Abstract
MicroRNAs (miRNAs) contribute to a wide variety of human diseases by regulating gene expression, leading to imbalances in gene regulatory networks. To discover novel hepatocellular carcinoma (HCC)-related miRNA-target axes and to elucidate their functions, we here performed a systematic investigation combining biological data acquisition and integration, miRNA-target prediction, network construction, functional assay and clinical validation. As a result, a total of 117 HCC differentially expressed miRNAs were identified, and 728 high confident target genes of these miRNAs were collected. Then, the interaction network of target genes was constructed and 221 key nodes with topological importance in the network were identified according to their topological features including degree, node-betweenness, closeness and K-coreness. Among these key nodes, Cyclin D1 had the highest node-betweenness, implying its bottleneck role in the network. Luciferase reporter assay confirmed that miRNA-19a, which was one of HCC downregulated miRNAs, directly targeted Cyclin D1 in HCC cells. Moreover, miR-19a might play inhibitory roles in HCC malignancy via regulating Cyclin D1 expression. Further clinical evidence also highlighted the prognostic potential of miR-19a/Cyclin D1 axis in HCC. In conclusion, this systematic investigation provides a framework to identify featured miRNAs and their target genes which are potent effectors in the occurrence and development of HCC. More importantly, miR-19a/Cyclin D1 axis might have promising applications as a therapeutic target and a prognostic marker for patients with HCC.
Keywords: cyclin D1, gene regulatory network, hepatocellular carcinoma, microRNA, microRNA-19a, prognosis
Introduction
Hepatocellular carcinoma (HCC) represents the most prevalent primary liver malignant disease and ranks the third cause of cancer-related deaths.1 It has an increasing incidence and accounts for approximately 700000 deaths annually all over the world.2 It is a multi-step process in hepatocarcinogenesis containing chronic hepatitis, cirrhosis, dysplastic nodules and malignant tumors. Over the past decades, many efforts have been made to improve the overall survival rate of HCC. Both liver transplantation and surgical resection are considered potentially curative treatments for early-stage HCC. However, more than two-thirds of HCC patients occur recurrence after surgical hepatic resection and more than four-fifths of HCC patients are unresectable due to fast infiltrating growth, early metastasis, high-grade malignancy.3 Growing clinical observations show that HCC patients with the same clinicopathologic characteristics often display different clinical outcome, implying that there may be several complex molecular and cellular events involved in the development and aggressive progression of HCC.4 Thus, it is extremely important to clarify the molecular mechanisms underlying hepatocarcinogenesis and to seek optimal biomarkers suitable for early diagnosis, efficient therapy and patients' prognosis.
MicroRNAs (miRNAs) constitute a group of short non-coding RNA molecules with 18-25 nucleotides in length.5 Based on miRBase (release 21), the human genome encodes 1881 (2588) precursor (mature) miRNAs, which potentially target the majority of the human genes.6 Functionally, miRNAs play key roles in diverse biological processes including cell development, proliferation, differentiation and apoptosis via regulating gene expression post-transcriptionally.7 Growing evidence shows the aberrant expression of miRNAs in various cancer cells. According to the functions of the target genes, miRNAs either act as oncomiRs or as tumor suppressors. As oncomiRs, they are upregulated and have a stimulating role for cancer progression; As tumor suppressors, they are downregulated and inhibit the expression of oncogenic targets.8 This dual role makes miRNAs play an important role in modulating cell proliferation, invasion, metastasis, survival and tumor angiogenesis.9 Especially, several miRNAs have been reported to be differentially expressed in HCC and may be pathogenically relevant,10–12 implying the potential roles of miRNAs as novel molecules or targets for HCC therapy. However, our understanding of miRNA expression patterns as potential biomarkers for diagnosis, prognosis, disease progression and personalized therapy is just emerging.
