Abstract
Evolution has provided eukaryotes with mechanisms that impede immature and/or aberrant ribosomes to engage in translation. These mechanisms basically either prevent the nucleo-cytoplasmic export of these particles or, once in the cytoplasm, the release of associated assembly factors, which interfere with the binding of translation initiation factors and/or the ribosomal subunit joining. We have previously shown that aberrant yeast 40S ribosomal subunits containing the 20S pre-rRNA can engage in translation. In this study, we describe that cells harbouring the dob1–1 allele, encoding a mutated version of the exosome-assisting RNA helicase Mtr4, accumulate otherwise nuclear pre-60S ribosomal particles containing the 7S pre-rRNA in the cytoplasm. Polysome fractionation analyses revealed that these particles are competent for translation and do not induce elongation stalls. This phenomenon is rather specific since most mutations in other exosome components or co-factors, impairing the 3′ end processing of the mature 5.8S rRNA, accumulate 7S pre-rRNAs in the nucleus. In addition, we confirm that pre-60S ribosomal particles containing either 5.8S + 30 or 5.8S + 5 pre-rRNAs also engage in translation elongation. We propose that 7S pre-rRNA processing is not strictly required for pre-60S r-particle export and that, upon arrival in the cytoplasm, there is no specific mechanism to prevent translation by premature pre-60S r-particles containing 3′ extended forms of mature 5.8S rRNA.
Keywords: Ribosome biogenesis, pre-rRNA processing, RNA exosome, RNA helicase, Mtr4/Dob1, Translation, yeast
Abbreviations
- DAPI
4,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- pre-rRNA
precursor rRNA
- NRD
Non-functional rRNA decay
- r-subunits
ribosomal subunits
- r-particles
ribosomal particles
- r-proteins
ribosomal proteins
- TRAMP complexes
Trf/Air/Mtr4 complexes
Introduction
In eukaryotes, ribosome biogenesis is a compartmentalized, multi-component and multi-step process; most of the biogenesis reactions take place in the nucleolus but later events occur in the nucleoplasm and the cytoplasm.1,2 Although the basic outline of ribosome biogenesis is reasonably well conserved throughout eukaryotes,3 this process has been best studied in the yeast Saccharomyces cerevisiae.1,2,4-6 In the yeast nucleolus, the mature 18S, 5.8S and 25S rRNAs are transcribed as a single large precursor rRNA (pre-rRNA) by RNA polymerase I, from which the external and internal spacer fragments are removed by endo- and exonucleolytic processing reactions.7,8 A pre-5S rRNA is transcribed independently by RNA polymerase III, which undergoes exonucleolytic processing at its 3′ end.8 Concomitant to pre-rRNA processing, pre-rRNAs are extensively modified by base and 2′-O-ribose methylation and pseudouridylation9,10; moreover, pre-rRNAs are subjected to diverse structural rearrangements and folding reactions while they associate/dissociate with trans-acting factors and assemble with most ribosomal proteins (r-proteins) to form nucleolar pre-ribosomal particles.2 On their maturation path from the nucleolus to the nucleoplasm, pre-ribosomal particles undergo a series of compositional changes, which finally enable them to recruit export factors and be transported to the cytoplasm,11-13 where both ribosomal subunits (r-subunits) acquire the competence to engage in translation.14 Cytoplasmic maturation of pre-40S ribosomal particles (r-particles) involves processing of the 20S pre-rRNA to mature 18S rRNA,15 the dissociation and recycling of late trans-acting factors and the assembly of few r-proteins.16,17 Cytoplasmic maturation of pre-60S r-particles involves processing of the 6S pre-rRNA to mature 5.8S rRNA18 (see Fig. S1), and, as for pre-40S particles, the release and recycling of late trans-acting factors and the assembly of the remaining r-proteins.19-21
Translation by aberrantly assembled r-subunits might have severely deleterious consequences to cells, among them, sequestering of translation factors, stalling of translation elongation and reduced fidelity in protein synthesis. Thus, evolution has provided cells with different surveillance mechanisms to monitor and eliminate defective ribosomes (for reviews, see refs. 22,23). First, incorrectly assembled pre-ribosomal particles are efficiently retained in the nucle(ol)us and subjected to degradation by a process involving the exosome and the Trf/Air/Mtr4 (TRAMP) complexes.24-26 Second, the release and/or activity of some of the abovementioned late trans-acting factors is clearly impaired when bound to improperly assembled pre-ribosomal particles that escaped the nuclear retention mechanism,27,28 which is also expected to induce the degradation of these particles. Moreover, r-subunit precursors containing point mutations in essential rRNA sites (i.e. peptidyl-transferase center, decoding site) are recognized and degraded by a distinct pathway, the so-called Non-functional rRNA decay (NRD) pathway (see ref. 23). Degradation of aberrant 40S r-subunits by NRD occurs in P-bodies, requires that these particles engage in translation and involves Dom34, Hbs1, Ski7, Xrn1 and the cytoplasmic exosome.29,30 In contrast, degradation of aberrant 60S r-subunits by NRD does not depend on their engagement in translation and involves the Mms1-Rtt101 E3 ubiquitin ligase, the Cdc48 complex, the proteasome and the cytoplasmic exosome.31,32
Despite the existence of all these surveillance mechanisms, different examples of relatively stable aberrant r-subunits able to engage in translation have been suggested in yeast. For example, we have previously shown that there is an accumulation of cytoplasmic 40S r-subunits containing the 20S pre-rRNA in both ubi3Δub and rpl3[W255C] mutant cells.28,33 Interestingly, these subunits associate into polysomes and they do apparently not stall on mRNAs, similarly as also reported for ltv1Δ pfa1Δ and ltv1Δ prp43–414 double mutants,34 strongly suggesting that, in these cases, the particles are efficient in translation initiation and elongation. Loss of function of trans-acting factors such as Fap7,35 Rio1 and Nob136 or r-proteins S0 or S1437,38 also lead to accumulation of 40S r-particles containing 20S pre-rRNA. However, in these cases, the particles are not or only poorly competent for translation elongation since a much higher 18S rRNA/20S pre-rRNA ratio could be observed in polysomal fractions compared to 80S fractions. The fact that pre-40S can engage in initiation of translation has been clearly shown for particles accumulating upon depletion of Rio1 and Nob1.36 Therefore, it can be concluded that compositional and/or structural differences may exist between immature cytoplasmic pre-40S r-particles from wild-type and different mutant cells.34 On the other hand, in the rrp6Δ mutant, immature 60S r-subunits containing the otherwise nuclear 5.8S + 30 pre-rRNAs are exported to the cytoplasm and are able to enter into polysomes.39 In wild-type cells, exported pre-60S r-particles contain the long or the short form of 6S pre-rRNA.18 In the cytoplasm, the nuclease Ngl2 processes these precursors to 5.8S pre-rRNAs.40 Since, 6S pre-rRNAs are not fully processed to 5.8S rRNAs in the viable ngl2Δ mutant, pre-60S r-particles containing 5.8S+5 pre-rRNAs are predicted to engage in translation.18,40
Here, we have studied the export and translation ability of pre-60S r-particles containing 7S pre-rRNAs. Our results show that, in distinct yeast mutants impaired in 3′ end maturation of 5.8S rRNA, a fraction of these otherwise nuclear particles could be exported to the cytoplasm and incorporated into polysomes. Export of these particles is independent of the TRAMP complex. Moreover, these particles are likely able to complete translation since no evidence for an increase in the stall frequency on mRNAs could be found. We propose that 7S pre-rRNA processing is not strictly required for pre-60S r-particle export and that, upon arrival in the cytoplasm, there is neither an impairment of the assembly of the last r-proteins nor a specific mechanism to prevent translation by immature pre-60S r-particles containing 3′ extended forms of mature 5.8S rRNA.
