Abstract
Plant microRNAs (miRNAs) are predominantly 21 nucleotides (nt) long but non-canonical lengths of 22 and 20 nt are commonly observed in diverse plant species. While miRNAs longer than 21 nt can be attributed to the neglect of unpaired bases within asymmetric bulges by the ruler function of DICER-LIKE 1 (DCL1), how 20-nt miRNA is generated remains obscure. Analysis of small RNA data revealed that 20-nt miRNA can be divided into 3 main groups featured by atypical 3′ overhangs or shorter duplex regions. Asymmetric bulges or mismatches at specific positions are commonly observed within each group and were shown to be crucial for 20-nt miRNA formation. Analysis of DCL1 cleavage sites on 20-nt miRNA precursors suggests that these determinants might alter precursor structure or trigger 3′-end decay of mature miRNA. The results herein advance our understanding of miRNA biogenesis and demonstrate that the effect of asymmetric bulges on miRNA length could be position-dependent.
Keywords: asymmetric bulge, DICER-LIKE 1, microRNA duplex, microRNA length, mismatch
Abbreviations
- AGO
ARGONAUTE protein
- DCL1
DICER-LIKE 1
- DCR
DICER
- fry1
fiery1 mutant
- HEN1
HUA ENHANCER 1
- miRNA*
miRNA star strand
- miRNAs
microRNAs
- PARE
parallel analysis of RNA ends
- SDNs
SMALL RNA DEGRADING NUCLEASES
- siRNA
small interfering RNAs
- SPARE
specific PARE
Introduction
Primary microRNA (miRNA) precursors are transcribed by RNA polymerase II and contain fold-back structures.1,2 The precise production of a mature miRNA from a primary precursor requires 2 types of cleavage. The primary cut, which removes the unpaired ends, occurs at a distance about 11 bp in animals and 15 bp in plants from the junction of single-stranded RNA and the hairpin stem.3-6 In animals, this step happens in the nucleus and is conducted by a complex composed of Drosha and DGCR8 (also known as Pasha).6,7 Then, the resulting hairpin structure (pre-miRNA) is exported to the cytoplasm where it is further excised by other endoribonucleases called DICER (DCR) proteins.8-10 In plants, pre-miRNA is processed by DICER-LIKE proteins (DCLs) to release mature miRNA duplexes in the nucleus.11,12 Plants lack Drosha homologs and the primary cut is also performed by DCL proteins.11 Plant miRNA precursors have more variable loop sizes and stem lengths than animal miRNA precursors.5 The sequential processing direction of plant miRNA precursors by DCL1 can be loop-to-base, base-to-loop, or bidirectional, dependent on the precursor structure.13,14
A canonical plant miRNA duplex is composed of a 19-bp duplex region and 2-nt 3’ overhangs at both termini. Both 3′ ends of a plant miRNA duplex are methylated by a conserved S-adenosyl-l-methionine-dependent RNA methyltransferase, HUA ENHANCER 1 (HEN1), which protects miRNAs from degradation and uridylation.15-17 In most cases, one strand of a miRNA duplex is selectively recruited to silencing slicers, ARGONAUTE (AGO) proteins to perform its function while the other strand, the miRNA star strand (miRNA*) or passenger strand, is degraded.18,19 The AGO-retained strand, also known as the guide strand, usually has higher abundance than the miRNA* and forms base pairing with target genes. However, growing evidence has suggested that miRNA* also can be loaded into AGO proteins and inhibit gene expression.20,21 A new nomenclature system which refers to miRNAs according to their location on pre-miRNA (miR-5p for the 5′ arm and miR-3p for the 3′ arm) has been adopted to replace the old nomenclature of miRNA and miRNA* in miRBase.22
The length of small RNAs is largely determined by DCR or DCL proteins which function like “molecular rulers.” The crystal structure of Giardia intestinalis DCR protein shows that the distance from the PAZ domain which binds the 3′ overhang of pre-miRNA to the catalytic ribonuclease III domain is equivalent to the length of small RNAs produced by this DCR protein.23 In contrast to the 3′ counting rule derived from the study of G. intestinalis DCR, human DCR possesses a motif recognizing the 5′-phosphorylated end of pre-miRNA and determines small RNA length from the 5′ end (the 5′ counting rule).24 Although the crystal structure of plant DCL proteins is not available, the conserved pattern of DCR/DCL protein domains across kingdoms suggests that they also act as molecular rulers.25,26 Arabidopsis thaliana has 4 DCL proteins which have some unique and redundant functions in small RNA pathways.27 The majority of plant miRNAs are 21-nt long and the biogenesis of plant miRNAs depends on DCL1,28-31 implying that plant DCL1 functions as a 21-nt molecular ruler. In addition to the properties of DCR or DCL proteins, miRNA length is also affected by DCR-binding proteins and pre-miRNA structure. The binding of different spliced forms of DCR partner protein Loquacious to DCR1 causes a shift in cleavage sites on pre-miRNA, resulting in a change in miRNA length from 21 to 23 nt accompanied by an alternation in target specificity.32 One miRNA precursor can produce isoforms of various lengths yet the dominant length varies even for miRNA within the same family.30,33,34 For instance, A. thaliana MIR168a predominantly produces mature miRNA of 21 nt while MIR168b produces 21-nt and 22-nt miRNA species at comparable levels.35 Asymmetric bulges have been shown to be responsible for the production of longer miRNAs in plants and animals as the unpaired bases within bulges are not measured by DCR or DCL proteins.29,30,34,36 In A. thaliana, both 21-nt and 22-nt miRNAs associated with AGO1 are able to guide cleavage on their targets while 22-nt miRNAs are highly enriched among triggers that initiate the biogenesis of secondary small interfering RNAs (siRNAs) from their targets.29,30
Besides 22 nt, 20 nt is a non-canonical miRNA length commonly observed in many plant species.28 However, how plants produce miRNAs shorter than 21 nt is still a mystery since the biogenesis of 20-nt miRNAs is also dependent on DCL1.31,37 We hypothesized that miRNA precursors contain sequence or structural determinants for the formation of 20-nt miRNAs. In this study, we identified 20-nt miRNA families from 4 plant species through bioinformatics analysis of small RNA data and classified them based on the length of duplex regions and 3′ overhangs. Structural analysis of these 20-nt miRNA duplexes provided clues for the determinants of 20-nt miRNA biogenesis which were further validated experimentally. Based on the results of bioinformatics analysis and experimental assays, herein, we propose 3 models for the production of 20-nt miRNAs.
Results
20-nt miRNA duplexes contain an atypical duplex region or overhang
To study the molecular basis of 20-nt miRNA formation in plants, we started with the identification of 20-nt miRNAs from 4 species: one bryophyte, Physcomitrella patens (ppt); 2 eudicots, Arabidopsis thaliana (ath) and Arabidopsis lyrata (aly); and one monocot, Oryza sativa (rice; osa). As the length of miRNA annotated in miRBase might be incorrect, we re-annotated the miRNA length with small RNA sequencing data (Fig. 1A). In total, we identified 68 20-nt miRNAs (including miRNA*) belonging to 18 families from the 4 plant species (Table S1). Because miRNA is produced as a duplex from a hairpin precursor, we then identified the complementary strand of 20-nt miRNA to reconstruct the duplex which could provide clues about how 20-nt miRNA is generated (Fig. 1A). A typical DCL1-produced miRNA duplex consists of a 19-bp duplex region and 2-nt overhangs at both 3′ ends (Fig. 1B). Almost all the 20-nt miRNAs paired with a 21 or 22-nt strand and thus form duplexes that deviated from the canonical 21-nt miRNA duplex in terms of the length of duplex regions or overhangs. Based on the length of duplex regions and overhangs, the 20-nt miRNAs could be classified into 3 major groups (see below).
Figure 1.
