Abstract
MicroRNA (miRNA) comprise a large family of non-protein coding transcripts which regulate gene expression in diverse biological pathways of both plants and animals. We recently used a systematic proteomic approach to generate a protein interactome map of the human miRNA pathway involved in miRNA biogenesis and processing. The interactome expands the number of candidate proteins in the miRNA pathway and connects the network to other cellular processes. Functional analyses identified TRIM65 and at least 3 other proteins as novel regulators of the miRNA pathway. Biochemical studies established that TRIM65 forms stable complexes with TNRC6 proteins and these molecules co-localize in P-body-like structures. Gain of function and RNAi analyses reveal that TRIM65 negatively regulates miRNA-driven suppression of mRNA translation by targeting TNRC6 proteins for ubiquitination and degradation. The potential molecular mechanisms which regulate TRIM65 catalytic activity are discussed.
Keywords: interactome, proteomics, RNA-induced silencing complex, tripartite motif proteins, ubiquitin E3 ligase, TNRC6
Abbreviations
- TRIM65
Tripartite Motif-Containing 65
- MiRNA
microRNA
- AGO
Argonaute
- RISC
RNA-induced silencing complex
- TNRC6
Trinucleotide repeat containing 6
- DGCR8
DiGeorge syndrome critical region gene 8
- TARBP2
TAR (HIV-1) RNA binding protein 2
- HCIP
High confidence interacting protein
- MOV10
Moloney leukemia virus 10
- AP-MS
Affinity purification coupled with mass spectrometry
- IMP-1
IGF2 mRNA-binding protein 1
- PDCD4
Programmed cell death 4
- PTEN
Phosphatase and tensin homolog
Introduction
RNA-mediated gene silencing is an evolutionarily conserved mechanism in which small RNAs induce inactivation of cognate mRNA sequences. MicroRNA (miRNA) represents a class of small RNAs found in plants, animals and some viruses. MiRNAs function like rheostats to rapidly and reversibly fine tune transcription ensuring proper development and homeostasis. MiRNAs have emerged as key regulators of diverse aspects of biology, including differentiation, cell death, metabolism and cellular defense.1-3 Some miRNAs regulate their mRNA target through direct cleavage, however in mammals they most often cause translational repression and mRNA destabilization. Disruption of miRNA function has consequences on cellular homeostasis, thereby contributing to disease pathogenesis, including neurodegenerative, cardiovascular or inflammatory disorders and cancer.4-7
MiRNAs are transcribed from endogenous genes by RNA polymerase II. A nuclear microprocessor complex containing a catalytic RNase III-type enzyme (DROSHA) crops primary miRNA transcripts to release a pre-miRNA hairpin structure of roughly 70 nucleotides. The pre-miRNA is exported to the cytosol where it is processed into a mature miRNA by another RNase III enzyme known as DICER, which is also the core of a large protein complex. The single-stranded mature miRNA contains about 22 nucleotides and is loaded into an effector RNA-induced silencing complex (RISC) where miRNAs silence complementary target mRNA. The RISC ribonucleoprotein complex contains Argonaute (AGO) and TNRC6 (also known as GW182) family members. The miRNA-loaded AGO complex serves as an address label for complementary or partially complementary mRNAs that become silenced by RISC-associated executioner complexes. The effector machinery controlling translational inhibition by miRNA can involve mRNA decapping and deadenylation but the mechanisms are still debated.8 The central constituents of the canonical miRNA-mediated gene silencing pathway are now outlined, but these components are incorporated in large protein complexes containing sets of incompletely defined cofactors. The molecules and molecular mechanisms which regulate miRNA function remain incompletely understood.
MicroRNA Interactome
Most proteins function by collaborating with other proteins in molecular networks that form dynamic machine-like structures.9,10 Methods to purify and identify the components of these protein-protein complexes have improved greatly in recent years and computational tools to organize and map the data evolved in parallel. Proteomic strategies have been successfully applied to define interaction networks and detect novel interactions or unexpected relationships which predict new players in a variety of fundamental biological processes.11-14 Recently, we described a discovery-type affinity purification mass spectrometry (AP-MS) approach to identify additional cofactors which regulate miRNA function in human cells.15 AP-MS relies on the affinity of a bait protein with its interaction partners to pull down protein complexes which are subsequently identified by high resolution tandem mass spectrometry. To facilitate protein purification, epitope tagged molecules are stably expressed in the cells of interest. Then protein complexes are eluted from antibody-conjugated resins under standardized conditions. Developments in computational mass spectrometry have allowed automated analysis of large amounts of MS data. Thus, AP-MS based proteomics has become one of the most versatile techniques for delineating biological pathways on a global scale.16
Several AP-MS studies have provided important insights to advance our understanding of multimolecular complexes and allowed initial characterization of nodes along the miRNA pathway. Gregory, et al17 used AP-MS to identify the double-stranded-RNA-binding protein, DGCR8 as a key component of a multiprotein microprocessor complex. Subsequently, epitope tagged DICER was shown to associate with TARBP2 and AGO proteins in human HEK293 cells.18,19 Using tagged TARBP2 as bait established reciprocal associations with DICER. The network was then extended to include ElF6 and MOV10.18,20 Similar studies examined mammalian AGO clade proteins as baits; these efforts identified associations with DICER, MOV10, TNRC6, and TARBP2 among others.19,21-24 Thus, there have been multiple efforts to examine one or a small set of proteins to characterize the network of protein associations in the miRNA pathway. To investigate how miRNA associated proteins are coordinated into a systematic circuit, we extended this approach to include 40 genes with known or suspected involvement in various phases of the miRNA pathway.15 Each gene was fused with the FLAG epitope and stably expressed in HEK293 cells (Fig. 1). Protein complexes were retrieved by elution from anti-FLAG immunoaffinity columns. Following gel digestion with trypsin, peptides were analyzed by LC-MS/MS. For statistical comparisons controls included datasets from >100 proteins isolated under identical conditions. Computational analyses of MS profiles were used to exclude nonspecific binding molecules and assemble a model network of high confidence interacting proteins (HCIP). Filtering out false positive associations depends on adequate reference control data sets. However, the rigorous algorithms used to limit false positives often result in a high false negative rate. Our recent studies characterized a human miRNA pathway interactome comprising 499 unique interactions between 40 baits and 363 high confidence interacting proteins.15 Comparing with community databases (BioGRID, IntAct and STING plus curated literature), we found 55 known interactors and 444 interactions which were not present in these databases. 93% (50 of 54) of the novel associations tested were validated by co-immunoprecipitation in HEK293T cells or by reciprocal interaction in AP-MS.15 However, this physical map does not represent a complete interaction network. Interactors may be omitted due to the high stringency of the algorithm, cell-type dependence and the low concentrations of some interacting proteins. Yet, the list of interacting proteins serves as a community resource and a guide for future efforts to identify miRNA cofactors. The interactome includes candidates which may directly or indirectly participate in miRNA activity and suggests how the miRNA machinery may be interwoven with other cellular processes. Additional functions of bait proteins which are unrelated to miRNA may be overlaid upon the miRNA circuit.