Results
A fraction of 7S pre-rRNAs accumulating in the dob1–1 mutant is cytoplasmic
Formation of the 3′ end of mature 5.8S rRNAs from the 7S pre-rRNAs is a very complicated multi-step process (Fig. S1), which requires the action of different nucleases (for a review, see8). The initial step consists in the 3′–5′ trimming of 7S pre-rRNAs to the 5.8S + 30 pre-rRNAs, which is carried out by the exonuclease activity of Rrp44, a subunit of the exosome.41,42 Next, 5.8S + 30 pre-rRNAs are processed to 6S pre-rRNAs by the exonuclease Rrp6, either in an exosome-dependent or -independent manner.39,43 In wild-type cells, these 2 reactions occur in the nucleus (Fig. S1) and pre-60S r-particles containing 6S pre-rRNA species are then exported to the cytoplasm.18 There, the 6S pre-rRNAs are successively processed to 5.8S + 5 pre-rRNAs by the 3′–5′ exonucleases Rex1, Rex2 and Rex344 and then to mature 5.8S rRNAs by the nuclease Ngl2.40
In the nucleus, processing of 7S pre-rRNAs requires additional co-factors, such as the RNA helicase Mtr4/Dob145,46 and the non-essential Rrp47/Lrp1 and Mpp6.47-49 Our group identified in the past MTR4/DOB1 as the gene complementing the thermo-sensitive dob1–1 mutation. This mutation leads to a deficit in 60S r-subunits due to the accumulation of 7S pre-rRNAs, which is accompanied by a mild reduction in the levels of mature 5.8S rRNAs.46 Interestingly and in contrast to other 60S r-subunit biogenesis mutants, dob1–1 mutant cells are specifically dependent on a high dosage of TIF3,46 which encodes the yeast translation factor eIF4B.50,51 Furthermore, the dob1–1 mutant shows a similar hypersensitivity to the protein synthesis inhibitor paromomycin as different translation initiation factor mutants.46 These results suggested that the dob1–1 mutation impairs translation, which is likely due to the synthesis of malfunctioning but translation-competent 60S r-subunits rather than the consequence of a general 60S r-subunit deficit.
To test this hypothesis, and taking into account the pre-rRNA processing phenotypes of the dob1–1 mutant, we first determined whether the dob1–1 mutant accumulates aberrant pre-60S r-particles containing 7S pre-rRNA in the cytoplasm. To this end, we performed immunoprecipitation experiments in wild-type and dob1–1 cells expressing TAP-tagged constructs of proteins specific for nuclear (Nop7-TAP), nucleo-cytoplasmic (Arx1-TAP), exclusively cytoplasmic (Lsg1-TAP) pre-60S r-particles, and for mature 60S r-subunits (L24A-TAP). Precipitated RNAs were analyzed by northern hybridization. As shown in Figure 1A, and as expected, 7S pre-rRNAs were specifically enriched upon precipitation of the Nop7-TAP and the Arx1-TAP baits in wild-type cells compared to the untagged control, but were not significantly enriched upon precipitation with the cytoplasmic Lsg1-TAP and L24A-TAP baits. In contrast, in dob1–1 cells, the Lsg1-TAP and L24A-TAP baits were able to efficiently precipitate 7S pre-rRNAs (Fig. 1B). No 27S pre-rRNAs were detected upon affinity-purification from extracts of the Lsg1-TAP bait in either wild-type or dob1–1 cells (Fig. S2A and data not shown). Moreover, no apparent nuclear retention of an Lsg1-GFP construct was observed in the dob1–1 strain (Fig. S2B). Altogether, these results indicate that pre-60S r-particles containing 7S pre-rRNA are being exported in the dob1–1 mutant.
Figure 1.