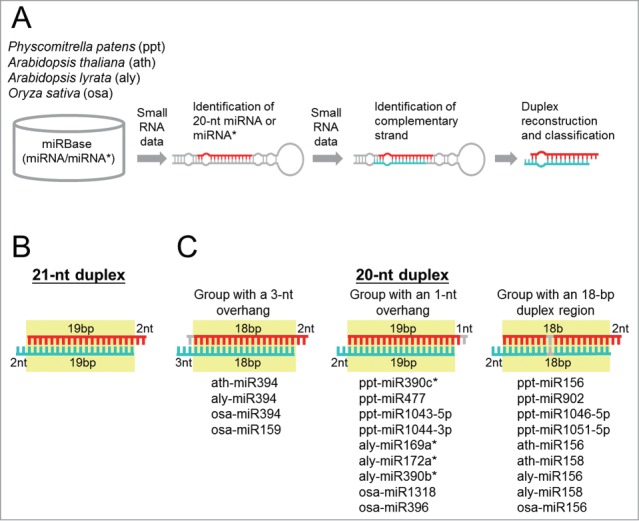
An atypical duplex region or overhang characterizes 20-nt miRNA duplexes. (A) Flowchart for the identification of 20-nt miRNA duplexes. The detailed procedures are described in the Materials and Methods. (B) Structure of a canonical 21-nt duplex. The length of the duplex region and the overhang is indicated in base pairs (bp) and nucleotides (nt), respectively in (B) and (C). The duplex region is highlighted in yellow and the nucleotides in asymmetric bulges are not plotted and contribute to the length of the duplex region in (B) and (C). (C) Three groups of 20-nt miRNA duplexes. The 20-nt miRNA strand and the complementary strand are highlighted in red and turquoise, respectively. The nucleotides presumed missing are indicated in gray. miRNA families assigned to each group are listed.
Duplexes in the first group possess an 18-bp duplex region and an unusual 3-nt overhang in the 21-nt strand, implying a shortening at the 5′ end of the 20-nt strand (Fig. 1C, left panel). Only miR394 from the 2 Arabidopsis species and rice and several loci of rice miR159 belong to this group. miR394 is 20-nt long in most vascular plants although a 21-nt variant could be found in few species.28 In contrast to miR394, miR159 is 21-nt long in most plant species suggesting that 20-nt miR159 in rice is an exception.
The second group is composed of miRNA with a typical 19-bp duplex region but an atypical 1-nt overhang in the 20-nt strand which suggests a shortening at the 3′ end of the 20-nt strand (Fig. 1C, middle panel). Eight miRNA families including 3 miRNA* were found that belong to this group. Among them, ppt-miR477 represents the largest family with abundant expression (Table S1). Unlike the predominant expression of 20-nt species across small RNA libraries in the other 2 groups, the ratio between 20- and 21-nt forms in this group varied among libraries derived from different tissues or developmental stages (Table S1). For instance, the abundance of 20-nt ppt-miR477 was either comparable to or higher than that of the 21-nt form in libraries derived from tissues containing gametophores or sporophytes of P. patens. However, the 21-nt species was the dominant form in libraries generated mainly from protonemata. miR477 is absent in Arabidopsis and rice but has been identified in poplar, orange, and tobacco as 21-nt long. The only 20-nt miRNA in this group which could be identified in more than one species was the star strand of ppt-miR390c and aly-miR390b. ath-miR390b*, which was not annotated in miRBase release 18, is also 20-nt long.
The third group includes miR156 from all 4 species, miR158 from the 2 Arabidopsis species and miR902, miR1046-5p, and miR1051-5p specifically from P. patens (Fig. 1C, right panel). miRNA duplexes within this group are characterized by an atypical 18-bp duplex region whereas the length of 3′ overhangs is normal (Fig. 1C, right panel). One-base-pair shortening in the duplex region may imply that both strands of a 20-nt miRNA duplex become shorter simultaneously. However, none of the 20-nt miRNA complementary strands in this group is 20 nt in length. This could be explained by the presence of asymmetric bulges in the complementary strand (see below). Besides the 4 plant species analyzed in this study, the unique length of 20 nt is preserved in miR156 in many other vascular species including ferns.28
20-nt miRNA duplexes with a 3-nt overhang: an asymmetric bulge in the overhang is the key determinant
The expression of ath-miR394 with a predominant length of 20 nt was shown to depend on DCL1 which mainly produces 21-nt plant miRNAs.37 We assumed that the 20-nt miRNAs or their precursors possess an altered RNA structure. The altered structure may interfere with DCL1 processing or promote RNA decay, leading to the production of miRNAs of abnormal length. The following experiments were designed to evaluate how sequence features contribute to the production of 20-nt miRNAs.
An asymmetric bulge previously revealed to be responsible for longer miRNA produced is conserved in the miR394 family in many plant species (Fig. 2A; Fig. S1).29,30,34 The bulge was found specifically in the 3′ overhang of miR394*. However, miR394* is 21-nt long rather than the 22-nt reported previously. Besides miR394 precursors, osa-miR159cdef precursors also have a bulge in the overhang of miR159* (Fig. 2A; Fig. S2). Therefore, we hypothesized that a bulge in the overhang is the key determinant for this group. Small RNA northern blot analysis showed that the length of miRNA produced from the bulge deleted construct (ath-MIR394aΔB) increased presumably to 21 nt compared to the endogenous 20-nt miR394 (Fig. 2B). A previous study showed that eliminating a bulged base reduced the length of the miRNA strand containing the bulge but did not alter the length of the complementary strand.36 In contrast to the previous report, deleting the bulge from miR394a* only increased the length of miR394a but did not alter the length of miR394a* itself (Fig. 2B). Besides 20-22 miRNAs, small RNAs of 24 nt were detected in this and some other blots in this study. Small RNAs of 24 nt can be produced from miRNA precursors through the DCL3/RDR2/Pol IV pathway but not depend on DCL1 processing.38 The excess production of miRNA precursors in transient expression assays might enhance the production of 24-nt small RNAs.
Figure 2.
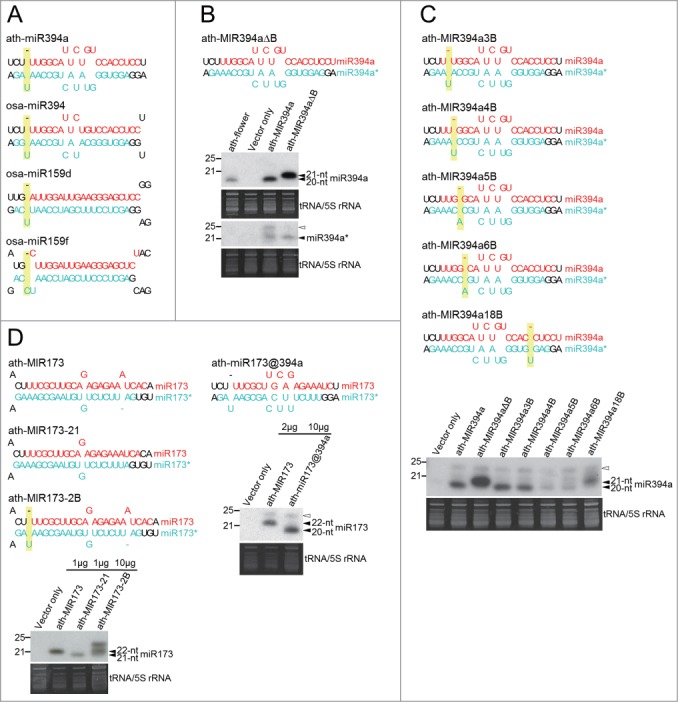
An asymmetric bulge in the overhang is the determinant of a 20-nt miRNA duplex with a 3-nt overhang. (A) 20-nt miRNA duplexes with a 3-nt overhang all contain an asymmetric bulge in the overhang of the complementary strand. The 20-nt miRNA strand and the complementary strand are highlighted in red and turquoise, respectively. If a 20-nt miRNA is in the 3′ arm of the pre-miRNA, the duplex is rotated 180 degrees. Asymmetric bulges in the overhang of mature miRNAs are highlighted in yellow. (B) An asymmetric bulge in the overhang is required for the production of 20-nt ath-miR394a. The length of miR394a and miR394a* derived from the original and modified precursors transiently overexpressed in N. benthamiana was analyzed with small RNA northern blot. Similar experimental procedures were used to determine miRNA length in (C) and (D). The migration of endogenous miR394a was determined by RNA extracted from A. thaliana flowers (ath-flower). (C) The effect of an asymmetric bulge on ath-miR394a length is position-dependent. Asymmetric bulges which were introduced to different positions of ath-MIR394a are highlighted in yellow. (D) The precursor of ath-miR394a is sufficient for the production of artificial 20-nt miRNA. An asymmetric bulge was introduced to the overhang of miR173*. Artificial 20-nt miR173 was expressed using the backbone of ath-MIR394a. Open arrowheads indicate the 24-nt siRNA resulted from transient overexpression of miRNA in N. benthamiana. Equal amounts of total RNA were loaded into each lane unless indicated otherwise. One representative small RNA RNA gel blot derived from 3 independent transient expression assays is shown.