Figure 1.
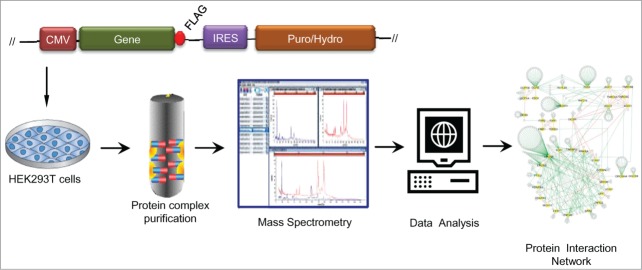
Construction and analysis of the miRNA pathway interactome. Schematic illustration describing the experimental pipeline including production of FLAG-tagged gene constructs, selection of HEK293T stable cell lines, protein complex affinity purification, identification of protein interactors by mass spectrometry, statistical data analysis and integration of high confidence interacting proteins into an interaction map. Details of the 499 protein interactions in the miRNA pathway interactome are presented in Li, et al. 15
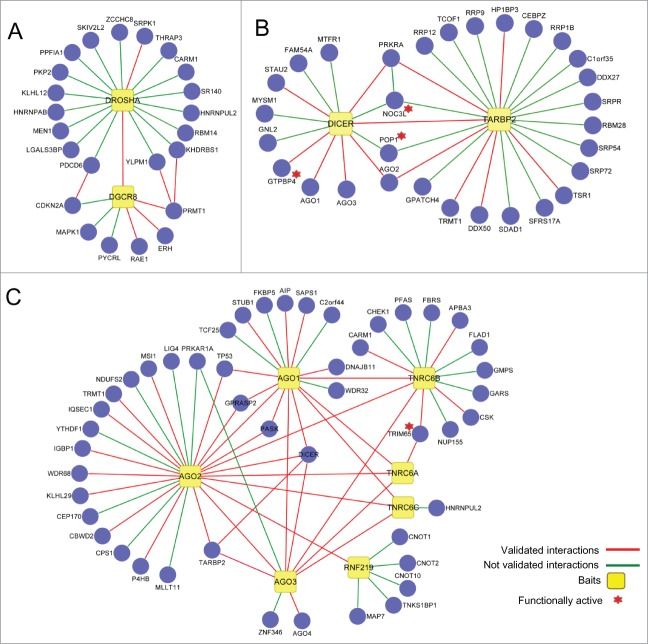
The landscape view of the miRNA interactome revealed extensive connectivity.15 Distinct subnetworks are also observed. For example, DROSHA and DGCR8 form an isolated microprocessor subnetwork consistent with their predominant nuclear localization (Fig. 2A). Five out of 7 HCIP associated with DGCR8 (CDKN2A, MAPK1, DROSHA, RAE1, PRMT1) and 16 out of 18 DROSHA interacting HCIP (SRPK1, THRAP3, CARM1, SR140, HNRNPUL2, RBM14, KHDRBS1, DGCR8, MEN1, HNRNPAB, PDCD6, PKP2, PPFIA1, SKIV2L2, YLPM1, ZCCHC8) have predominant nuclear localization, while the other proteins are more broadly distributed. The reported functions of these HCIP cover transcriptional regulation, RNA splicing, and RNA export.
Figure 2.
Subnetwork maps of the miRNA pathway interactome. (A) DROSHA-DGCR8 microprocessor module. (B) DICER-TARBP2 segment. A complete listing of HCIP for PRKRA is supplied in reference 15. (C) AGO-TNRC6 effector network. Line colors indicate validated interactions. Stars denote novel functionally validated targets.
The cytosolic endoribonuclease, DICER is located in the hub of the miRNA pathway. DICER along with its cofactor, TARBP2 process pre-miRNA and help assemble and load mature miRNAs into AGO complexes.18,25,26 Thirteen HCIP associate with DICER in the network, 7 of which (GTPBP4, POP1, NOC3L, MTFR1, FAM54A, GNL2, MYSM1) were not previously reported (Fig. 2B). Five DICER interactors (NOC3L, POP1 and 3 previously known interactors, AGO2, PRKRA and TARBP2) form a highly interconnected subnetwork. TARBP2 associates with at least 23 distinct partners, although it is not clear how many of these interactions are direct or if some depend upon RNA bridging.
The CCR4-NOT deadenylase associates with the RISC,27 but the precise connections between the complexes are not well understood. RNF219, an unstudied ring finger protein, bridges 4 subunits of the CCR4-NOT deadenylase complex (CNOT1, CNOT2, CNOT10, TNKS1BP1) to the AGO subnetwork (Fig. 2C). Taken together, the MPI comprises at least 3 major clusters corresponding to the well-established core components, DROSHA, DICER and AGO.