Growing evidence shows that miRNAs function in a multiple-to-multiple relationship with their target genes, suggesting that a specific miRNA can regulate the expression of up to thousand mRNAs, and a specific mRNA can be regulated by multiple miRNAs.13 A concept as miRNA-mediated gene regulatory network has been proposed based on this theory, implying that the interference of miRNAs in physiological or pathological processes is quite complicated. Due to the limitations of existing experimental approaches when clarifying complex biological systems composed of perturbing miRNA and target expressions and multiple layers of regulation, it is very difficult to investigate miRNA regulatory mechanisms and functions.14 To address this problem, a systematic approach, which can unravel the underlying mechanisms by which miRNAs exert their functions, becomes increasingly appealing. Since this approach can provide a systematic and comprehensive perspective on roles of miRNAs in gene regulatory networks, we in the current study performed a systematic investigation combining biological data acquisition and integration, miRNA-target prediction, network construction and analysis, functional assay and clinical validation, to discover novel HCC-related miRNA-target axes and to elucidate their functions.
Results & Discussion
As shown in Figure 1, the technical strategy of this study included four steps: first, five miRNA expression profiles were collected according to our literature retrieval and a list of the differential expressed miRNAs in HCC were obtained from data integration; Second, high confident target genes of HCC differentially expressed miRNAs were collected from miRTarBase and their relationships with HCC progression were analyzed based on GO annotation system and KEGG pathway database; Third, miRNA-mediated gene regulatory network was constructed and crucial miRNA-target interactions were screened according to their network topological importance; And finally, the function and clinical significance of crucial miRNA-target interactions were experimentally validated. Our results were described as follows.
Figure 1.
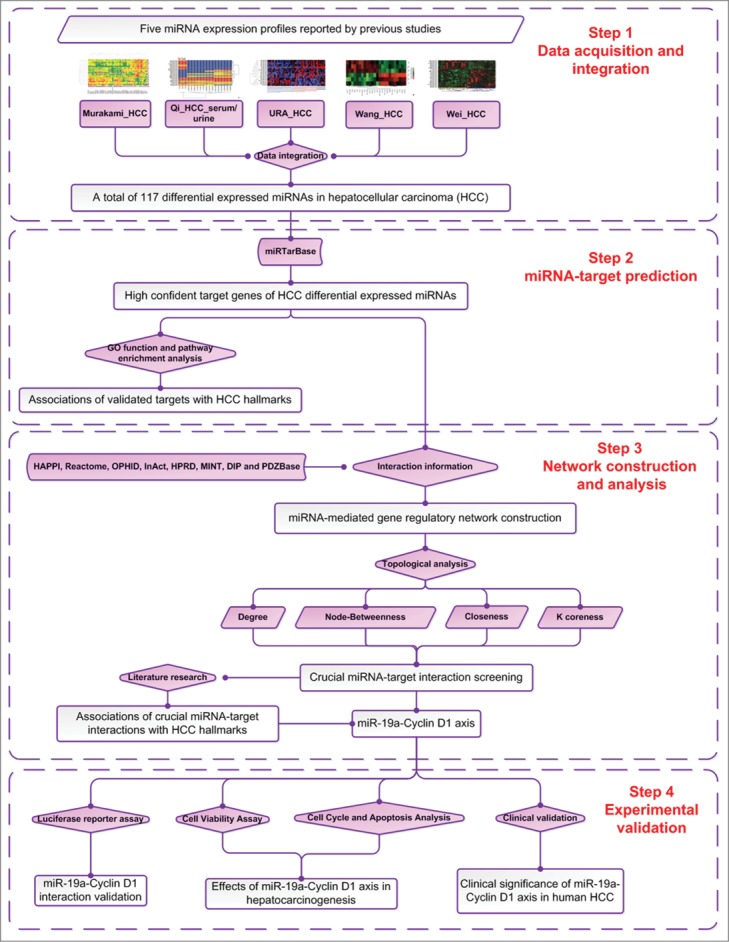
A schematic diagram of this systematic investigation to discover novel hepatocellular carcinoma (HCC)-related miRNA-target axes and to elucidate their functions.