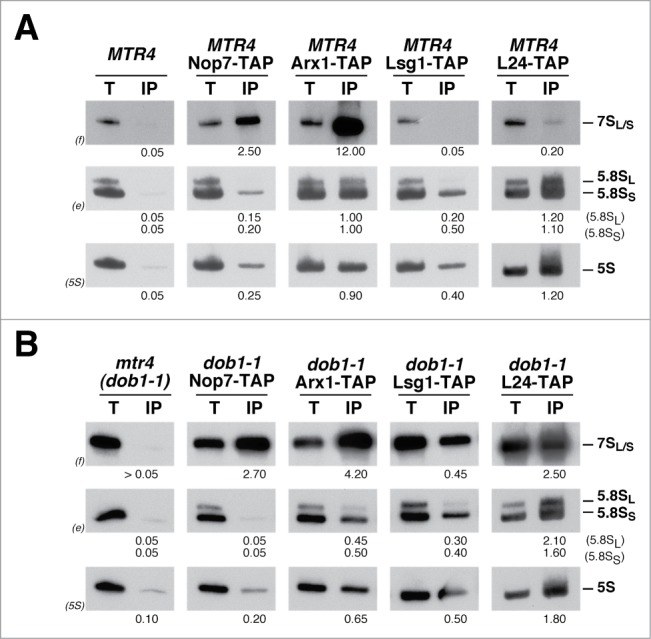
Pre-60S r-particles containing 7S pre-rRNAs are exported to the cytoplasm in the dob1–1 mutant. Isogenic wild-type (MTR4) and dob1–1 (mtr4) strains were grown at 30°C in YPD medium to mid-log phase. Immunoprecipitation experiments were carried out using IgG-Sepharose and whole-cell extracts from wild-type (A) and dob1–1 (B) control cells (no TAP bait) or cells expressing TAP-tagged Nop7, Arx1, Lsg1, and L24A. RNA was extracted from the beads (lanes IP) or from an amount of total extract corresponding to 1/100 of that used for the immunoprecipitations (lanes T), separated on a 7% polyacrylamide-8M urea gel, transferred to a nylon membrane and subjected to northern hybridization with the 3 probes indicated in parentheses to detect 7S pre-rRNAs and mature 5.8S and 5S rRNAs. Probes are described in Figure 2 and Supplemental Table 2. Signal intensities were measured by phosphorimager scanning; values (below each IP lane) refer to the percentage of each RNA recovered after purification.
To further confirm that 7S pre-rRNAs are associated with exported pre-60S r-particles in the dob1–1 mutant, we studied the localization of different pre-rRNAs by fluorescence in situ hybridization (FISH) with probes complementary to the D-A2 region of ITS1 (probe FISH ITS1), and the E-C2 (probe FISH ITS2–1) and C2-C1 regions of ITS2 (probe FISH ITS2–2). The first probe hybridizes to 35S, 33S, 32S and 20S pre-rRNAs, the second one to 35S, 33S, 32S, all 27S pre-rRNAs and all 3′ extended pre-5.8S precursors, and the third one to 35S, 33S, 32S, all 27S pre-rRNAs and the 5′ extended pre-25S precursors, respectively (Fig. 2A). None of these probes hybridize with mature rRNAs. Before performing the FISH experiments, we checked that dob1–1 cells do not display an enlarged nucleoplasm or nucleus (Fig. S3). When probes FISH ITS1 and FISH ITS2–2 were used, similar intensity and distribution of the signals were observed in both wild-type and dob1–1 cells (Fig. 2B, C). However, when the FISH ITS2–1 probe was used, a stronger FISH signal was observed in dob1–1 mutant cells than in wild-type cells, which is consistent with the 7S pre-rRNA accumulation detected in this strain. More importantly, there is a clear appearance of cytoplasmic ITS2–1 signal in dob1–1 cells, indicating that 3′ extended forms of 5.8S rRNA, most likely corresponding to 7S pre-rRNAs (Fig. 1B), accumulate in the cytoplasm of dob1–1 mutant cells. However, still a substantial amount of 3′ extended forms of 5.8S rRNA accumulates in the nucleus of dob1–1 mutant cells.
Figure 2.
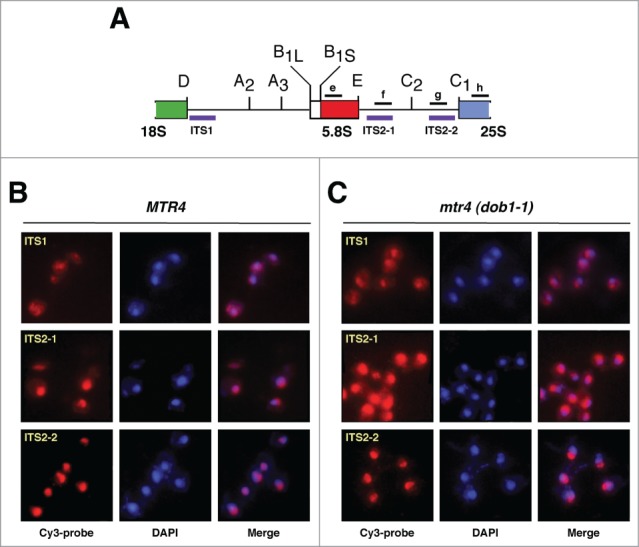
Detection of pre-ribosomal particles by fluorescence in situ hybridization (FISH). (A) Primary structure of the 2 internal transcribed spacers, ITS1 and ITS2, of the 35S pre-rRNA. Processing sites are shown. The location of the 3 Cy-3 labeled probes used for FISH (purple) and of the oligonucleotides probes used for northern hybridization (black) is indicated. (B and C) Wild-type (MTR4) and dob1–1 (mtr4) cells were grown at 30°C in YPD medium to mid-log phase. Distinct pre-rRNAs were detected by FISH using Cy-3 labeled ITS1, ITS2–1 or ITS2–2 probes (red; purple in merge panels). Cells were further stained with DAPI to visualize the nucleoplasm (blue). All images shown were captured using identical exposure times and processed in the same manner.
Cytoplasmic pre-60S r-particles containing the 7S pre-rRNA get incorporated into translating ribosomes
To assess whether the cytoplasmic, 7S pre-rRNA containing pre-60S r-particles present in dob1–1 cells are incorporated into translating ribosomes, we analyzed the distribution of the 7S pre-rRNAs in polysome profiles of dob1–1 mutant cell extracts by sucrose gradient fractionation and northern blotting and compared it to that of wild-type cells (Fig. 3). In wild-type cells, 7S pre-rRNAs co-migrated exclusively with the 60S r-subunit peak, which is indicative of its presence in nuclear pre-60S r-particles. In contrast, 7S pre-rRNAs were not only associated with the 60S r-subunit peak, but also co-sedimented significantly with polysomes in the dob1–1 mutant.
Figure 3.