RNAi knockdown was used for screening the effects of 66 HCIP, which lacked known roles in the miRNA-pathway, on let-7a miRNA function in HeLa cells.28 Six candidate genes with multiple hits were identified. Knockdown of 4 genes (POP1, NOC3L, PURA and TRIM65) also regulated let-7a reporter activity in A549 cells.15 POP1 associates with DICER and TARBP2 in the miRNA pathway interactome (Fig. 2B). POP1 is a subunit of the RNase P and RNase MRP endoribonuclease complexes. Eukaryotic POP1 coordinates with other subunits including RPP38 and RPP30 to bind predicted RNA stem loop structures.29 These ribonuclease complexes are involved removing the 5’ leader sequences from tRNAs and may also participate in generation of miRNAs by a non-canonical biogenesis pathway.30,31
RNAi screening indicated NOC3L (also known as FAD24) contributes to miRNA biogenesis or processing. NOC3L is an oligonucleotide binding protein involved in adipogenesis and glucose metabolism.32,33 Let-7 family miRNAs also play crucial roles in adipose tissue differentiation and glucose homeostasis,34,35 but potential links between NOC3L and miRNAs were previously unknown. NOC3L interacts with PRKRA (also termed PACT), DICER and TARBP2. PRKRA is structurally and functionally related to TARBP2. PRKRA and TARBP2 molecules have similar size, share domain structure, bind double stranded RNA and both are present in ∼500 kDa complexes that contain DICER and AGO.36,37 PRKRA enhances strand selection for selected miRNAs37 and depletion of PRKRA strongly affects accumulation of mature miRNA in vivo.36 However, the underlying mechanisms for these activities are not understood. DICER may differentially employ PRKRA and TARBP2 proteins to form or stabilize discrete RISC assemblies.38 NOC3L is uniquely positioned to coordinate miRNA processing (Fig. 2B).
PURA interacts with LIN28A and LIN28B in our network.15 LIN proteins were first characterized as controllers of developmental timing in C. elegans.39 LIN family proteins act as suppressors of let-7 miRNA biogenesis in undifferentiated cells. In humans, LIN28 is selectively activated in a subset of poorly differentiated tumors.40 PURA is an evolutionarily conserved cellular protein which binds single-stranded oligonucleotides with purine-rich repeats. PURA participates in processes of transcription and RNA transport,41-43 but there were no prior reports connecting PURA with the miRNA pathway. Interestingly, NOC3L and PURA primarily display nuclear localization, suggesting they may participate in a nuclear RISC-like complex or in pre-miRNA processing.44 In contrast, to the above positive regulators of miRNA activity, TRIM65 negatively regulates let-7a activity. The molecular basis for TRIM65 activity is discussed below. Finally, RNAi depletion of GTPBP4 and PRKRIR increased let-7a reporter levels in HeLa cells, but they do not significantly regulate reporter activity in the A549 human lung cell line.15 Further screening of additional cell types and miRNA systems is required to validate these results and identify other cofactors involved in miRNA pathways. In summary, a sampling of HCIP (66 of 363) identified 6 genes capable of regulating let-7a miRNA activity, 2 of which may be cell type dependent.
TRIM65 and the miRNA Pathway
TRIM65, a previously unidentified regulator of miRNA processing, is a member of the tripartite motif family. TRIM family members share a characteristic N-terminal tripartite motif composed of an ordered combination of 3 domains consisting of a zinc finger of the RING type followed by one or 2 B-box segments and a coiled coil (CC) region (Fig. 3). The RING segment is a rigid cross-braced structure in which the zinc coordination sites are interleaved.45 B-box domains also adopt a cross-brace conformation and bind zinc atoms.46,47 TRIM65 possesses only one B-box, a B-box2 type domain.48 The coiled coil domain contains hyper-helical regions, which are common building blocks in protein chemistry. The CC domain is often associated with protein-protein interactions and oligomerization. Each domain within the TRIM motif also exists independently in many proteins. The patterned conjunction of domains within the TRIM motif has been preserved from worms to humans. Not only has the TRIM motif been evolutionarily conserved, but the success of the tripartite motif architecture is witnessed by the large expansion of TRIM genes in vertebrate species. Humans encode about 70 members of the TRIM family. The success of the TRIM motif in metazoan evolution suggests that a common biochemical function underlies this structure. The TRIM module is followed by a diverse group of domains in the C-terminal segment. At least 11 types of C-terminal domains are found among human TRIM proteins49; the variability in these C-terminal segments is predicted to contribute to function by recruiting unique classes of binding partners.
Figure 3.
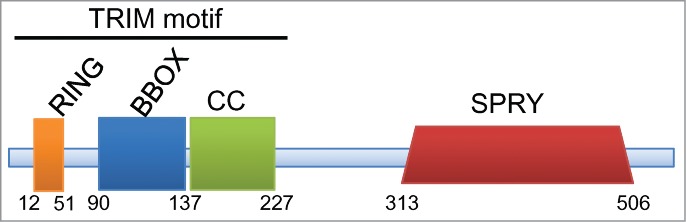
Schematic representation of TRIM65. The full length TRIM65 peptide contains 517 amino acids. The N-terminus includes the multi-domain tripartite (TRIM) motif. Diagram shows positions of the RING finger, B box zinc finger, coiled coil and SPRY domains. The RING domain imparts E3 ubiquitin ligase activity.