Differentially expressed miRNAs in HCC progression control broad biological functions
A total of 117 HCC differentially expressed miRNAs, including 64 upregulated and 53 downregulated miRNAs, were obtained from five miRNA expression profiles reported by previous studies.15-19 To elucidate the functions of these miRNAs, 728 high confident target genes for them were collected from miRTarBase. Please see detail information on these validated targets in Supplementary Table 1.
The dysregulation directions of these differentially expressed miRNAs during HCC progression imply their promotive or suppressive progression potentials. However, their functions in HCC are still largely unknown. Fortunately, enrichment analysis of the validated target genes of these HCC differentially expressed miRNAs could give us a global clue of their functional roles in HCC progression. Here, the enrichment analysis based on Gene Ontology (GO) annotation system, which uses a controlled and hierarchical vocabulary to assign function to genes or gene products in any organism, was performed. As shown in Figure 2A, the validated target significantly controlled many biological processes directly relevant to cancers, such as cell differentiation, cell proliferation, apoptosis, cell cycle, cell death and cell migration. Notably, 573 in 728 (78.71%) validated target genes are significantly involved in the regulation of cell differentiation which has been demonstrated to be significantly correlated with the invasive proclivity and the tumor recurrence of HCC patients.20 These results implied several potential regulatory roles of these HCC differentially expressed miRNAs with no previously reported involvement in hepatocarcinogenesis.
Figure 2.
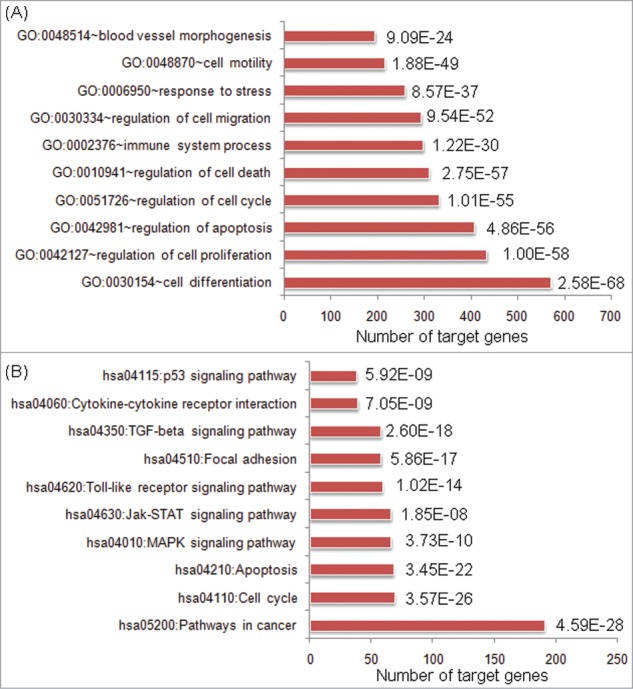
Top ten enriched gene ontology (GO) biological processes (A) and KEGG pathways (B) involved by validated target genes of hepatocellular carcinoma (HCC) differentially expressed microRNAs (miRNAs). P values were marked beside the bars.
On the other hand, among the pathways enriched by validated target genes for HCC differentially expressed miRNAs, we concentrated on cell cycle, apoptosis, MAPK signaling pathway, Jak-STAT signaling pathway, Toll-like receptor signaling pathway, focal adhesion, TGF-beta signaling pathway and p53 signaling pathway (Fig. 2B), which all have broad effects on cell behavior. Among them, cell cycle, apoptosis, MAPK signaling pathway and p53 signaling pathway are involved in multiple steps of cell viability, and evidences had shown that dysregulations of the cell viability components may lead to tumor formation. These results suggested that the differentially expressed miRNAs in HCC progression may control broad biological functions associated with cancers.