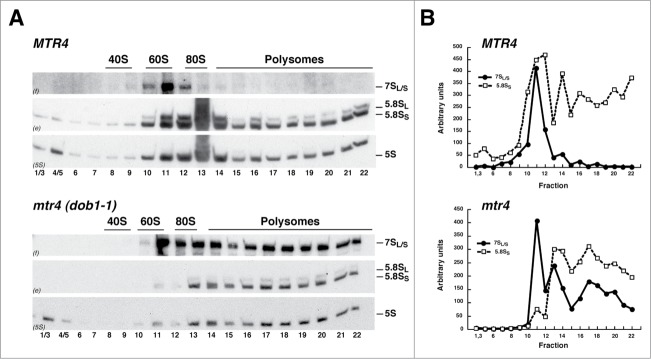
Pre-60S r-particles containing 7S pre-rRNAs engage in translation in the dob1–1 mutant. Wild-type (MTR4) and dob1–1 (mtr4) cells were grown at 30°C in YPD medium to mid-log phase. (A) Cells extracts were prepared and 10 A260 units of each extract were resolved in 7–50% sucrose gradients and fractionated. RNA was extracted from each fraction and analyzed by northern blotting using probes f, e and 5S, which reveal 7S pre-rRNAs and mature 5.8S and 5S rRNAs, respectively. The position of free 40S and 60S r-subunits, 80S ribosomes and polysomes, obtained from the recorded A254 profiles, are shown. (B) Signal intensities of the 7S pre-rRNAs (black dot, continuous line) and 5.8SS rRNA (white squares, dashed line) were determined for each fraction by phosphorimager scanning and represented as arbitrary units.
To confirm that the 7S pre-rRNA containing particles sedimenting with polysomes in the dob1–1 mutant were not simply aggregates, cell extracts of both wild-type and dob1–1 cells were prepared under polysome run-off conditions, by omission of cycloheximide, either in standard buffer or in a buffer lacking Mg2+, which causes dissociation of 80S ribosomes into 40S and 60S r-subunits.52 Under these conditions, the bulk of 7S pre-rRNAs and 5.8S rRNAs was shifted to positions of the 80S and 60S fractions in the dob1–1 mutant, similarly as observed for the 5.8S rRNAs in wild-type cells (Fig. S4). This result clearly reveals that the fractions containing 7S pre-rRNA were indeed associated with polysomes. Moreover, as shown in Figure 3, when the 7S pre-rRNAs levels were quantified within the different fractions from standard sucrose gradients and compared to those of 5.8SS rRNA in the same fractions in both wild-type and dob1–1 cells, we found that 7S pre-rRNA levels were particularly high in the 60S r-subunit peak compared to those of 5.8SS rRNA in the dob1–1 mutant, which indicates that this peak consists of both nuclear and cytoplasmic pre-60S r-particles containing the 7S precursor. However and remarkably, the 7S/5.8SS RNA ratio remains constant along the 80S and polysome fractions in the dob1–1 mutant. Altogether, these findings clearly reveal that the cytoplasmic pre-60S r-particles containing 7S pre-rRNA observed in dob1–1 mutant cells are competent for both translation initiation and elongation.
Analysis of mutants affected in 3′ end maturation of 5.8S rRNA
As abovementioned, 3′ end maturation of 5.8S rRNA in yeast is a multi-step and multi-component pathway (see Fig. S1). Thus, we next determined whether immature, translation-competent pre-60S r-particles containing 3′ extended forms of 5.8S rRNA are present in other mutants affecting this maturation process (see Table S1). Total RNA was extracted from these mutant strains, either grown at 30°C or shifted to 37°C, and subjected to northern blot analysis with probes hybridizing to the different precursors of mature 5.8S rRNAs. As shown in Figure 4, the csl4–1, mtr3–1, and rrp4–1 mutants, as previously reported,53,54 accumulated 7S pre-rRNAs. The strains lacking Rrp6 and Rrp4739,47 were selected since they accumulated slightly 7S pre-rRNAs and significantly 5.8S + 30 pre-rRNAs. Finally, the ngl2Δ mutant40 accumulated the 5.8S + 5 pre-rRNA species and did not produce completely matured 5.8S rRNAs.
Figure 4.
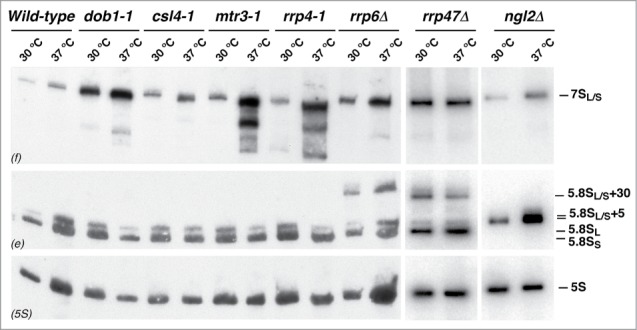
Pre-rRNA processing of the 7S pre-rRNAs in different mutants affected in 5.8S rRNA maturation. A wild-type strain and the indicated mutants were grown at 30°C in YPD medium to mid-log phase or shifted for 4 h to 37°C. Total RNA was prepared and 5 μg was separated on a 7% polyacrylamide-8M urea gel, transferred to a nylon membrane and subjected to northern hybridization with the 3 probes indicated in parentheses to detect 7S pre-rRNAs and mature 5.8S and 5S rRNAs. Note that probe e also reveals 5.8S + 30 and 5.8S + 5 pre-rRNAs in the rrp6Δ and rrp47Δ, and ngl2Δ mutants, respectively.