To examine whether the position of an asymmetric bulge in the overhang is critical for the formation of 20-nt miR394a, we moved the bulge to the duplex region. In constructs containing a bulge located 3 or 4-nt away from the 3′ end of miR394a* (ath-MIR394a3B and ath-MIR394a4B), miR394a still remained 20-nt long (Fig. 2C). When a bulge was moved to the positions 5 or 6 nt away from the 3′ end of miR394a* (ath-MIR394a5B and ath-MIR394a6B), the accumulation of miR394a was strongly abolished in these 2 constructs (Fig. 2C). We suspect that a bulge at positions close to the less complementary region of the miR394a duplex might distort the hairpin structure of miRNA and interfere with DCL1 recognition. In the construct containing a bulge at the 5′ end of miR394* (ath-MIR394a18B), miR394a formed a 21-nt miRNA the same as the construct without a bulge (ath-MIR394aΔB) (Fig. 2C). The analysis of predicted pre-miRNA structure from original and modified ath-MIR394a constructs indicated an association between the production of 20-nt miRNA and the formation of a 3-nt overhang in ath-MIR394a, ath-MIR394a3B, and ath-MIR394a4B (Fig. S3). A 3-nt overhang remains to form in the pre-miRNA of ath-MIR394a3B and ath-MIR394a4B because one or 2 base pairs are not sufficient to hold an asymmetric bulge. The result demonstrates that the effect of asymmetric bulges on miRNA length is not equivalent and could be dependent on their relative position in the miRNA duplex.
To test whether a bulge in the overhang is sufficient to reduce miRNA length, we used a 22-nt miRNA, ath-miR173, and introduced a bulge at the equivalent position of ath-MIR173 (ath-MIR173-2B). However, this change reduced miRNA expression and resulted in multiple lengths without a dominant 21-nt miR173 which was demonstrated previously with the use of the 21-nt construct (ath-MIR173-21) (Fig. 2D, left panel).30 Next, we tested if ath-MIR394a could serve as a backbone for producing artificial 20-nt miRNA. A 22-nt miRNA, miR173, could be expressed in 20-nt form with the use of ath-MIR394a (ath-miR173@394a) although at a lower expression level (Fig. 2D, right panel). This result suggests that the precursor of ath-miR394a is required and sufficient for the production of 20-nt miRNA while the accumulation of 20-nt miRNA might be influenced by sequences of mature miRNA. In A. thaliana, AGO proteins preferentially bind to small RNAs with specific 5′ terminal nucleotides and of certain lengths.39 Loading to AGO proteins is crucial for stabilizing miRNAs.40,41 It is possible that the change of miRNA length revealed in this study might alter AGO loading and affect miRNA accumulation. Interestingly, although miR394 and miR159 target different genes, their 5′ and 3′ terminal sequences are highly similar (Fig. 2A).
A shorter miRNA could be attributed to abnormal processing of pre-miRNA by DCL1 or RNA decay of mature miRNA. To differentiate between these 2 possibilities, we compared miRNA termini to DCL1 cleavage sites on miRNA precursors with the use of publicly available high-throughput sequencing data. Cleavage sites of DCL1, processing direction, and the number of cuts to release mature miRNA could be inferred based on sequencing data of miRNA processing intermediates generated by parallel analysis of RNA ends (PARE) or degradome sequencing.13,42 Previous studies have shown that DCL1 processes ath-MIR394 from base to loop while a loop-to-base processing is conserved for miR159/miR319 families.13,42,43 Notably, more than 2 DCL1 cuts are required to release miR394 and miR159. According to rice PARE data,44 2 major DCL1 cuts indeed frame the 5′ and 3′ termini of 20-nt osa-miR159f, indicating that miR159f is produced as 20-nt long, not degraded to a length of 20 nt (Fig. 3A). PARE data could not reveal DCL1 cleavage sites flanking ath-miR394 because it is generated through a sequential base-to-loop mechanism. We thus performed cycle RT-PCR to determine the cleavage sites of DCL1 on ath-MIR394a with the RNA extracted from fiery1 (fry1) mutant in which exoribonuclease activity is reduced and the accumulation of miRNA processing intermediates is enhanced.45 We were able to detect the 5′ terminus of 20-nt ath-miR394a and the 3′ terminus of 21-nt ath-miR394a*, confirming that DCL1 cleavage results in a 3-nt overhang (Fig. 3B). Taken together, these results demonstrate the distinct effect of an asymmetric bulge in the overhang on miRNA length.
Figure 3.
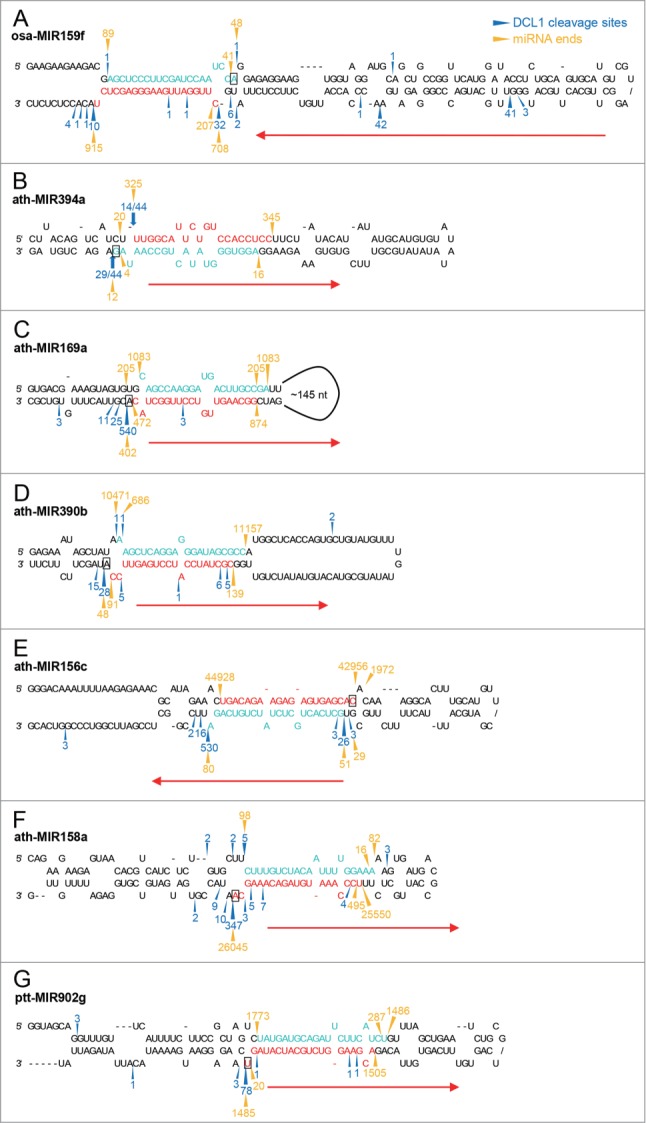
Comparison of miRNA termini with DCL1 cleavage sites. (A) Two major DCL1 cleavage sites revealed by rice PARE data define 20-nt osa-miR159f. The positions and counts of DCL1 cleavage on osa-MIR159f were based on rice PARE data published by Li et al.44 (B) Cycle RT-PCR reveals DCL1 cleavage sites consistent to the 5′ end of 20-nt miR394 and the 3′ end of 21-nt miR394* which form a 3-nt overhang. (C and D) DCL1 cleavage sites are mapped to the position 1 nt downstream of 20-nt ath-miR169a* (C) and ath-miR390b* (D) 3′ ends. The positions and counts of DCL1 cleavage on ath-MIR169a and ath-MIR390b were based on A. thaliana SPARE data published by Bologna et al.13 (E–G) Major DCL1 cleavage sites are consistent to the 5′ and 3′ termini of 20-nt miRNA duplexes with an 18-bp duplex region. The positions and counts of DCL1 cleavage on ath-MIR156c (E), ath-MIR158a (F), and ppt-MIR902g (G) were based on A. thaliana SPARE data and P. patens degradome data published by Bologna et al. and Addo-Quaye et al., respectively.13,42 Sequences of 20-nt miRNAs and their complementary strands identified with small RNA data are highlighted in red and turquoise, respectively. Blue arrowheads and numbers indicate DCL1 cleavage sites and counts based on degradome or SPARE data. Blue arrows and numbers indicate the positions and frequency of DCL1 processing sites detected by cycle RT-PCR. Gold arrowheads and numbers indicate the termini and counts of the 2 most abundant small RNA sequences overlapping with the annotated miRNAs for each strand. Red arrows indicate the directions of DCL1 processing and the nucleotides presumably associated with the putative 3′ binding pocket of DCL1 are indicated with rectangles.