Network analysis
The interaction information of validated target genes of HCC differentially expressed miRNAs were used to construct candidate miRNAs-mediated gene regulatory network, which consists of 4141 interaction pairs. Four topological features, ‘Degree,’ ‘Node betweenness’ and ‘Closeness’ and ‘K coreness’ (defined in ‘Materials and methods’ section) were chosen to identify topological crucial targets of HCC differentially expressed miRNAs. Among these topological features, ‘Degree’, ‘Node betweenness’ and ‘Closeness’ centralities can measure a node's topological importance in the network. The larger a node's degree/betweenness/closeness centrality is, the more important the node is in the network.21 After calculating the value of the four features for each node, the median values of ‘Degree,’ ‘Node betweenness’ and ‘Closeness’ and ‘K coreness’ were 8, 20.51, 0.04 and 6, respectively. Therefore, we determined that nodes with ‘Degree’>8, ‘Node betweenness’>20.51, ‘Closeness’>0.04 and ‘K coreness’>6 were topological important nodes. As a result, 221 crucial targets of HCC differentially expressed miRNAs with topological importance were identified. Please see topological features of these crucial targets in Supplementary Table S2.
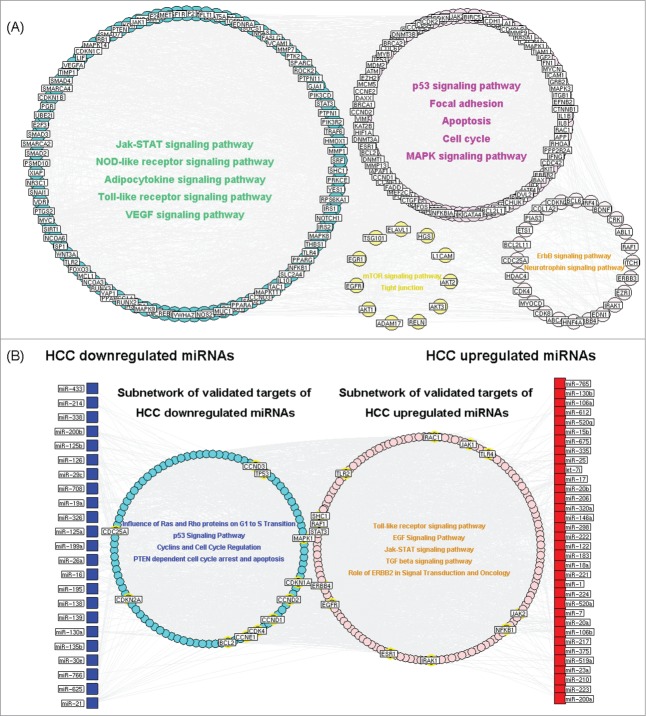
Modularity has been reported to be another important aspect of an interaction network.22 Nodes that are highly interconnected within the network are usually involved in the same biological modules or pathways. Using Markov clustering algorithm, we divided the interaction network of crucial targets of HCC differentially expressed miRNAs into four functional modules containing 97, 89, 24, and 11 nodes, respectively (Fig. 3A). The functional enrichment analysis based on GO annotation system and KEGG pathway of crucial targets of HCC differentially expressed miRNAs in four modules highlighted their involvements in hepatocarcinogenesis. The biggest functional module was significantly associated with Jak-STAT signaling pathway, Toll-like receptor signaling pathway and VEGF signaling pathway. The other modules were respectively involved in p53 signaling pathway, cell cycle, apoptosis, focal adhesion, mTOR signaling pathway and ErbB signaling pathway.
Figure 3.
(A) Interaction network of crucial target genes of hepatocellular carcinoma (HCC) differentially expressed miRNAs. Using Markov clustering algorithm, the network was divided into four functional modules. (B) Interaction network of HCC differentially expressed miRNAs and the corresponding crucial target genes. Pathways which were significantly enriched by crucial target genes of HCC downregulated and upregulated miRNAs in the two subnetworks were identified.