Gene expression profiling and immunohistochemistry indicate trim65 genes are universally expressed in human tissues. Anti-TRIM65 antibodies display diffuse staining patterns on human cell lines with concentrations in P-body-like structures.15 The human trim65 gene (NCBI reference sequence NM 173547) consists of 6 exons and is located on chromosome 17q25.1. Expression of TRIM family proteins is often complicated by the occurrence of multiple alternate splice transcripts including some which display opposing function.50–52 The dominant expression of a single TRIM65 isoform in most cell lines tested should simplify analysis. However, we recently cloned an additional TRIM65 isoform which lacks exon 5 (predicting a 22 amino acid deletion of residues 307 through 328) from the T98 glioblastoma cell line. The distribution and function of this shortened isoform are unknown. Genes homologous to trim65 exist among mammals, chicken and pufferfish but orthologs were not found in invertebrates.48 Genome-wide association studies of cerebral white matter lesions link trim65 to subcortical vascular dementia, a common cause of disability in the elderly.53-55 Aside from these linkage studies little was known about TRIM65.
Comparisons of genetic relatedness based on sequence similarities within the core TRIM motif, indicate TRIM65 branched from an ancient subgroup of TRIM proteins, including many which express β-propeller-like NHL structures at their C-terminus.48 This ancient family of TRIM-NHL proteins includes several molecules which physically interact with the RISC and mediate a regulatory role on miRNA activity. TRIM-NHL proteins are expressed in worms (NHL-2 and LIN41), flies (Mei-P26) and vertebrates (TRIM32 and TRIM71).56 The Drosophila mei-P26 gene was the first TRIM homolog implicated in regulation of the miRNA pathway.57 Mei-P26 and a RING-less TRIM-NHL-like protein termed BRAT interact directly or indirectly with AGO proteins and control miRNA expression in the Drosophila ovarian stem cell lineage.57 TRIM71 regulates translational repression and colocalizes with RISC associated molecules including AGO, MOV10 and TNRC6B.58 TRIM71 interactions with RISC-associated molecules are largely RNA-dependent. 58 Mammalian TRIM71 mediates AGO ubiquitination, but the functional consequences of TRIM71-mediated AGO ubiquitination are unclear.58-61 LIN41 is the C. elegans ortholog of TRIM71; LIN41 was shown to immunoprecipitate with DICER.62 While Mei-P26 and TRIM71/LIN41 inhibit miRNA-mediated repression, NHL-2 and TRIM32 enhance silencing of miRNA target genes. TRIM32 inhibits proliferation and induces differentiation of murine neural progenitor cells.63 The mechanistic basis for this activity may include regulation of miRNA activity, as TRIM32 interacts with AGO proteins and enhances the activity of the known stem cell regulator, let-7a miRNA.63 The C. elegans NHL-2 protein also regulates let-7 miRNAs and interacts with the CGH-1 helicase which in turn associates and colocalizes with AGO and TNRC6 orthologs in P-body-like cytoplasmic structures.64 However, the molecular mechanisms by which these evolutionarily conserved TRIM proteins negatively or positively modulate miRNA activity are still largely unknown.
TRIM65 is the latest member of the TRIM family shown to regulate miRNA activity. Unlike the TRIM family members described above, TRIM65 possesses a SPRY domain (domain in SPla and the RYanodine receptor) at the C-terminus (Fig. 3). The SPRY domain structure is characterized as a bent β-sandwich formed by 2 anti-parallel β-sheets.65 The SPRY domain is thought to serve as a protein interaction module.66 Several TRIM family members carrying the SPRY domain were found to participate in innate immunity.52 The role of the SPRY domain in TRIM65 activity has not been determined.
Studies of the miRNA interactome noted that TRIM65 bound to TNRC6 proteins. Domain mapping experiments demonstrated that the TRIM65 CC domain is sufficient for TNRC6A and TNRC6B association.15 This association was stable in the presence of RNase, suggesting direct protein-protein interactions. TNRC6A and its paralogs are part of the multimeric RISC effector complexes.27,67-69 Endogenous TRIM65 is found in punctate cytosolic structures which partially co-localize with endogenous TNRC6A in HeLa and HEK293 cells. After treatment with MG132 these TRIM65-containing P-body-like structures appear enlarged and concentrated.15 It is unknown if TRIM65 participates in assembly/disassembly of the RISC.
Based on its intact RING domain, we predicted TRIM65 was an ubiquitin E3 ligase and that TNRC6 proteins were substrates. In vitro ubiquitination assays demonstrated bacteria-derived TRIM65-GST effectively delivered ubiquitin to TNRC6A. In the contrast, a TRIM65 ligase defective mutant containing a disrupted RING domain failed to catalyze ubiquitination.15 To examine the role of TRIM65 in ubiquitination of endogenous TNRC6, TRIM65 and its RING mutant were transfected into HeLa cells treated with the MG132 proteasome inhibitor. Overexpression of TRIM65 enhances TNRC6A ubiquitination in vivo, while the TRIM65 mutant had little effect on TNRC6A ubiquitination.15 Furthermore, TRIM65 overexpression results in degradation of endogenous TNRC6A protein when MG132 proteasome inhibitor is absent.15 In contrast, TNRC6A protein levels show no substantial change when cells are transfected with the TRIM65 ligase mutant. TNRC6B or TNRC6C protein levels are also reduced when co-expressed with TRIM65.15 The effects on TNRC6 were specific as the level of other proteins in the RISC machinery was not affected. Knockdown with siRNA was used to corroborate the role of TRIM65. TNRC6A ubiquitination is reduced and its half-life is prolonged after silencing TRIM65.15 Furthermore, there is evidence of constitutive TNRC6A ubiquitination,15,70 which may be due to endogenous TRIM65 activity. The combined experiments suggest TRIM65 is a cognate E3 ligase which selectively targets TNRC6 proteins for ubiquitin-mediated degradation by the proteasome. Yet details of the ubiquitination process including the type of ubiquitin linkage, the E2 conjugating enzymes involved and the TNRC6 ubiquitin acceptor sites remain unidentified.