We next assessed the ability of the corresponding pre-60S r-particles containing the above pre-5.8S rRNA species from the different mutants to engage in translation. Similarly as done for the dob1–1 mutant, cell extracts were prepared under polysome-preserving conditions from the wild-type strain and the different mutants and subjected to sucrose gradient fractionation; then, RNA was prepared from individual fractions, separated by electrophoresis and analyzed by northern blotting. As shown in Figure 5, pre-60S r-particles containing 7S pre-rRNAs were predominantly found in 60S fractions but not in polysomes in extracts from the csl4–1, mtr3–1, and rrp4–1 mutants grown at 30°C. The 7S pre-rRNAs were neither found in polysomes from the rrp47Δ and ngl2Δ strains and very faintly in polysome-containing fractions in extracts from the rrp6Δ mutant (Fig. 5). Interestingly, when the rrp4–1 and rrp6Δ mutants were subjected to a shift at 37°C for 4 h, 7S pre-rRNAs did associate into polysomes (Fig. 5 and data not shown). In contrast, 7S pre-rRNA containing pre-60S r-particles from the csl4–1, mtr3–1, and rrp47Δ mutants subjected to a similar shift did not enter polysomes (Fig. 5 and data not shown). We further studied the rrp4–1 mutant to determine whether 7S pre-rRNA species were exported at 30°C and 37°C. To this end, we performed immunoprecipitation experiments in rrp4–1 cells expressing TAP-tagged constructs of either Nop7 or Lsg1 and analyzed the precipitated RNAs by northern hybridization. As shown in Figure 6, at either 30°C or 37°C, 7S pre-rRNAs were specifically enriched upon precipitation of the Nop7-TAP bait; however, these precursors were only significantly enriched upon precipitation with the cytoplasmic Lsg1-TAP bait upon the shift to 37°C.
Figure 5.
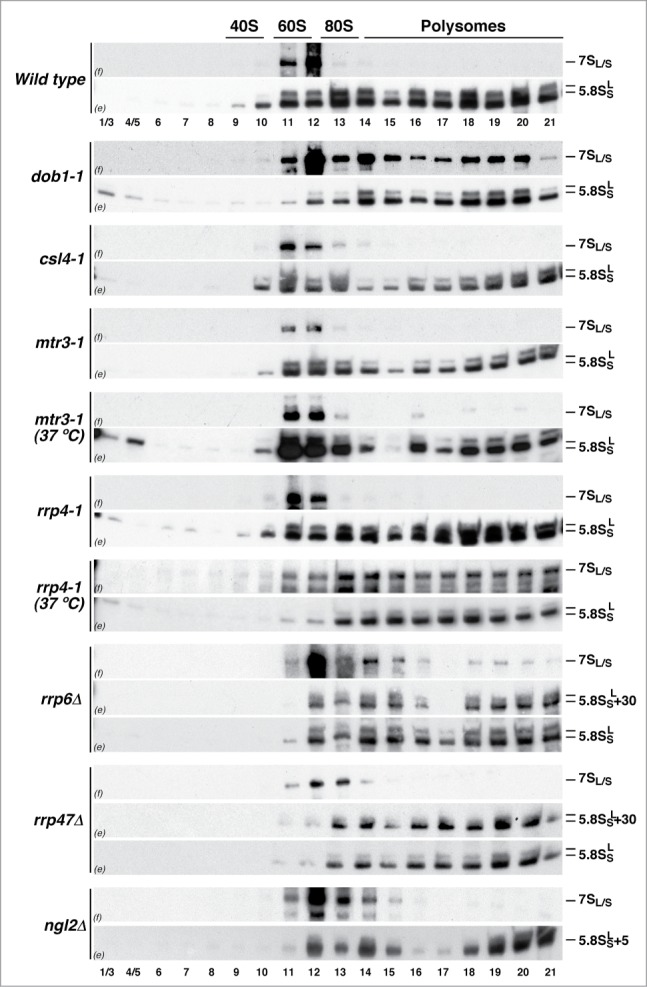
Ability of pre-60S r-particles containing 3′ end extended precursors of 5.8S rRNAs to engage in translation in different mutant strains. A wild-type strain and the indicated mutants were grown at 30°C in YPD medium to mid-log phase. The mtr3–1 and rrp4–1 mutants were also shifted for 4 h to 37°C. Cell extracts were prepared and 10 A260 units of each extract were resolved in 7–50% sucrose gradients and fractionated. RNA was extracted from each fraction and analyzed by northern blotting using probes f and e, which reveal 7S pre-rRNAs and mature 5.8S rRNAs, respectively. Note that probe e also reveals 5.8S + 30 and 5.8S + 5 pre-rRNAs in the rrp6Δ and rrp47Δ, and ngl2Δ mutants, respectively. The position of free 40S and 60S r-subunits, 80S ribosomes and polysomes, obtained from the recorded A254 profiles, are indicated.
Figure 6.
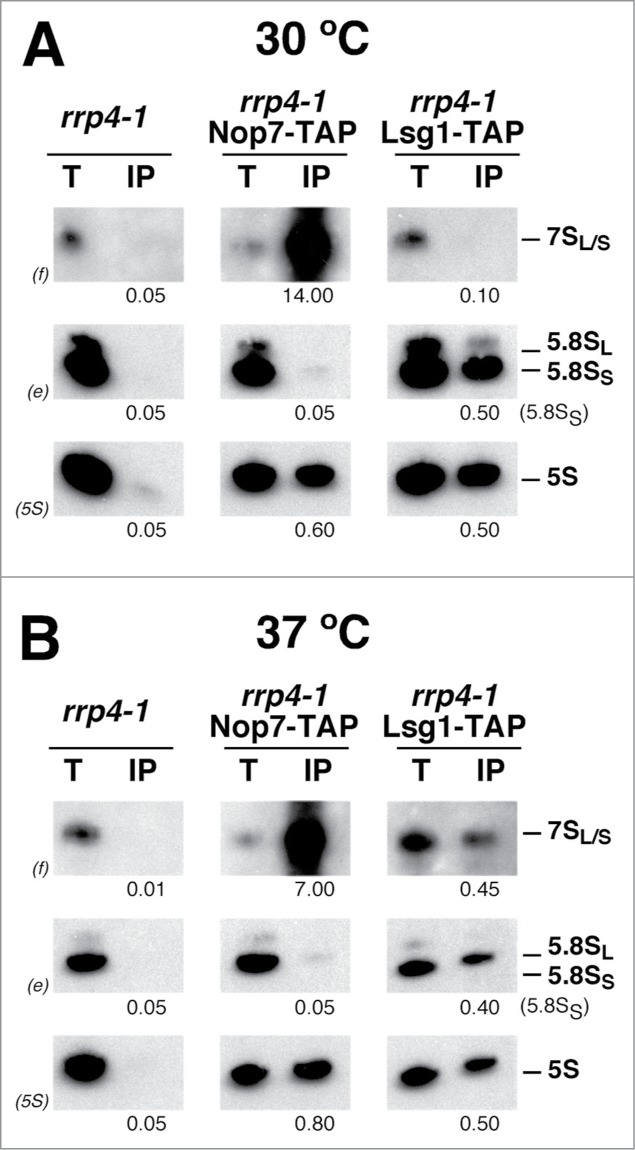
Pre-60S r-particles containing 7S pre-rRNAs are exported to the cytoplasm in the rrp4–1 mutant at the non-permissive temperature. The rrp4–1 strain was grown at 30°C in YPD medium to mid-log phase (A) or shifted for 4 h to 37°C (B). Immunoprecipitation experiments were carried out using IgG-Sepharose and whole-cell extracts from untagged or TAP-tagged Nop7 and Lsg1 cells. RNA was extracted from the beads (lanes IP) or from an amount of total extract corresponding to 1/100 of that used for the immunoprecipitations (lanes T), separated on a 7% polyacrylamide-8M urea gel, transferred to a nylon membrane and subjected to northern hybridization with the probes indicated in parentheses to detect 7S pre-rRNAs and mature 5.8S and 5S rRNAs. Signal intensities were measured by phoshorimager scanning; values (below each IP lane) refer to the percentage of each RNA recovered after purification.