20-nt miRNA duplexes with a 1-nt overhang: a mismatch at the duplex 3′ end results in shortening of the miRNA 3′ end
None of the miRNA duplexes in the group with a 1-nt overhang have asymmetric bulges but all of them contain at least one mismatch (Fig. 4A). Interestingly, in this group, the majority of the duplexes have a mismatch at position 18 or 19 of the 20-nt miRNAs. Therefore, we assumed that a mismatch close to the 3′ terminus of a miRNA duplex might be the determinant for a 20-nt miRNA duplex with a 1-nt overhang. We selected ppt-miR477a to test this hypothesis since the ppt-miR477 family represents the most abundant species in this group. The duplex of ppt-miR477a has 2 mismatches at positions 8 and 19. We modified the nucleotides in the miR477a* to make mismatching base pairs into matching base pairs at position 19 (ppt-MIR477a19p) and position 8 (ppt-MIR477a8p) as a control (Fig. 4B). Unlike miR156 or miR394 which is predominantly found in a 20-nt form, endogenous ppt-miR477 is heterogeneous, found in 20-nt and 21-nt forms (Table S1). When transiently expressing ppt-MIR477a in N. benthamiana, both 20-nt and 21-nt forms could be detected while the ratio between the 2 forms was variable depending on the age of infiltrated N. benthamiana (Fig. S4). Since the predominant length of endogenous ppt-miR477 also varied among tissues (Table S1), the length variation we observed in transient expression assays might reflect the nature of ppt-miR477 in different tissues or at different developmental stages. As shown in Figure 4B, matching the base pair at position 19 in ppt-MIR477a19p successfully transformed miR477a into predominantly 21 nt in length. On the other hand, matching the base pair at position 8 in ppt-MIR477a8p did not change miR477a length. Although the ratio between 21- and 20-nt ppt-miR477a varied in leaves of different ages, the impact of structural changes on miR477a length was consistent (Fig. S4). The length of miR477a* was not altered along with the increase of miR477a length in ppt-MIR477a19p, indicating that the 3′ terminal mismatch has an asymmetric effect on miRNA/miRNA* length (Fig. 4B).
Figure 4.
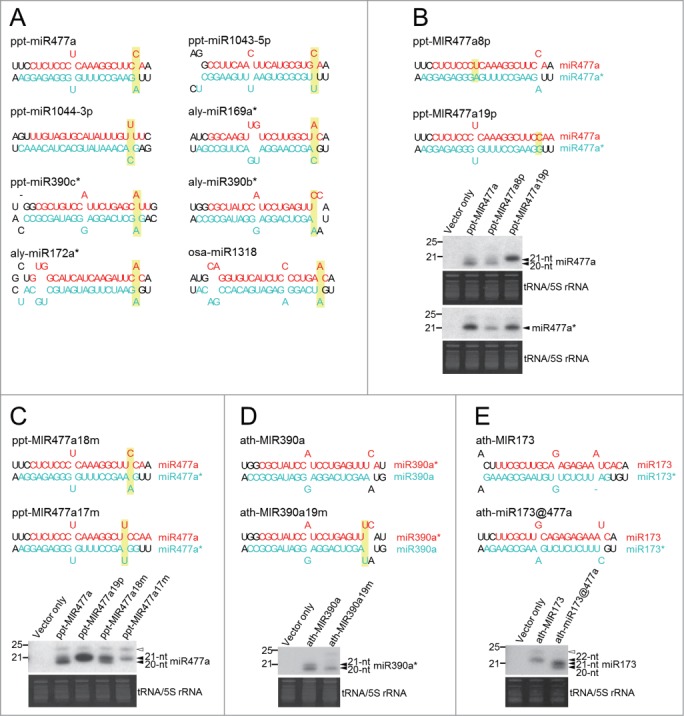
A mismatch at the duplex 3′ end is the determinant of a 20-nt miRNA duplex with a 1-nt overhang. (A) 20-nt miRNA duplexes with a 1-nt overhang contain a mismatch at the duplex 3′ end. The 20-nt miRNA strand and the complementary strand are highlighted in red and turquoise, respectively. If a 20-nt miRNA is in the 3′ arm of a pre-miRNA, the duplex is rotated 180 degrees. Mismatches at the duplex 3′ end are highlighted in yellow. (B) A mismatch at the duplex 3′ end is required for the production of 20-nt ppt-miR477a. The length of miR477a and miR477a* derived from the original and modified precursors transiently overexpressed in N. benthamiana was analyzed with small RNA northern blot. Mismatches which were eliminated from the original precursor or introduced to different positions in (B), (C) and (D) are highlighted in yellow. Similar experimental procedures were performed to determine miRNA length in (C) and (D). (C) The effect of a mismatch on ppt-miR477a length is position-dependent. (D) The 20-nt form becomes dominant when introducing a mismatch to position 19 of ath-miR390a*. (E) Heterogeneous expression of 20-nt and 21-nt artificial ath-miR173 is generated from the backbone of ptt-MIR477a. Open arrowheads indicate the 24-nt siRNA resulting from transient overexpression of miRNA in N. benthamiana. A representative small RNA RNA gel blot derived from 3 independent transient expression assays is shown.
The constraint on mismatch position within this group raised the question of whether only mismatches at positions 18 and 19 could result in the production of 20-nt miRNA. Moving the terminal mismatch to position 18 (ppt-MIR477a18m) led to an increase in the 21-nt form but a notable amount of the 20-nt form was still detected (Fig. 4C). On the other hand, when a mismatch was moved to position 17 (ppt-MIR477a17m), the 20-nt form was barely detectable while the 21-nt form became the predominant species.
Based on the sequencing data we analyzed, endogenous ath-miR390a* is predominantly 21-nt while ath-miR390b* is 20-nt. Mismatches are located at positions 20 and 19 of the star strand of ath-miR390a and ath-miR390b respectively. Although both 21-nt and 20-nt miR390a* were observed when we transiently expressed ath-MIR390a in N. benthamiana, introducing a mismatch at position 19 (ath-MIR390a19m) considerably decreased the level of the 21-nt form, leading to predominant expression of the 20-nt form (Fig. 4D). We also tested the production of artificial 20-nt miRNA with the backbone of ppt-MIR477a. Although the 20-nt species could be detected, the predominant form is 21-nt long when expressing ath-miR173 from ppt-MIR477a (ath-miR173@477a) (Fig. 4E). In summary, a mismatch at the duplex 3′ end is the key determinant of the biogenesis of 20-nt miRNA duplexes with a 1-nt overhang but the level of 20-nt species may be affected by developmental stages or miRNA sequences.
To understand how a non-canonical 1-nt overhang is generated, we then used publicly available high-throughput sequencing data of processing intermediates of miRNA precursors to map DCL1 cleavage sites. As ath-miRNA390b* and ath-miRNA169a* are also 20-nt long, we thus examined specific PARE (SPARE) data for A. thaliana miRNA.13 SPARE data of ath-MIR169a and ath-MIR390b indicate that DCL1 predominantly makes a cut 1-nt downstream of the 20-nt miRNA 3′ end and theoretically results in the production of a typical 2-nt overhang (Fig. 3C, D). Therefore, a non-canonical 1-nt overhang in this group is likely to be the consequence of a 3′-to-5′ RNA decay after the miRNA is released from its precursor. Plant small RNAs are methylated at the 2′ hydroxyl group of the 3′ terminal nucleotide by HEN1.16 In hen1 mutant, plant miRNAs are truncated and tailed with poly U.15,17 We thus suspected that 20-nt miRNAs might be the consequence of 3′ truncation due to the lack of methylation on the 3′ end. To test this possibility, we carried out β-elimination treatment after which unmethylated miRNAs run faster than methylated miRNAs due to the loss of the terminal nucleotide.46 As expected, ath-miR156c migrated faster in the hen1-1 mutant (Fig. S5). However, the migration of endogenous ppt-miR477 and ppt-miR390c* was not changed after the β-elimination treatment, indicating that these 20-nt miRNAs are methylated in planta (Fig. S5). Therefore, the mechanism responsible for trimming miRNA to 20-nt long in this group remains to be determined.
20-nt miRNA duplexes with an 18-bp duplex region: a position-specific asymmetric bulge measured by DCL1 leads to 20-nt miRNA
All the 20-nt miRNAs in the group with an 18-bp duplex region contained at least one asymmetric bulge in the complementary strand (Fig. 5A). However, the length of these complementary strands with a single 1-nt bulge is not 22-nt but 21-nt. There are 2 1-nt bulges in ath-miR156c* while other miR156* and other 20-nt miRNA complementary strands mostly have a single bulge in the 3′ half. Therefore, we removed the bulge in the 3′ half of ath-miR156c* to test its effect on miR156c length (Fig. 5B). Small RNA RNA gel blot showed that the dominant length of miR156c shifted to 21 nt while the length of miR156c* was not changed although one base was deleted from miR156c* in ath-MIR156cΔB (Fig. 5B). To further confirm that a bulge is the determinant for this group, we removed the only bulge from ath-miR158a* and observed the same size increase of miR158a to 21 nt from the construct ath-MIR158aΔB (Fig. 5C).