TRIM65 Relieves miRNA Mediated Translational Repression
As TRIM65 regulates the ubiquitination and stability of TNRC6 proteins it was predicted that it should have a corresponding role in regulating miRNA activity. Silencing TRIM65 with siRNA reduces let-7a reporter activity; ectopic expression of TRIM65 has the reciprocal effect. In contrast, a mutant TRIM65 construct in which the RING domain was disrupted to destroy E3 ubiquitin ligase activity, failed to modulate reporter activity.15
TRIM65 knockdown with siRNAs was also used to evaluate the effect of TRIM65 depletion on the endogenous miRNA-guided mRNA silencing machinery. Two miR-21 targeted genes, PDCD4 and PTEN71,72 and one let-7-targeted gene, IMP-173 were examined after knockdown of TRIM65. Depletion of TRIM65 increases TNRC6A protein levels resulting in concomitant reduction of PDCD4, PTEN and IMP-1 (Fig. 4). Real-time PCR quantification was used to monitor mRNA decay rates of PDCD4, PTEN and IMP-1 after transcription was stopped with Actinomycin D. Their mRNA abundance and half-life are not altered by silencing TRIM65, indicating TRIM65 regulates the expression of protein-coding genes at the post-transcriptional level.15 As expected, knockdown of TNRC6A or/and TNRC6B leads to increases of PDCD4, PTEN and IMP-1 expression (Fig. 4). Collectively, the data suggest a model in which TRIM65 relieves miRNA-mediated translational repression by ubiquitination and degradation of TNRC6 (Fig. 5).
Figure 4.
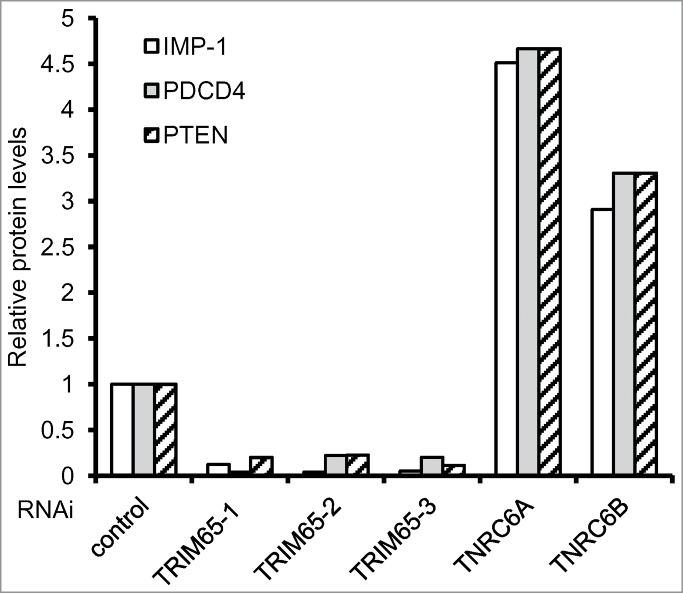
TRIM65 regulates gene expression. Control or validated siRNAs against TRIM65 (3 independent sequences), TNRC6A and TNRC6B were transfected into HeLa cells. After 48 hr endogenous IMP-1, PDCD4, PTEN and β-actin levels were examined by Western blot. Band intensity was quantitated and normalized to β-actin.
Figure 5.
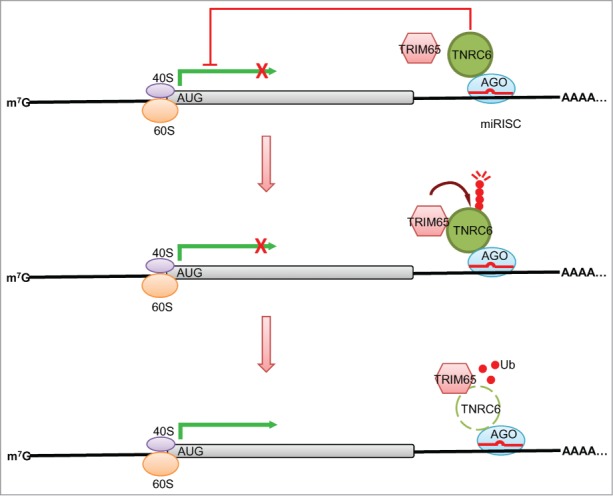
Model of the successive steps in TRIM65-mediated regulation of the miRNA pathway. MiRNA effector complexes containing AGO and TNRC6 direct mature miRNAs to recognize and repress expression of target genes. TRIM65, an E3 ubiquitin ligase, associates with TNRC6 proteins in AGO containing miRNA effector complexes. The signals which control assembly of TRIM65-TNRC6 complexes or activation of E3 ligase activity remain unknown. TRIM65-mediated ubiquitination of TNRC6 substrates results in their degradation by the proteasome. Without the proper set of cofactors AGO effector complexes have impaired silencing function and translation proceeds in the absence of inhibition by miRNAs.
