin Y, Kielkopf CL. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry 2008; 47:5503-14; PMID:18435545; http://dx.doi.org/ 10.1021/bi7022392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy 2008; 16:1833-40; PMID:18797453; http://dx.doi.org/ 10.1038/mt.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res 2011; 39:9329-38; PMID:21813458; http://dx.doi.org/ 10.1093/nar/gkr586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Kariko K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 2012; 20:948-53; PMID:22334017; http://dx.doi.org/ 10.1038/mt.2012.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell 2010; 7:618-30; PMID:20888316; http://dx.doi.org/ 10.1016/j.stem.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Boros G, Miko E, Muramatsu H, Weissman D, Emri E, Rozsa D, Nagy G, Juhász A, Juhász I, van der Horst G, et al. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: A new approach with future therapeutic potential. J Photochem Photobiol B 2013; 129:93-9; PMID:24211294; http://dx.doi.org/ 10.1016/j.jphotobiol.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 2010; 38:5884-92; PMID:20457754; http://dx.doi.org/ 10.1093/nar/gkq347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Aspden JL, Jackson RJ. Differential effects of nucleotide analogs on scanning-dependent initiation and elongation of mammalian mRNA translation in vitro. RNA 2010; 16:1130-7; PMID:20423978; http://dx.doi.org/ 10.1261/rna.1978610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Gutgsell NS, Deutscher MP, Ofengand J. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 2005; 11:1141-52; PMID:15928344; http://dx.doi.org/ 10.1261/rna.2550105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 1998; 4:1407-17; PMID:9814761; http://dx.doi.org/ 10.1017/S1355838298981146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. O'Connor M, Gregory ST. Inactivation of the RluD pseudouridine synthase has minimal effects on growth and ribosome function in wild-type Escherichia coli and Salmonella enterica. J Bacteriol 2011; 193:154-62; PMID:21037010; http://dx.doi.org/ 10.1128/JB.00970-10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Ejby M, Sorensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc Nat Acad Sci U S A 2007; 104:19410-5; PMID:18032607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kipper K, Sild S, Hetenyi C, Remme J, Liiv A. Pseudouridylation of 23S rRNA helix 69 promotes peptide release by release factor RF2 but not by release factor RF1. Biochimie 2011; 93:834-44; PMID:21281690; http://dx.doi.org/ 10.1016/j.biochi.2010.12.018 [DOI] [PubMed] [Google Scholar]
- 188. Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009; 15:1716-28; PMID:19628622; http://dx.doi.org/ 10.1261/rna.1724409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 2007; 28:965-77; PMID:18158895 [DOI] [PubMed] [Google Scholar]
- 190. Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 2009; 37:7665-77; PMID:19820108; http://dx.doi.org/ 10.1093/nar/gkp816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagacé C, Stokkers PC, Hommes DW. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS genetics 2011; 7:e1001283; PMID:21298027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, Coghlan G, Lamont RE, Liu S, Wiechert J, Cattini PA. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am J Hum Genet 2009; 84:728-39; PMID:19463982; http://dx.doi.org/ 10.1016/j.ajhg.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature genetics 1998; 19:32-8; PMID:9590285; http://dx.doi.org/ 10.1038/ng0598-32 [DOI] [PubMed] [Google Scholar]
- 194. Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res 2011; 39:2445-57; PMID:21087996; http://dx.doi.org/ 10.1093/nar/gkq1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 1999; 402:551-5; PMID:10591218; http://dx.doi.org/ 10.1038/990141 [DOI] [PubMed] [Google Scholar]
- 196. Thumati NR, Zeng XL, Au HH, Jang CJ, Jan E, Wong JM. Severity of X-linked dyskeratosis congenita (DKCX) cellular defects is not directly related to dyskerin (DKC1) activity in ribosomal RNA biogenesis or mRNA translation. Human mutation 2013; 34:1698-707; PMID:24115260; http://dx.doi.org/ 10.1002/humu.22447 [DOI] [PubMed] [Google Scholar]
- 197. Patton JR, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J Biol Chem 2005; 280:19823-8; PMID:15772074; http://dx.doi.org/ 10.1074/jbc.M500216200 [DOI] [PubMed] [Google Scholar]
- 198. Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 2006; 443:302-7; PMID:16943774; http://dx.doi.org/ 10.1038/nature05151 [DOI] [PubMed] [Google Scholar]
- 199. Li S, Duan J, Li D, Yang B, Dong M, Ye K. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 2011; 25:2409-21; PMID:22085967; http://dx.doi.org/ 10.1101/gad.175299.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett 2002; 514:17-25; PMID:11904174 [DOI] [PubMed] [Google Scholar]
- 201. Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 2005; 24:2403-13; PMID:15962000; http://dx.doi.org/ 10.1038/sj.emboj.7600718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 2004; 24:5797-807; PMID:15199136; http://dx.doi.org/ 10.1128/MCB.24.13.5797-5807.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA biology 2010; 7:192-204; PMID:20215871; http://dx.doi.org/ 10.4161/rna.7.2.11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Huet T, Miannay FA, Patton JR, Thore S. Steroid receptor RNA activator (SRA) modification by the human pseudouridine synthase 1 (hPus1p): RNA binding, activity, and atomic model. PloS One 2014; 9:e94610; PMID:24722331; http://dx.doi.org/ 10.1371/journal.pone.0094610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Ero R, Leppik M, Liiv A, Remme J. Specificity and kinetics of 23S rRNA modification enzymes RlmH and RluD. RNA 2010; 16:2075-84; PMID:20817755; http://dx.doi.org/ 10.1261/rna.2234310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Ramamurthy V, Swann SL, Paulson JL, Spedaliere CJ, Mueller EG. Critical aspartic acid residues in pseudouridine synthases. J Biol Chem 1999; 274:22225-30; PMID:10428788; http://dx.doi.org/ 10.1074/jbc.274.32.22225 [DOI] [PubMed] [Google Scholar]
- 207. Spedaliere CJ, Hamilton CS, Mueller EG. Functional importance of motif I of pseudouridine synthases: mutagenesis of aligned lysine and proline residues. Biochemistry 2000; 39:9459-65; PMID:10924141; http://dx.doi.org/ 10.1021/bi001079n [DOI] [PubMed] [Google Scholar]
- 208. Chan CM, Huang RH. Enzymatic characterization and mutational studies of TruD–the fifth family of pseudouridine synthases. Arch Biochem Biophys 2009; 489:15-9; PMID:19664587; http://dx.doi.org/ 10.1016/j.abb.2009.07.023 [DOI] [PubMed] [Google Scholar]