Figure 5.
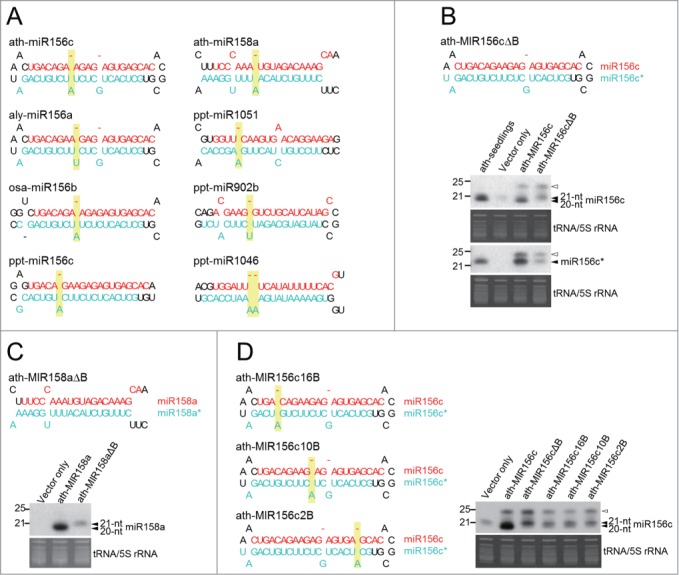
A position-specific asymmetric bulge is the determinant of a 20-nt miRNA with an 18-bp duplex region. (A) miRNAs of 20-nt with an 18-bp duplex region all possess an asymmetric bulge at neighboring positions. The 20-nt miRNA strand and the complementary strand are highlighted in red and turquoise, respectively. If a 20-nt miRNA is in the 3′ arm of a pre-miRNA, the duplex is rotated 180 degrees. Asymmetric bulges located in the 3′ half of the 20-nt miRNA complementary strand are highlighted in yellow. (B) An asymmetric bulge in the 3′ half of miR156c* is required for the formation of 20-nt ath-miR156c. The length of miR156c and miR156c* derived from the original and modified precursors transiently overexpressed in N. benthamiana was analyzed with small RNA northern blot. The migration of endogenous miR156c was determined by RNA extracted from A. thaliana seedlings (ath-seedlings). Similar experimental procedures were performed to determine miRNA length in (C) and (D). (C) An asymmetric bulge in the 3′ half of miR158a* is required for the formation of 20-nt ath-miR158a. (D) The effect of an asymmetric bulge on ath-miR156c length is position-dependent. Asymmetric bulges introduced in modified constructs are highlighted in yellow. Open arrowheads indicate the 24-nt siRNA resulted from transient overexpression of miRNA in N. benthamiana. A representative small RNA RNA gel blot derived from 3 independent transient expression assays is shown.
Asymmetric bulges in the 3′ half of 20-nt miRNA complementary strands are mainly located between positions 11 and 14 (Fig. 5A). We then examined the effect of bulge position on miRNA length by moving the bulge to the 5′ half, the middle and after position 14. Although none of the 3 positions resulted in the predominant and strong expression of the 20 nt similar to the native precursor, the proportion of 20-nt miRNA to 21-nt miRNA was relatively higher in the constructs with a bulge in the middle and after position 14 (ath-MIR156c10B and ath-MIR156c16B) than in the construct with a bulge close to the 5′ end of miR156c* (ath-MIR156c2B) (Fig. 5D). This result suggests that different positions of asymmetric bulges may exert distinct effects on miRNA length.
The analysis of SPARE and degradome data provided further insights into how DCL1 processes miRNA precursors within this group. DCL1 cleavage sites on ath-MIR156c suggested by SPARE data are consistent with the both termini of 22-nt ath-miR156c* (Fig. 3E). This implies that one of 2 1-nt bulges in ath-miR156c* is measured by DCL1 otherwise the length of ath-miR156c* would become 23-nt. The removal of the adenine bulge from ath-miR156c* did not alter its length as previously reported for 22-nt miRNA (Fig. 5B),36 suggesting that this bulge is measured by DCL1. ath-miR158a and ppt-miR902g are in the 3′ arm of the precursors which DCL1 processes in the base-to-loop direction (Fig. 3F, G). The primary cuts of DCL1 to release ath-miR158a and ppt-miR902g indeed match the 3′ termini of these 2 20-nt miRNAs and thus exclude the possibility of 3′ RNA decay resulting in shorter miRNA (Fig. 3F, G).
Although 21-nt ath-miR390a could be expressed in 20-nt form with the backbone of ath-MIR156c (ath-miR390a@156c) (Fig. 6A, B), introducing an asymmetric bulge to the duplex region of ath-MIR390a (ath-MIR390a-12B) to mimic the duplex structure in this group failed to produce 20-nt ath-miR390a (Fig. 6A, C). We thus suspected that the position of an asymmetric bulge relative to the putative 3′ binding pocket of DCL1 might be crucial for the effect on miRNA length. Although both loop-to-base and base-to-loop DCL1 processing was found in this group, 20-nt miRNAs are excised from the arm presumably associated with the putative 3′ binding pocket of DCL1 while asymmetric bulges are present in the opposite arm (Figs. 3E–G). However, ath-MIR390a generated mature miR390a from the arm away from the putative 3′ binding pocket of DCL1 through short base-to-loop processing.13 We thus inverted the miRNA duplex of ath-MIR390a (ath-MIR390a-Inv) to relocate miR390a to the arm associated with the putative 3′ binding pocket of DCL1 (Fig. 6A). The introduction of an asymmetric bulge to the inverted ath-MIR390a (ath-MIR390a-Inv-12B) did indeed convert 21-nt ath-miR390a to 20-nt long (Fig. 6A, D). This result implies that specific interaction between an asymmetric bulge and DCL1 protein might be required for the formation of 20-nt miRNA in this group.
Figure 6.
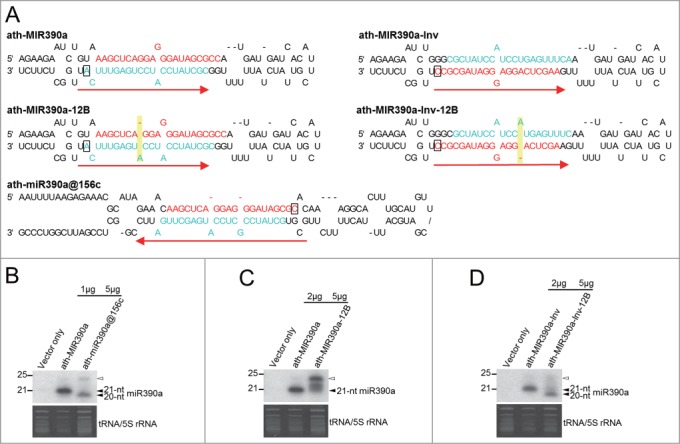
The position of an asymmetric bulge relative to the putative 3′ binding pocket of DCL1 is crucial for the biogenesis of 20-nt miRNA. (A) Foldbacks of ath-MIR390a, ath-miR390a@156c, ath-MIR390a-12B, ath-MIR390a-Inv and ath-MIR390a-Inv-12B. The sequences of miR390a and miR390a* in each foldback are highlighted in red and turquoise, respectively. Red arrows indicate the directions of DCL1 processing and the nucleotides presumably associated with the putative 3′ binding pocket of DCL1 are indicated with rectangles. Asymmetric bulges which were introduced to ath-MIR390a are highlighted in yellow. (B) The precursor of ath-miR156c is sufficient for the production of artificial 20-nt ath-miR390a. (C) The introduction of an asymmetric bulge to the duplex region of ath-MIR390a failed to result in predominant expression of 20-nt miRNA. (D) An asymmetric bulge needs to be in the arm away from the putative 3′ binding pocket of DCL1 to convert ath-MIR390a into a 20-nt miRNA precursor. Amounts of total RNA loaded into each lane are indicated. Open arrowheads indicate the 24-nt siRNA resulted from transient overexpression of miRNA in N. benthamiana. A representative small RNA northern blot derived from 3 independent transient expression assays is shown.