These results suggest that the pre-60S r-particles containing 7S pre-rRNAs might not be similar in the different mutants. Thus, particles unable to exit to the cytoplasm are expected to either retain factors that prevent their export or lack factors that prepare them for export competence. In addition, for those able to arrive in the cytoplasm, there might be no apparently specific mechanism to prevent their engagement in translation.
Discussion
With this study, we provide, to the best of our knowledge, the first report on cytoplasmic pre-60S r-particles containing 7S pre-rRNAs. These particles, which are normally restricted to the nucleus in wild-type cells, are exported to the cytoplasm in certain mutant conditions where 7S pre-rRNA processing is affected, such as in dob1–1, rrp4–1, and rrp6Δ mutant cells. Whether these particles are being actively exported to the cytoplasm and/or arrive there as a result of leaking passively through nuclear pores remains to be elucidated. Moreover, these aberrant cytoplasmic pre-60S r-particles are competent for translation, as indicated by their occurrence in polysome fractions. Importantly, the ratio between 7S pre-rRNA and mature 5.8S and 5S rRNAs was comparable in all the polysome fractions in these mutants; thus, indicating that these particles efficiently perform translation elongation without being stalled on the mRNAs. The TRAMP complexes have been described as major nuclear co-factors for RNA degradation of pre-rRNA precursors present in defective nuclear pre-60S r-particles.26,55 Thus, if a leakage process for export would operate, it would be expected that inactivation of the TRAMP complex would increase the bulk of cytoplasmic pre-60S r-particles in mutants affected in 7S pre-rRNA processing. However, when we assessed the effects of the absence of Trf4 alone or in combination with Air1 in the csl4–1, mtr3–1, and rrp4–1 mutants, we only detected a slight increase in the amount of pre-60S particles engaged in translation (Fig. S5). Therefore, these results argue in favor of an active export for pre-60S r-particles containing 7S pre-rRNA in these mutants. Indeed, a predominant leakage process for export is unlikely since pre-60S containing 7S pre-rRNAs do accumulate in other mutants impaired in 5.8S rRNA maturation, but are efficiently retained in the nucleus (for an example, see Figs. 4 and 6). It remains a challenging task to understand at a molecular level what promotes the retention or induces the export of pre-60S r-particles containing 7S pre-rRNAs in the different mutants. In this respect, the dob1–1 mutation changes the glutamate codon (GAG) at amino acid position 796 to a lysine codon (AAG). Strikingly, E796 is located within the major helix of the so-called KOW domain in the “arch” of Mtr4.56,57 This domain has been shown to contribute to RNA binding,57 suggesting that it might be required to present RNA substrates, including 7S pre-rRNAs, to the Mtr4 helicase core and, thus, to the exosome. Therefore, we envisage that the acquisition of export competence by pre-60S r-particles may rely, among other mechanisms, on the non-recruitment or efficient release of the exosome from the particles.
Pre-60S r-particles containing 7S pre-rRNA are not the sole aberrant pre-60S r-particles able to engage in translation. In a previous work, it has been reported that a fraction of pre-60S r-particles containing 5.8S + 30 pre-rRNAs can enter into polysomes in the rrp6Δ mutant39; our study confirms this finding and furthermore reveals that this also occurs in the rrp47Δ mutant. Moreover, since the ngl2Δ strain is viable and has very little effect on cellular growth (40 and our own results), pre-60S r-particles containing 5.8S + 5 pre-rRNA were predicted to be able to engage in translation, an assumption that we have herein corroborated. Altogether, these results indicate that the 3′-end extension of mature 5.8S rRNA, as previously presumed,18 does not significantly hamper the structure or the role of the relevant functional sites of the ribosome during translation (see Fig. S6). We propose that in wild-type conditions nuclear export acts as an efficient barrier preventing immature pre-60S r-particles containing 3′ extended forms of mature 5.8S rRNA to enter the pool of translating ribosomes in the cytoplasm. Therefore, if these particles escape to the cytoplasm, they obligatorily engage in translation without affecting elongation efficiency. Further experiments are required to elucidate whether differences do nevertheless exist for the translation of specific mRNAs when carried out either by normal or “aberrant” 7S pre-rRNA containing ribosomes.
Materials and Methods
Yeast strains, media and microbiological methods
All yeast strains used in this study are listed in Supplemental Table 1. Growth and handling of yeast and standard media were done following established procedures.58
Sucrose gradient centrifugation
Cell extract for polysome and r-subunit analyses were performed according to Foiani et al.,59 as previously described52 using an ISCO UA-6 system equipped to continuously monitor A254. Fractions of 0.5 ml were collected from the gradients and RNA was extracted from the different fractions and analyzed as described below.