Proteins of the TNRC6 family are central components of the RISC and coordinate downstream silencing events. The molecular mechanisms directing assembly, stability and remodeling of TNRC6 complexes are incompletely understood.74,75 Unrestricted TRIM65 activity could destroy TNRC6 stability, in turn impairing RISC stability, miRNA activity and perturbing cellular homeostasis. Thus, the molecular mechanisms which switch TRIM65 E3 ligase activity on or off are critical for understanding miRNA regulation. To investigate the relationship among TNRC6, TRIM65 and other components of the RISC, cell lysates from cells stably expressing FLAG-TRIM65 were fractionated by centrifugation over a sucrose gradient. AGO and TNRC6 proteins sediment across a broad range of sucrose densities,19,23 FLAG-TRIM65 was primarily located in a low sucrose density fraction with a subset of TNRC6 and AGO proteins. In contrast, FXR1 and PABP1 did not sediment with TRIM65, but were found in the higher density AGO and TNRC6 containing fractions.15 The results emphasize the heterogeneity of RISC effector complexes; it is not clear if TRIM65 containing AGO/TNRC6 complexes are active or inactive in miRNA-mediated gene silencing. In addition to TRIM65 monomers, more slowly migrating oligomeric structures were observed by SDS-PAGE under reducing conditions. The oligomeric TRIM65 bands survive heating to 95°C for 5 minutes in SDS sample buffer. TRIM65 oligomers were shown to sediment in the same fractions as monomeric TRIM65. Co-immunoprecipitation suggests that TNRC6A interacts with both TRIM65 monomers and oligomers.15 Co-expression of HA- and FLAG-tagged TRIM65 demonstrates the capacity for TRIM65 self-association. The apparent molecular weight of the TRIM65 oligomers is consistent with a homo-trimer.15 Although most TRIM proteins form dimers, TRIM21 is an example of another SPRY containing TRIM protein which forms homo-trimers.76 The domain(s) responsible for TRIM65 oligomerization is unknown. Examination of GFP fusion proteins containing isolated RING, B-box, CC or SPRY domains failed to identify an individual region sufficient for self-association.15
Close inspection of the monomeric TRIM65 band revealed the appearance of a doublet. Digestion with a serine/threonine phosphatase, calf-intestinal alkaline phosphatase, suggested the presence of TRIM65 phosphoproteins. To date phosphorylated TRIM65 was only detected in the monomeric fraction of cell derived TRIM65. The apparent reciprocal relationship between phosphorylation and oligomerization requires further examination. Since non-phosphorylated bacterially derived TRIM65 can conjugate ubiquitin on TNRC6A in vitro,15 phosphorylation may not be required for TRIM65 enzyme activity. It remains possible that phosphorylation inhibits TRIM65 catalytic activity or that phosphorylation participates in regulating the balance of TRIM65 monomers and trimers. As the activity and stability of many proteins are regulated posttranslationally, further biochemical analysis is required to understand whether oligomerization or posttranslational modifications modulate TRIM65 E3 ligase and miRNA activity.
Conclusions and Future Perspectives
Development of mass spectrometry-based proteomics coupled with improvements in computational analyses allow system-wide studies of protein interactions. The resulting lists of interacting proteins include potential new members of system pathways and suggest avenues of communication with other signaling modules. Results can depend on cell type and additional evidence is required to integrate novel interaction partners into the overall organization of miRNA circuits. Applying this screening approach to the human miRNA pathway (Version 1.0) identified a repertoire of ∼500 protein-protein interactions. Unraveling which candidates participate in canonical or alternative miRNA pathways remains a challenge. The components which process and present miRNA often overlap with molecules involved in regulating processing of other non-coding RNAs including siRNA and piRNA. Thus, candidates identified in this screen may include constituents of related pathways. Future directions include deciphering the crosstalk between the miRNA network and other cellular processes. Identification of TRIM65 as a new regulator of miRNA activity established the utility and power of the proteomic discovery approach.
The past decade witnessed discovery of multiple molecules critical for miRNA biogenesis and processing. We are beginning to appreciate that dysregulation of miRNA expression or function contributes to aberrant gene expression patterns associated with various diseases including cancer.39,77-81 Thus, there are many translational motives for unraveling the molecular mechanisms which regulate miRNA pathways. TRIM65 represents a previously unidentified regulator of miRNA processing. TRIM65 is a ubiquitously expressed ubiquitin E3 ligase that selectively targets TNRC6 family proteins for proteasomal degradation. As TNRC6 proteins are essential components of the RISC and miRNA effector machinery, their degradation negatively regulates miRNA-mediated gene silencing. Tight control of miRNA activity is required for cellular homeostasis. Thus, TRIM65 activity cannot proceed in an uncontrolled manner. Future research will elucidate whether TRIM65 functions to regulate specific miRNAs or if it has a more global effect on the miRNA pathway. The fundamental mechanisms regulating assembly of TRIM65 complexes and TRIM65 E3 ligase activity remain to be determined. Understanding the molecular mechanisms underlying TRIM65 E3 ligase activity and identifying the intrinsic and extrinsic factors which regulate TRIM65 activity in different contexts are critical outstanding issues. Exploring these unresolved issues may offer opportunities to regulate miRNA activity in disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH grants AI089829 and AI099860.