Discussion
In this study, 3 models for the biogenesis of 20-nt miRNAs in plants were established (Fig. 7). Our data suggested that position-specific asymmetric bulges and mismatches are the key determinants for shorter miRNA formation. Asymmetric bulges may alter the structure of pre-miRNAs and result in a 20-nt fragment between 2 DCL1 cuts (Figs. 7A, B). Mismatches at the duplex 3′ end, on the other hand, may cause RNA decay after mature miRNA is released from pre-miRNA (Fig. 7C). These RNA structures could be found in 20-nt miRNAs across species, suggesting that the mechanisms are evolutionarily conserved.
Figure 7.
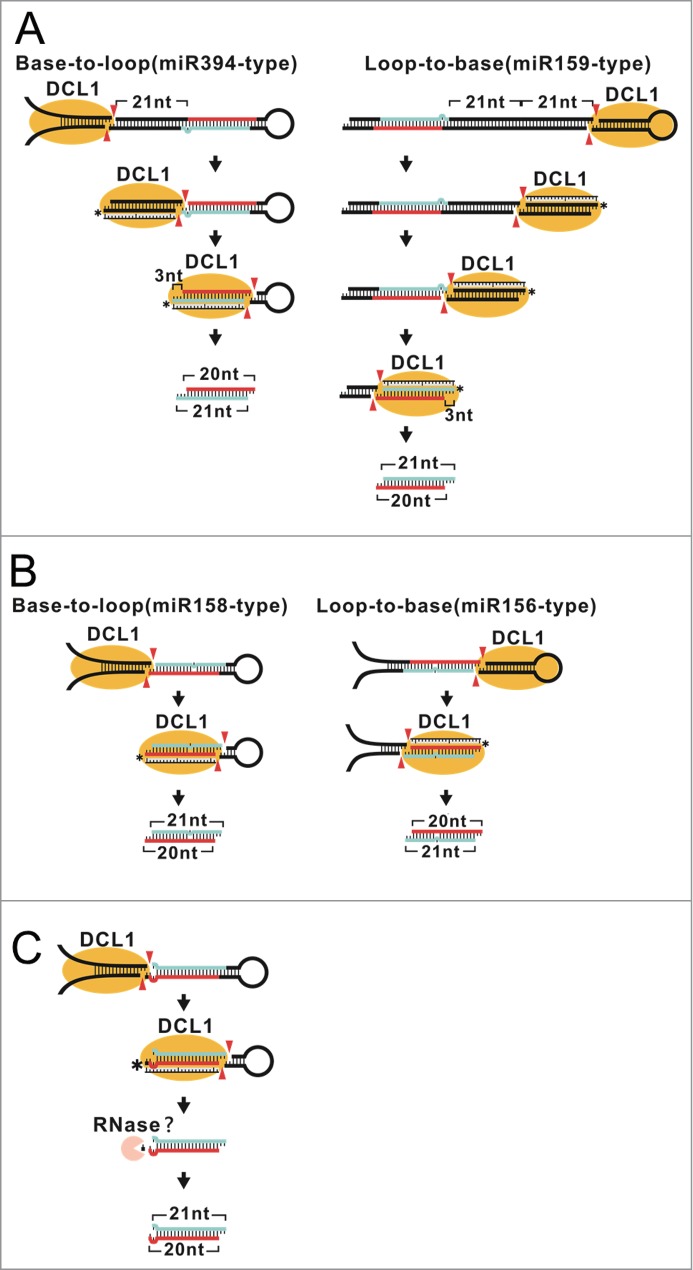
Three models for 20-nt miRNA biogenesis in plants. (A) A model for the formation of a 20-nt miRNA duplex with a 3-nt overhang. DCL1 produces 20-nt miRNA in the 5′ arm in a base-to-loop direction while 20-nt miRNA in the 3′ arm is in a loop-to-base direction. The asymmetric bulge in the 3′ overhang of a 20-nt miRNA duplex is released after the primary DCL1 cut to free mature miRNA. The expanded bulge results in the formation of an atypical 3-nt overhang. DCL1 measures a length of 21 bp from the terminus of the 3′ overhang, resulting in a 20 nt/21 nt miRNA duplex. (B) A model for the formation of 20-nt miRNA with an 18-bp duplex region. A DCL1-measured asymmetric bulge results in a 20-nt miRNA on the strand containing the asymmetric bulge and a 21-nt miRNA on the complementary strand. (C) A model for the formation of a 20-nt miRNA duplex with a 1-nt overhang. A 21-nt miRNA is released after DCL1 cleavages. A mismatch at the duplex 3′ end results in an abnormal 3′ overhang that may trigger 3′ terminal trimming and lead to a 20-nt miRNA. The putative 3′ binding pocket of DCL1 is indicated with an asterisk and DCL1 cuts are indicated with red arrowheads. The mature sequences of 20-nt miRNA and its complementary strand are highlighted in red and turquoise, respectively.
The effect of asymmetric bulges on miRNA length revealed in this study contrasts with previous reports. Previously, it was shown that a miRNA strand with a 1-nt asymmetric bulge becomes 22-nt long while the complementary strand still remains 21-nt.29,30 The analysis of human miRNAs also indicated that miRNA precursors with an asymmetric structure generally produce longer miRNAs than those without an asymmetric structure.34 However, this study demonstrated that asymmetric bulges can result in the production of 20-nt miRNAs through 2 different mechanisms. In these cases, an asymmetric bulge does not increase the length of the miRNA strand with a bulge while decreasing the length of the complementary strand. The distinct effect of asymmetric bulges on the length of a miRNA might be explained by their position or geometry. Most asymmetric bulges in 22-nt miRNAs are located in the duplex region and are not opened until mature miRNA duplexes are unwound. In the case of 20-nt miRNA duplexes with a 3-nt overhang, asymmetric bulges are in the overhang and are released after the primary DCL1 cut to release the mature miRNA (Fig. 7A). Instead of the production of 22-nt miRNA from the strand containing asymmetric bulges, asymmetric bulges in the overhang lead to the production of 20-nt miRNA from the complementary strand. Based on this result, plant DCL1 protein is more likely to adopt the 3′ counting rule when processing pre-miRNA with a 3-nt overhang. Notably, this model explains the biogenesis of 20-nt miR159 and miR394 which both require more than 2 DCL1 cuts to release mature miRNAs. Because ath-MIR173 precursor which has a short stem could not be modified to produce shorter miRNAs (Fig. 2D), the sequential DCL1 cleavage on the long stem of miRNA precursors might be also crucial for the formation of 20-nt miRNA with a 3-nt overhang.
The molecular basis behind the formation of 20-nt miRNAs with an 18-bp duplex region but normal 3′ overhangs is more difficult to explain as their asymmetric bulges are also present in the duplex region like in 22-nt miRNAs (Fig. 5A). Although a bulged base looks identical in secondary structure, previous studies have shown that a bulged base can adopt distinct arrangements in tertiary structure.47 A crystal structure of an RNA duplex with a looped-out adenine does not deviate from a regular A-form RNA helix much,48 supporting the previous model of 22-nt miRNA formation that a looped-out bulge does not change the length of a miRNA duplex.29 With this model, a looped-out bulge will lead to longer mature miRNA on the strand containing the bulge. By contrast, a nuclear magnetic resonance study of a 24-nt hairpin showed that an unpaired adenine could be stacked between adjacent bases, leading to conformational distortion around the bulged base.49 It is thus tempting to postulate that the bulged adenine in miR156c and miR158a duplexes might have a configuration different from bulged bases in 22-nt miRNA duplexes. The measurement of asymmetric bulges in the miR156c and miR158a duplexes by DCL1 did not result in longer miRNA on the strand with an asymmetric bulge but led to shorter miRNA on the complementary strand (Fig. 7B). The constrained position of bulges in this group relative to the putative 3′ binding pocket of DCL1 protein implies that specific RNA-protein interaction may influence the geometry of bulges.