RNA analyses
Total RNA was extracted by the acid-phenol method.60 RNA was also extracted from sucrose gradient fractions as exactly described.61 Equal amounts of total RNA (5 μg) or equal volumes of each fraction were loaded on 1.2% agarose-6% formaldehyde or on 7% polyacrylamide-8M urea gels as described.62 Northern hybridization was performed as previously described.46 Signal intensities were quantified using a FLA-5100 imaging system and Image Gauge (Fujifilm). The sequences of the oligonucleotides used for northern hybridization are listed in Supplemental Table 2.
Fluorescence microscopy
Fluorescence in situ hybridization (FISH) was carried out in fixed cells as previously described,33,63 using Cy3-labeled ITS1- or ITS2-specific probes (see Table S2). Cells were also stained with DAPI (4,6-diamidino-2-phenylindole) to visualize DNA. Images were acquired using an Olympus BX61 fluorescence microscope equipped with a digital camera. Images were analyzed using the Cell Sens software.
The localization of GFP-tagged Lsg1 was inspected by fluorescence microscopy as previously described.64
RNA precipitations
Extracts from distinct TAP-tagged assembly factors were used to affinity-purified pre-ribosomal particles on the road to mature ribosomes by IgG-Sepharose beads as previously described.65 Pre-rRNAs copurifying with these particles were recovered from the beads by phenol-chloroform extraction, as described in,65 and assayed by northern hybridization as indicated above.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank M. Carballo and J.L. Ribas of the Biology and Microscopy facilities from the University of Seville (CITIUS) for help with the phosphorimager analysis and fluorescence microscopy, respectively. We also thank F.J. Espinar-Marchena for handling structures of the yeast ribosome and A. van Hoof, and A. Morillon for yeast strains.
Funding
This study was supported by grants from the Spanish Ministry of Science and Innovation (MICINN) and ERDF (BFU2013–42958-P; BFU2010–15690) and the Andalusian Government (BIO-271, P08-CVI-03508) to J.d.l.C. and the Swiss National Science Foundation (PP00P3_123341 and PP00P3_146344/1) to D.K. J.J.G.-G. was a recipient of a FPI Ph.D. fellowship from the MICINN.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 2008; 65:2334-59; PMID:18408888; http://dx.doi.org/ 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013; 195:643-81; PMID:24190922; http://dx.doi.org/ 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 2003; 313:17-42; PMID:12957375; http://dx.doi.org/ 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- 4.de la Cruz J, Kressler D, Linder P. Ribosomal subunit assembly In: Olson MOJ, ed. Nucleolus. Georgetown: Kluwer academic; LandesBioscience/eurekah.com, 2004:258-85 [Google Scholar]
- 5.Gerhardy S, Menet AM, Pena C, Petkowski JJ, Panse VG. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 2014; 123:327-44; PMID:24817020; http://dx.doi.org/ 10.1007/s00412-014-0463-z [DOI] [PubMed] [Google Scholar]
- 6.de la Cruz J, Karbstein K, Woolford JL Jr.. Functions of ribosomal proteins in assembly of eukaryotic ribosome in vivo. Annu Rev Biochem 2015; 84:93-129; PMID:25706898; http://dx.doi.org/ 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 2015; 6:225-42; PMID:25346433; http://dx.doi.org/ 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip Rev RNA 2015; 6:191-209; PMID:Can't; http://dx.doi.org/ 10.1002/wrna.1267 [DOI] [PubMed] [Google Scholar]
- 9.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 2006; 34:D158-D62; PMID:16381836; http://dx.doi.org/ 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 2015; 22:11-9; PMID:25565028; http://dx.doi.org/ 10.1038/nsmb.2939 [DOI] [PubMed] [Google Scholar]
- 11.Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes. PLoS Genet 2012; 8:e1002915; PMID:22956913; http://dx.doi.org/ 10.1371/journal.pgen.1002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassler J, Klein I, Schmidt C, Kallas M, Thomson E, Wagner MA, Bradatsch B, Rechberger G, Strohmaier H, Hurt E, et al.. The conserved Bud20 zinc finger protein is a new component of the ribosomal 60S subunit export machinery. Mol Cell Biol 2012; 32:4898-912; PMID:23045392; http://dx.doi.org/ 10.1128/MCB.00910-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta 2010; 1803:673-83; PMID:19879902; http://dx.doi.org/ 10.1016/j.bbamcr.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 14.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 2010; 35:260-6; PMID:20137954; http://dx.doi.org/ 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J Biol Chem 1973; 248:1412-6; PMID:4568815 [PubMed] [Google Scholar]
- 16.Hector RD, Burlacu E, Aitken S, Bihan TL, Tuijtel M, Zaplatina A, Cook AG, Granneman S. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res 2014; 42:12138-54; PMID:25200078; http://dx.doi.org/ 10.1093/nar/gku815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 2011; 333:1449-53; PMID:21835981; http://dx.doi.org/ 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson E, Tollervey D. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol Cell Biol 2010; 30:976-84; PMID:20008552; http://dx.doi.org/ 10.1128/MCB.01359-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 2010; 39:196-208; PMID:20670889; http://dx.doi.org/ 10.1016/j.molcel.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Mateos M, García-Gómez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JPG. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res 2009; 37:7519-32; PMID:Can't; http://dx.doi.org/ 10.1093/nar/gkp806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, Unterweger S, Rechberger G, Pertschy B, Bergler H. Rlp24 activates the AAA-ATPase Drg1 to initiate cytoplasmic pre-60S maturation. J Cell Biol 2012; 199:771-82; PMID:23185031; http://dx.doi.org/ 10.1083/jcb.201205021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol 2013; 23:242-50; PMID:23375955; http://dx.doi.org/ 10.1016/j.tcb.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine DL. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci 2010; 35:267-77; PMID:20097077; http://dx.doi.org/ 10.1016/j.tibs.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 24.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005; 121:713-24; PMID:15935758; http://dx.doi.org/ 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 25.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 2000; 28:1684-91; PMID:10734186; http://dx.doi.org/ 10.1093/nar/28.8.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 2006; 25:1534-46; PMID:16541108; http://dx.doi.org/ 10.1038/sj.emboj.7601035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bussiere C, Hashem Y, Arora S, Frank J, Johnson AW. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol 2012; 197:747-59; PMID:22689654; http://dx.doi.org/ 10.1083/jcb.201112131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Gómez JJ, Fernández-Pevida A, Lebaron S, Rosado IV, Tollervey D, Kressler D, de la Cruz J. Final pre-40S maturation depends on the functional integrity of the 60s subunit ribosomal protein L3. PLoS Genet 2014; 10:e1004205; PMID:Can't; http://dx.doi.org/ 10.1371/journal.pgen.1004205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell 2006; 24:619-26; PMID:17188037; http://dx.doi.org/ 10.1016/j.molcel.2006.10.008 [DOI] [PubMed] [Google Scholar]