References
- 1. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci U S A 2013; 110:7056-61; PMID:23569256; http://dx.doi.org/ 10.1073/pnas.1219385110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012; 13:239-50; PMID:22436747; http://dx.doi.org/ 10.1038/nrm3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012; 148:1172-87; PMID:22424228; http://dx.doi.org/ 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008; 456:980-4; PMID:19043405; http://dx.doi.org/ 10.1038/nature07511 [DOI] [PubMed] [Google Scholar]
- 6. Dangwal S, Thum T. microRNA therapeutics in cardiovascular disease models. Annu Rev Pharmacol Toxicol 2014; 54:185-203; PMID:24111539; http://dx.doi.org/ 10.1146/annurev-pharmtox-011613-135957 [DOI] [PubMed] [Google Scholar]
- 7. Zwiers A, Kraal L, van de Pouw Kraan TC, Wurdinger T, Bouma G, Kraal G. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol 2012; 188:1573-7; PMID:22262659; http://dx.doi.org/ 10.4049/jimmunol.1101494 [DOI] [PubMed] [Google Scholar]
- 8. Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and Biochemical Insights to the Role of the CCR4-NOT Complex and DDX6 ATPase in MicroRNA Repression. Mol Cell 2014; 54:751-65; PMID:24768538; http://dx.doi.org/ 10.1016/j.molcel.2014.03.036 [DOI] [PubMed] [Google Scholar]
- 9. Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell 2012; 150:1068-81; PMID:22939629; http://dx.doi.org/ 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006; 440:637-43; PMID:16554755; http://dx.doi.org/ 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- 11. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 2004; 6:97-105; PMID:14743216; http://dx.doi.org/ 10.1038/ncb1086 [DOI] [PubMed] [Google Scholar]
- 13. Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol 2012; 14:93-105; PMID:22119785; http://dx.doi.org/ 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 2011; 35:426-40; PMID:21903422; http://dx.doi.org/ 10.1016/j.immuni.2011.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S, Wang L, Fu B, Berman MA, Diallo A, Dorf ME. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc Natl Acad Sci U S A 2014; 111:6970-5; PMID:24778252; http://dx.doi.org/ 10.1073/pnas.1322545111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell 2011; 145:787-99; PMID:21620140; http://dx.doi.org/ 10.1016/j.cell.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432:235-40; PMID:15531877; http://dx.doi.org/ 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- 18. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005; 436:740-4; PMID:15973356; http://dx.doi.org/ 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 2008; 14:2580-96; PMID:18978028; http://dx.doi.org/ 10.1261/rna.1351608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature 2007; 447:823-8; PMID:17507929; http://dx.doi.org/ 10.1038/nature05841 [DOI] [PubMed] [Google Scholar]
- 21. Frohn A, Eberl HC, Stohr J, Glasmacher E, Rudel S, Heissmeyer V, Mann M, Meister G. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol Cell Proteomics 2012; 11:1442-56; PMID:22918229; http://dx.doi.org/ 10.1074/mcp.M112.017756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol 2005; 15:2149-55; PMID:16289642; http://dx.doi.org/ 10.1016/j.cub.2005.10.048 [DOI] [PubMed] [Google Scholar]
- 23. Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 2007; 8:1052-60; PMID:17932509; http://dx.doi.org/ 10.1038/sj.embor.7401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol 2009; 29:4144-55; PMID:19470757; http://dx.doi.org/ 10.1128/MCB.00380-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Vito C, Riggi N, Cornaz S, Suva ML, Baumer K, Provero P, Stamenkovic I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer cell 2012; 21:807-21; PMID:22698405; http://dx.doi.org/ 10.1016/j.ccr.2012.04.023 [DOI] [PubMed] [Google Scholar]
- 26. Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 2009; 41:365-70; PMID:19219043; http://dx.doi.org/ 10.1038/ng.317 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006; 20:1885-98; PMID:16815998; http://dx.doi.org/ 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev 2004; 18:504-11; PMID:15014042; http://dx.doi.org/ 10.1101/gad.1184404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJ, van Venrooij WJ. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA 1999; 5:512-24; PMID:10199568; http://dx.doi.org/ 10.1017/S1355838299982079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43:892-903; PMID:21925378; http://dx.doi.org/ 10.1016/j.molcel.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lygerou Z, Pluk H, van Venrooij WJ, Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J 1996; 15:5936-48; PMID:8918471 [PMC free article] [PubMed] [Google Scholar]
- 32. Johmura Y, Watanabe K, Kishimoto K, Ueda T, Shimada S, Osada S, Nishizuka M, Imagawa M. Fad24 causes hyperplasia in adipose tissue and improves glucose metabolism. Biological & pharmaceutical bulletin 2009; 32:1656-64; PMID:19801824; http://dx.doi.org/ 10.1248/bpb.32.1656 [DOI] [PubMed] [Google Scholar]
- 33. Tominaga K, Johmura Y, Nishizuka M, Imagawa M. Fad24, a mammalian homolog of Noc3p, is a positive regulator in adipocyte differentiation. J Cell Sci 2004; 117:6217-26; PMID:15564382; http://dx.doi.org/ 10.1242/jcs.01546 [DOI] [PubMed] [Google Scholar]
- 34. Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med 2011; 236:997-1004; PMID:21844119; http://dx.doi.org/ 10.1258/ebm.2011.011101 [DOI] [PubMed] [Google Scholar]
- 35. Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 2011; 108:21075-80; PMID:22160727; http://dx.doi.org/ 10.1073/pnas.1118922109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J 2006; 25:522-32; PMID:16424907; http://dx.doi.org/ 10.1038/sj.emboj.7600942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 2013; 19:639-48; PMID:23531496; http://dx.doi.org/ 10.1261/rna.037424.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res 2013; 41:6568-76; PMID:23661684; http://dx.doi.org/ 10.1093/nar/gkt361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ambros V. The functions of animal microRNAs. Nature 2004; 431:350-5; PMID:15372042; http://dx.doi.org/ 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 40. Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell 2010; 140:445-9; PMID:20178735; http://dx.doi.org/ 10.1016/j.cell.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 41. Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol 1992; 12:5673-82; PMID:1448097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012; 149:1393-406; PMID:22658674; http://dx.doi.org/ 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 43. Wortman MJ, Hanson LK, Martinez-Sobrido L, Campbell AE, Nance JA, Garcia-Sastre A, Johnson EM. Regulation of PURA gene transcription by three promoters generating distinctly spliced 5-prime leaders: a novel means of fine control over tissue specificity and viral signals. BMC Mol Biol 2010; 11:81; PMID:21062477; http://dx.doi.org/ 10.1186/1471-2199-11-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Redfern AD, Colley SM, Beveridge DJ, Ikeda N, Epis MR, Li X, Foulds CE, Stuart LM, Barker A, Russell VJ, et al. RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci U S A 2013; 110:6536-41; PMID:23550157; http://dx.doi.org/ 10.1073/pnas.1301620110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399-434; PMID:19489725; http://dx.doi.org/ 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 46. Massiah MA, Matts JA, Short KM, Simmons BN, Singireddy S, Yi Z, Cox TC. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J Mol Biol 2007; 369:1-10; PMID:17428496; http://dx.doi.org/ 10.1016/j.jmb.2007.03.017 [DOI] [PubMed] [Google Scholar]