The group of 20-nt miRNA with a 1-nt overhang is distinct from the other 2 groups regarding the ratio of 20 to 21-nt abundance, DCL1 processing sites and structural determinants. Overall, this group shows less dominant expression of the 20-nt form compared to the other 2 groups (Table S1). An inconsistency between 20-nt miRNA 3′ termini and DCL1 cleavage sites was found in this group but not the other 2 groups (Fig. 3), suggesting that post-dicing trimming is responsible for the production of 20-nt miRNAs in this group (Fig. 7C). The key determinant for this group is a 1-bp mismatch at position 18 or 19 of the 20-nt miRNA strand (Fig. 4). A mismatch at these 2 positions leads to an atypical 3′ overhang whereas a mismatch at position 17 recovers a canonical 2-nt overhang. Beta-elimination assay indicated that both 20 and 21-nt miRNAs in this group were methylated (Fig. S5), excluding the possibility that the production of 20-nt miRNA is due to the truncation of unmethylated 21-nt miRNA. As HEN1 recognizes miRNA duplexes rather than single-stranded miRNA,50 methylated 20-nt miRNA further implies that RNA trimming might occur before the miRNA duplex is unwound and the responsible exoribonucleases are able to act on double-stranded RNA. Homologs of RNase T are likely the enzymes involved in post-dicing decay of the miRNA duplex as RNase T plays a role in the 3′ terminal trimming of tRNA (transfer RNA) and rRNA (ribosomal RNA) whose 5′ and 3′ termini form a duplex with a 3′ overhang.51-53 Moreover, the last base pair in the duplex region influences the length of a 3′ overhang generated by RNase T digestion.54 An A-T (or A-U) pair results in a 1-nt overhang while a G-C pair leads to a 2-nt overhang. According to the structural analysis of RNase T with a duplex DNA, an A-T base pair at the duplex end was melted which might expose an additional base for RNase T digestion. A mismatch at the miRNA duplex 3′ end may trigger the trimming of overhang through a similar mechanism (Fig. 7C). Although several SMALL RNA DEGRADING NUCLEASES (SDNs) contain a RNase T domain, their unique function in degrading single-stranded miRNA but not miRNA duplex, miRNA precursor and rRNA termini makes them less likely to be the enzymes involved in the model we proposed.55
Although 20-nt miRNA does not represent a major class of plant miRNAs, highly conserved or lineage-specific 20-nt miRNAs appear in most plant species.28 Within the Arabidopsis genus, the fraction of 20-nt miRNA families among conserved miRNAs is about 10% which is comparable to that of 22-nt miRNA families.56 However, 22-nt miRNA families are enriched to around 30% in less conserved miRNAs while 20-nt miRNAs slip to less than 5%. The opposite trends for 22-nt and 20-nt miRNA populations through evolution may imply that factors contributing to the birth and death of these 2 length classes are distinct. Our results may provide a molecular basis to support the notion that 22-nt miRNAs may arise more frequently than 20-nt species. Our present results together with previous studies show that asymmetric bulges are the main determinant for both 22-nt and 20-nt miRNA.29,30 However, bulge position is much more constrained in the 20-nt population than in the 22-nt population. Although mismatches could also lead to the formation of 20-nt miRNAs, the position of mismatches is also critical and only few positions are effective. Moreover, the difficulty involved in highly expressing 21-nt or 22-nt miRNAs in a 20-nt form suggests that there are other unidentified factors involved in 20-nt miRNA biogenesis or accumulation. As determinants for some 20-nt miRNA identified in this study are located in the passenger strand, these structural features are likely lost during evolution if a length of 20 nt has no beneficial consequence. However, miR156, miR158 and miR394 are predominantly 20-nt long in many land plants.28 Taken together, our results lend support to the possibility that a non-canonical 20-nt length might confer miRNA with unique properties and has thus been preserved through evolution.
Materials and Methods
Identification of 20-nt miRNA and the complementary strand
Small RNA sequencing data sets downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) were used to re-annotate P. patens, A. thaliana, A. lyrata and O. sativa miRNA and miRNA* retrieved from miRBase release 18 (http://www.mirbase.org/) (Table S2). Because the abundance of some miRNAs was low, we pooled the reads from multiple small RNA libraries for the following analysis. The reads of miRNA length isoforms which have the 5′ end defined in miRBase but variable 3′ ends allowing the length of 20-24 nt were compared. miRNAs of 20 nt were selected if the most abundant length species was 20-nt long. We further removed miRNA candidates with less than 5 reads, or reads that overlapped with repeats or transposons. Some miRNAs within a family share identical sequences for the first 20 nt which may cause mis-annotation of 20-nt miRNA loci with lower expression. To avoid this problem, we excluded the 20-nt miRNAs sharing identical reads with miRNAs annotated as 21 nt or longer within the same family. The abundance of 20-nt miRNA isoforms in each library is provided in Supplementary Table 1. To reconstruct the duplex of 20-nt miRNAs, we identified the complementary strand of 20-nt miRNAs with the use of small RNA sequencing data. Among the small RNAs which were generated from the opposite arm of 20-nt miRNAs and had at least 15 nt pairing to the sequence of 20-nt miRNAs, the one with the highest abundance was defined as the complementary strand.
Structural analysis of 20-nt miRNA duplexes and their precursors
The structure of 20-nt miRNA duplexes and their pre-miRNA was based on the prediction deposited in miRBase while the structure of modified pre-miRNA was predicted by the RNAfold program in the Vienna RNA Webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
miRNA expressing constructs
All miRNA precursors used in this study were driven by the CaMV 35S promoter and cloned into pCAMBIA1390. Overlapping PCR was used for modifying miRNA precursors. Primers for cloning the original miRNA precursors are listed in Supplemental Table 3.
Nicotiana benthamiana transient expression assay
The procedures for transient expression assay have been described previously.57 Leaves of 3- to 4-week-old N. benthamiana plants grown at 27°C with 14-hour light/10-hour dark period were infiltrated with Agrobacterium tumefaciens strain GV3101 or C58C1 carrying miRNA expressing constructs. Infiltrated tissues were collected after 2–3 days for total RNA extraction. At least 3 independent transient expression assays were performed to determine the length of miRNA produced by each construct.
Small RNA northern blot analysis
Total RNA of infiltrated tissues was extracted with PureLink® Plant RNA reagent (Ambion) following the standard protocol. For small RNA northern blot analysis, unless otherwise specified, 3–5 μg of total RNA was separated on 15% denaturing polyacrylamide TBE-urea gels (Invitrogen) and transferred to Hybond-N+ membranes (GE Healthcare). DNA oligonucleotides complementary to miRNA or miRNA* were used as probes in northern blot to determine miRNA length and expression from original or modified miRNA precursors. The sequences of probes used in this study are listed in Supplementary Table S3. The probes were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs) and hybridized to the membrane with ULTRAhyb-Oligo buffer (Ambion) at 37°C overnight. After washing 2 to 3 times with buffer containing 2× SSC and 0.1% SDS, the membrane was exposed to X-ray film for 1-7 days.
Mapping of DCL1 cleavage sites with PARE and degradome data
PARE and degradome data sets downloaded from GEO were used in the analysis of miRNA processing intermediates for mapping DCL1 cleavage sites (Table S4). PARE and degradome reads were mapped to miRNA precursor sequences and the 5′ end of reads represent DCL1 cleavage sites.
Cycle RT-PCR
Cycle RT-PCR was performed following the method described previously with minor modifications.13 Five micrograms of low molecular weight RNA isolated from A. thaliana fry1-6 mutant (SALK_020882) with the use of mirVana miRNA Isolation Kit (Ambion) was self-ligated with T4 RNA ligase (Ambion) at 4°C overnight. RNA was then precipitated and resuspended in nuclease-free water. Next, SuperScript III Reverse Transcriptase (Invitrogen) was used for first-strand cDNA synthesis with a loop-specific primer, cyc-RT-394-R. A pair of primers, cyc-RT-394-R and cyc-RT-394-F, listed in Supplemental Table 3, were used for PCR reactions. PCR products were then cloned into pJET1.2/blunt vector (Thermos Scientific) and sequenced.
Beta-elimination treatment
The treatment of β-elimination was performed as the protocol described previously with minor modifications.46 Low molecular weight RNA of P. patens grown on solid BCDAT medium at 25°C under continuous white light was isolated using mirVana miRNA Isolation Kit (Ambion). One to 8 micrograms of P. patens small RNA was dissolved in 70 µl borax/boric acid buffer, pH 8.6 (4.375 mM borax, 50 mM boric acid) and 10 µl 0.2 M sodium periodate (Sigma) was added. The reaction was incubated in the dark at room temperature for 10 min. Eight microliters of glycerol was then added to the reaction and incubation was continued in the dark for 10 min at room temperature. RNA was then precipitated and dissolved in 80 µl borax/boric acid buffer, pH 9.5 (33.75 mM borax, 50 mM boric acid, pH adjusted by NaOH), incubated for 90 min at 45°C, and then precipitated again. The pellet was washed with 70% ethanol, dissolved in 10 µl nuclease-free water, and analyzed by small RNA northern blot. Total RNA of 10-day-old A. thaliana hen1-1 seedlings grown at 22°C with 16-hour light/8-hour dark period was extracted using PureLink® Plant RNA reagent (Ambion) and used as a positive control of β-elimination treatment.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Shih-Long Tu and Dr. Chiung-Yun Chang at Academia Sinica for providing P. patens tissues for RNA extraction and Mr. Meng-Chun Lin for providing A. thaliana hen1-1 RNA. We also thank Mr. Cheng-Yu Hou for the help in preparing the figures and Ms. Miranda Loney in Academia Sinica for English editing of this paper.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by grants from the Taiwan Ministry of Science and Technology (NSC 100-2311-B-001-030-MY2) and Academia Sinica to Ho-Ming Chen. Ming-Hsuan Lu was supported by the Biological Programs for Selected Senior High School Students in Academia Sinica.