- 30.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 2009; 34:440-50; PMID:19481524; http://dx.doi.org/ 10.1016/j.molcel.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii K, Kitabatake M, Sakata T, Ohno M. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J 2012; 31:2579-89; PMID:22505030; http://dx.doi.org/ 10.1038/emboj.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev 2009; 23:963-74; PMID:19390089; http://dx.doi.org/ 10.1101/gad.1775609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lacombe T, García-Gómez JJ, de la Cruz J, Roser D, Hurt E, Linder P, Kressler D. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 2009; 72:69-84; PMID:19210616; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06622.x [DOI] [PubMed] [Google Scholar]
- 34.Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 2009; 284:35079-91; PMID:19801658; http://dx.doi.org/ 10.1074/jbc.M109.040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol 2005; 25:10352-64; PMID:16287850; http://dx.doi.org/ 10.1128/MCB.25.23.10352-10364.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J 2010; 29:80-92; PMID:19893492; http://dx.doi.org/ 10.1038/emboj.2009.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford CL, Randal-Whitis L, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res 1999; 59:704-10; PMID:9973221 [PubMed] [Google Scholar]
- 38.Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Léger-Silvestre I, Gas N, Woolford JL Jr.. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell 2004; 14:331-42; PMID:15125836; http://dx.doi.org/ 10.1016/S1097-2765(04)00215-1 [DOI] [PubMed] [Google Scholar]
- 39.Briggs MW, Burkard KTD, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8S rRNA 3′ end formation. J Biol Chem 1998; 273:13255-63; PMID:9582370; http://dx.doi.org/ 10.1074/jbc.273.21.13255 [DOI] [PubMed] [Google Scholar]
- 40.Faber AW, Van Dijk M, Raué HA, Vos JC. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′- end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 2002; 8:1095-101; PMID:12358428; http://dx.doi.org/ 10.1017/S1355838202021027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 2009; 16:56-62; PMID:19060898; http://dx.doi.org/ 10.1038/nsmb.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 2007; 14:15-22; PMID:17173052; http://dx.doi.org/ 10.1038/nsmb1184 [DOI] [PubMed] [Google Scholar]
- 43.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 2008; 36:6645-55; PMID:18940861; http://dx.doi.org/ 10.1093/nar/gkn743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 2000; 19:1357-65; PMID:10716935; http://dx.doi.org/ 10.1093/emboj/19.6.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuch B, Feigenbutz M, Makino DL, Falk S, Basquin C, Mitchell P, Conti E. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J 2014; 33:2829-46; PMID:25319414; http://dx.doi.org/ 10.15252/embj.201488757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 1998; 17:1128-40; PMID:9463390; http://dx.doi.org/ 10.1093/emboj/17.4.1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol 2003; 23:6982-92; PMID:12972615; http://dx.doi.org/ 10.1128/MCB.23.19.6982-6992.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol 2008; 28:5446-57; PMID:18591258; http://dx.doi.org/ 10.1128/MCB.00463-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al.. A panoramic view of yeast noncoding RNA processing. Cell 2003; 113:919-33; PMID:12837249; http://dx.doi.org/ 10.1016/S0092-8674(03)00466-5 [DOI] [PubMed] [Google Scholar]
- 50.Altmann M, Müller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J 1993; 12:3997-4003; PMID:8404865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coppolecchia R, Buser P, Stotz A, Linder P. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J 1993; 12:4005-11; PMID:8404866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kressler D, Rojo M, Linder P, de la Cruz J. Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Nucleic Acids Res 1999; 27:4598-608; PMID:10556316; http://dx.doi.org/ 10.1093/nar/27.23.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof A, Staples RR, Baker RE, Parker R. Function of the Ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol 2000; 20:8230-43; PMID:11027292; http://dx.doi.org/ 10.1128/MCB.20.21.8230-8243.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev 1996; 10:502-13; PMID:8600032; http://dx.doi.org/ 10.1101/gad.10.4.502 [DOI] [PubMed] [Google Scholar]
- 55.Dez C, Dlakic M, Tollervey D. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA 2007; 13:1516-27; PMID:17652137; http://dx.doi.org/ 10.1261/rna.609807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J 2010; 29:2205-16; PMID:20512111; http://dx.doi.org/ 10.1038/emboj.2010.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA 2010; 107:12139-44; PMID:20566885; http://dx.doi.org/ 10.1073/pnas.1004953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press, 1994 [Google Scholar]
- 59.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol 1991; 11:3203-16; PMID:2038326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Saccharomyces cerevisiae. Current Protocols in Molecular Biology. New York, N. Y.: John Wiley & Sons, Inc., 1994:13.0.1-13.14.17 [Google Scholar]
- 61.de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 1998; 4:1268-81; PMID:9769101; http://dx.doi.org/ 10.1017/S1355838298981158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venema J, Planta RJ, Raué HA. In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae In: Martin R, ed. Protein synthesis: Methods and Protocols. Totowa, N. J.: Humana Press, 1998:257-70 [DOI] [PubMed] [Google Scholar]
- 63.Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 2000; 14:830-40; PMID:10766739 [PMC free article] [PubMed] [Google Scholar]
- 64.Babiano R, de la Cruz J. Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 2010; 38:5177-92; PMID:20392820; http://dx.doi.org/ 10.1093/nar/gkq260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol Cell Biol 2005; 25:9269-82; PMID:16227579; http://dx.doi.org/ 10.1128/MCB.25.21.9269-9282.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