- 47. Massiah MA, Simmons BN, Short KM, Cox TC. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J Mol Biol 2006; 358:532-45; PMID:16529770; http://dx.doi.org/ 10.1016/j.jmb.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 48. Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol 2008; 8:225; PMID:18673550; http://dx.doi.org/ 10.1186/1471-2148-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol 2014; 426:1265-84; PMID:24333484; http://dx.doi.org/ 10.1016/j.jmb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res 2011; 31:145-58; PMID:21198351; http://dx.doi.org/ 10.1089/jir.2010.0111 [DOI] [PubMed] [Google Scholar]
- 51. Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential Roles of PML Isoforms. Frontiers Oncol 2013; 3:125; PMID:23734343; http://dx.doi.org/ 10.3389/fonc.2013.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, García-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 2013; 38:384-98; PMID:23438823; http://dx.doi.org/ 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fornage M, Debette S, Bis JC, Schmidt H, Ikram MA, Dufouil C, Sigurdsson S, Lumley T, DeStefano AL, Fazekas F, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol 2011; 69:928-39; PMID:21681796; http://dx.doi.org/ 10.1002/ana.22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Freudenberger P, Schmidt R, Schmidt H. Genetics of age-related white matter lesions from linkage to genome wide association studies. J Neurol Sci 2012; 322:82-6; PMID:22795385; http://dx.doi.org/ 10.1016/j.jns.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmidt H, Freudenberger P, Seiler S, Schmidt R. Genetics of subcortical vascular dementia. Exp Gerontol 2012; 47:873-7; PMID:22735669; http://dx.doi.org/ 10.1016/j.exger.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wulczyn FG, Cuevas E, Franzoni E, Rybak A. MiRNA need a TRIM regulation of miRNA activity by Trim-NHL proteins. Adv Exp Med Biol 2010; 700:85-105; PMID:21627033; http://dx.doi.org/ 10.1007/978-1-4419-7823-3_9 [DOI] [PubMed] [Google Scholar]
- 57. Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 2008; 454:241-5; PMID:18528333; http://dx.doi.org/ 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res 2013; 41:518-32; PMID:23125361; http://dx.doi.org/ 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 2009; 11:1411-20; PMID:19898466; http://dx.doi.org/ 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- 60. Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 2012; 3:923; PMID:22735451; http://dx.doi.org/ 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen J, Lai F, Niswander L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev 2012; 26:803-15; PMID:22508726; http://dx.doi.org/ 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 2006; 124:343-54; PMID:16439208; http://dx.doi.org/ 10.1016/j.cell.2005.11.036 [DOI] [PubMed] [Google Scholar]
- 63. Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009; 136:913-25; PMID:19269368; http://dx.doi.org/ 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 2009; 136:926-38; PMID:19269369; http://dx.doi.org/ 10.1016/j.cell.2009.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grutter C, Briand C, Capitani G, Mittl PR, Papin S, Tschopp J, Grütter MG. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS letters 2006; 580:99-106; PMID:16364311; http://dx.doi.org/ 10.1016/j.febslet.2005.11.076 [DOI] [PubMed] [Google Scholar]
- 66. D'Cruz AA, Babon JJ, Norton RS, Nicola NA, Nicholson SE. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci 2013; 22:1-10; PMID:23139046; http://dx.doi.org/ 10.1002/pro.2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 2009; 15:1433-42; PMID:19535464; http://dx.doi.org/ 10.1261/rna.1703809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 2005; 7:1267-74; PMID:16284622; http://dx.doi.org/ 10.1038/ncb1334 [DOI] [PubMed] [Google Scholar]
- 69. Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 2005; 7:1261-6; PMID:16284623; http://dx.doi.org/ 10.1038/ncb1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11:1143-9; PMID:19684575; http://dx.doi.org/ 10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- 71. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115:787-98; PMID:14697198; http://dx.doi.org/ 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- 72. Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2009; 28:73-84; PMID:18850008; http://dx.doi.org/ 10.1038/onc.2008.370 [DOI] [PubMed] [Google Scholar]
- 73. Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res 2008; 68:2587-91; PMID:18413726; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0264 [DOI] [PubMed] [Google Scholar]
- 74. Liu X, Jin DY, McManus MT, Mourelatos Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell 2012; 46:507-17; PMID:22503104; http://dx.doi.org/ 10.1016/j.molcel.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Flores O, Kennedy EM, Skalsky RL, Cullen BR. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res 2014; 42:4629-39; PMID:24464996; http://dx.doi.org/ 10.1093/nar/gkt1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol 2007; 44:2406-14; PMID:17118455; http://dx.doi.org/ 10.1016/j.molimm.2006.10.013 [DOI] [PubMed] [Google Scholar]
- 77. Chuang AY, Chuang JC, Zhai Z, Wu F, Kwon JH. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm bowel dis 2014; 20:126-35; PMID:24297055; http://dx.doi.org/ 10.1097/01.MIB.0000436954.70596.9b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Harada M, Luo X, Murohara T, Yang B, Dobrev D, Nattel S. MicroRNA regulation and cardiac calcium signaling: role in cardiac disease and therapeutic potential. Circ Res 2014; 114:689-705; PMID:24526675; http://dx.doi.org/ 10.1161/CIRCRESAHA.114.301798 [DOI] [PubMed] [Google Scholar]
- 79. Iliopoulos D. MicroRNA Circuits Regulate the Cancer-Inflammation Link. Sci signal 2014; 7:pe8; PMID:24667374; http://dx.doi.org/ 10.1126/scisignal.2005053 [DOI] [PubMed] [Google Scholar]
- 80. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin oncol 2014; 11:145-56; PMID:24492836; http://dx.doi.org/ 10.1038/nrclinonc.2014.5 [DOI] [PubMed] [Google Scholar]
- 81. Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 2009; 15:1443-61; PMID:19561119; http://dx.doi.org/ 10.1261/rna.1534709 [DOI] [PMC free article] [PubMed] [Google Scholar]