References
- 1.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol 2005; 138:2145-54; PMID:16040653; http://dx.doi.org/ 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004; 23:4051-60; PMID:15372072; http://dx.doi.org/ 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol 2010; 20:42-8; PMID:20015654; http://dx.doi.org/ 10.1016/j.cub.2009.10.073 [DOI] [PubMed] [Google Scholar]
- 4.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol 2010; 20:37-41; PMID:20015653; http://dx.doi.org/ 10.1016/j.cub.2009.10.076 [DOI] [PubMed] [Google Scholar]
- 5.Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr Biol 2010; 20:49-54; PMID:20005105; http://dx.doi.org/ 10.1016/j.cub.2009.10.072 [DOI] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006; 125:887-901; PMID:16751099; http://dx.doi.org/ 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al.. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003; 425:415-9; PMID:14508493; http://dx.doi.org/ 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- 8.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science 2004; 303:95-8; PMID:14631048; http://dx.doi.org/ 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- 9.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol 2004; 14:156-9; PMID:15134074; http://dx.doi.org/ 10.1016/j.tcb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834-8; PMID:11452083; http://dx.doi.org/ 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- 11.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A 2004; 101:12753-8; PMID:15314213; http://dx.doi.org/ 10.1073/pnas.0403115101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A 2005; 102:3691-6; PMID:15738428; http://dx.doi.org/ 10.1073/pnas.0405570102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bologna NG, Schapire AL, Zhai J, Chorostecki U, Boisbouvier J, Meyers BC, Palatnik JF. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res 2013; 23:1675-89; PMID:23990609; http://dx.doi.org/ 10.1101/gr.153387.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Zhou Y, Castillo-Gonzalez C, Lu A, Ge C, Zhao YT, Duan L, Li Z, Axtell MJ, Wang XJ, et al.. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 2013; 20:1106-15; PMID:23934148; http://dx.doi.org/ 10.1038/nsmb.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 2005; 15:1501-7; PMID:16111943; http://dx.doi.org/ 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 2006; 34:667-75; PMID:16449203; http://dx.doi.org/ 10.1093/nar/gkj474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai J, Zhao Y, Simon SA, Huang S, Petsch K, Arikit S, Pillay M, Ji L, Xie M, Cao X, et al.. Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 2013; 25:2417-28; PMID:23839787; http://dx.doi.org/ 10.1105/tpc.113.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 2005; 102:11928-33; PMID:16081530; http://dx.doi.org/ 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115:209-16; PMID:14567918; http://dx.doi.org/ 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393( *)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 2011; 42:356-66; PMID:21549312; http://dx.doi.org/ 10.1016/j.molcel.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA 2011; 17:312-26; PMID:21177881; http://dx.doi.org/ 10.1261/rna.2537911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008; 36:D154-8; PMID:17991681; http://dx.doi.org/ 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science 2006; 311:195-8; PMID:16410517; http://dx.doi.org/ 10.1126/science.1121638 [DOI] [PubMed] [Google Scholar]
- 24.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011; 475:201-5; PMID:21753850; http://dx.doi.org/ 10.1038/nature10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009; 457:405-12; PMID:19158786; http://dx.doi.org/ 10.1038/nature07755 [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee K, Campos H, Kolaczkowski B. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol 2013; 30:627-41; PMID:23180579; http://dx.doi.org/ 10.1093/molbev/mss263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 2005; 15:1494-500; PMID:16040244; http://dx.doi.org/ 10.1016/j.cub.2005.07.024 [DOI] [PubMed] [Google Scholar]
- 28.Montes RA, de Fatima Rosas-Cardenas F, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martinez N, Meyers BC, Green PJ, de Folter S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 2014; 5:3722; PMID:24759728; http://dx.doi.org/ 10.1038/ncomms4722 [DOI] [PubMed] [Google Scholar]
- 29.Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 2010; 17:997-1003; PMID:20562854; http://dx.doi.org/ 10.1038/nsmb.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci U S A 2010; 107:15269-74; PMID:20643946; http://dx.doi.org/ 10.1073/pnas.1001738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 2006; 25:3347-56; PMID:16810317; http://dx.doi.org/ 10.1038/sj.emboj.7601217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 2012; 151:533-46; PMID:23063653; http://dx.doi.org/ 10.1016/j.cell.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebhardt HA, Fedynak A, Fahlman RP. Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence 2010; 1:12; PMID:20534119; http://dx.doi.org/ 10.1186/1758-907X-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ. Structural basis of microRNA length variety. Nucleic Acids Res 2011; 39:257-68; PMID:20739353; http://dx.doi.org/ 10.1093/nar/gkq727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One 2009; 4:e6442; PMID:19649244; http://dx.doi.org/ 10.1371/journal.pone.0006442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci U S A 2012; 109:2461-6; PMID:22308502; http://dx.doi.org/ 10.1073/pnas.1200169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song JB, Huang SQ, Dalmay T, Yang ZM. Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol 2012; 53:1283-94; PMID:22619471; http://dx.doi.org/ 10.1093/pcp/pcs080 [DOI] [PubMed] [Google Scholar]
- 38.Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 2010; 38:6883-94; PMID:20621980; http://dx.doi.org/ 10.1093/nar/gkq590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al.. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008; 133:116-27; PMID:18342361; http://dx.doi.org/ 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 2004; 18:1187-97; PMID:15131082; http://dx.doi.org/ 10.1101/gad.1201404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 2011; 8:1149-57; PMID:21941127; http://dx.doi.org/ 10.4161/rna.8.6.17665 [DOI] [PubMed] [Google Scholar]
- 42.Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 2009; 15:2112-21; PMID:19850910; http://dx.doi.org/ 10.1261/rna.1774909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 2009; 28:3646-56; PMID:19816405; http://dx.doi.org/ 10.1038/emboj.2009.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J 2010; 62:742-59; PMID:20202174; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04187.x [DOI] [PubMed] [Google Scholar]
- 45.Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 2007; 19:3451-61; PMID:17993620; http://dx.doi.org/ 10.1105/tpc.107.055319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Vilkaitis G, Yu B, Klimasauskas S, Chen X. Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymol 2007; 427:139-54; PMID:17720483; http://dx.doi.org/ 10.1016/S0076-6879(07)27008-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure 2000; 8:R47-54; PMID:10745015; http://dx.doi.org/ 10.1016/S0969-2126(00)00110-6 [DOI] [PubMed] [Google Scholar]
- 48.Portmann S, Grimm S, Workman C, Usman N, Egli M. Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage. Chem Biol 1996; 3:173-84; PMID:8807843; http://dx.doi.org/ 10.1016/S1074-5521(96)90260-4 [DOI] [PubMed] [Google Scholar]
- 49.Borer PN, Lin Y, Wang S, Roggenbuck MW, Gott JM, Uhlenbeck OC, Pelczer I. Proton NMR and structural features of a 24-nucleotide RNA hairpin. Biochemistry 1995; 34:6488-503; PMID:7756280; http://dx.doi.org/ 10.1021/bi00019a030 [DOI] [PubMed] [Google Scholar]
- 50.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307:932-5; PMID:15705854; http://dx.doi.org/ 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Deutscher MP. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc Natl Acad Sci U S A 1995; 92:6883-6; PMID:7542780; http://dx.doi.org/ 10.1073/pnas.92.15.6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 1996; 86:503-12; PMID:8756732; http://dx.doi.org/ 10.1016/S0092-8674(00)80123-3 [DOI] [PubMed] [Google Scholar]
- 53.Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA 1999; 5:139-46; PMID:9917073; http://dx.doi.org/ 10.1017/S1355838299981669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsiao YY, Yang CC, Lin CL, Lin JL, Duh Y, Yuan HS. Structural basis for RNA trimming by RNase T in stable RNA 3′-end maturation. Nat Chem Biol 2011; 7:236-43; PMID:21317904; http://dx.doi.org/ 10.1038/nchembio.524 [DOI] [PubMed] [Google Scholar]
- 55.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 2008; 321:1490-2; PMID:18787168; http://dx.doi.org/ 10.1126/science.1163728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 2010; 22:1090-103; PMID:20407023; http://dx.doi.org/ 10.1105/tpc.110.073882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 2003; 33:949-56; PMID:12609035; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01676.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