Moreover, the interaction network of HCC differentially expressed miRNAs and the corresponding crucial target genes was constructed as shown in Figure 3B. We also identified pathways which were significantly enriched by crucial target genes of HCC downregulated and upregulated miRNAs in the two subnetworks. As a result, the crucial target genes of HCC downregulated miRNAs specially participated in the influence of Ras and Rho proteins on G1 to S Transition, p53 signaling pathway, cyclins and cell cycle regulation, cell cycle arrest and apoptosis, implying these HCC downregulated miRNAs might have suppressive progression potential by significantly control multiple steps of cell cycle. On the other hand, five cancer related pathways, including Toll-like receptor signaling pathway, EGF Signaling Pathway, Jak-STAT signaling pathway, TGF beta signaling pathway, Role of ERBB2 in Signal Transduction and Oncology were significantly enriched by crucial target genes of HCC upregulated miRNAs, suggesting their associations with hepatocarcinogenesis.
Experimental validation
Among 221 crucial target genes of HCC differentially expressed miRNAs, Cyclin D1 has the highest node betweenness (Table S2), suggesting it may function as a bottleneck in the miRNA-mediated gene regulatory network. From the view of network topology, growing evidence shows that ‘node betweenness’ (‘bottleneck-ness’) is a much more significant indicator of essentiality than ‘degree’ (‘hub-ness’) in networks, because bottlenecks often control most of the information flow in the network.23 Since ‘node betweenness’ may be a good predictor of essentiality, we here chose Cyclin D1 for detailed analysis.
As shown in Table S1, eight HCC differentially expressed miRNAs, including miR-106b, miR-15b, miR-16, miR-17, miR-195, miR-19a, miR-20a and miR-338, were experimentally validated to target Cyclin D1. According to our literature retrievals, Shen and colleagues reported that miR-106b expression was markedly upregulated in hepatoma cells and hepatoma tissues compared with immortalized normal liver epithelial cells and normal hepatic tissues, and its overexpression could modulate entry into the G(1)/S transitional phase by upregulating Cyclin D124; Chung and colleagues indicated that miR-15b expression in HCC tissues might predict a low risk of HCC recurrence and the modulation of miR-15b expression might be useful as an apoptosis-sensitizing strategy for HCC treatment25; The regulatory role of miR-15b on Cyclin D1 was validated by Sun and colleagues using glioma cells26; Accumulating studies have demonstrated that miR-16 may be related with the biological behavior of HCC and it may be considered as a potential indicator to estimate the tumor size or the recurrence of HCC27; The regulatory role of miR-16 on Cyclin D1 was also validated in various cancer cells, such as bladder cancer, non-small cell lung cancer, colorectal cancer, osteosarcoma, mantle cell lymphoma and prostate cancer28-31; Recent studies have reported the prognostic value of miR-17 in human HCC and miR-17 has been observed to inhibit breast cancer cellular proliferation through G1/S cell cycle arrest via binding to the Cyclin D132; Yu and colleague also found a Cyclin D1/miRNA-17/20 regulatory feedback loop in control of breast cancer cell proliferation33; Xu and colleagues implied that miR-195 may block the G(1)/S transition by repressing Rb-E2F signaling through targeting multiple molecules, including Cyclin D1, CDK6, and E2F334; Qin and colleagues reported that miR-19a could mediate the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells35; miR-20a has been found to be decreased in HCC and may correlate with HCC recurrence and prognosis36; The aberrant expression of miR-338 has been indicated to be associated with clinical aggressiveness, such as, tumor size, tumor-node-metastasis stage, vascular invasion and intrahepatic metastasis.37 These findings reveal the clinical significance of miR-106b, miR-15b, miR-16, miR-17, miR-195, miR-20a and miR-338 in human HCC and also validated their regulatory roles on Cyclin D1 in different cancer cells. However, the involvement of miR-19a-Cyclin D1 axis in HCC is still unclear. Therefore, we would like to perform further systematic experimental validation to address this problem.
Reverse correlation between miR-19a and cyclin D1 expression in human HCC tissues and cells
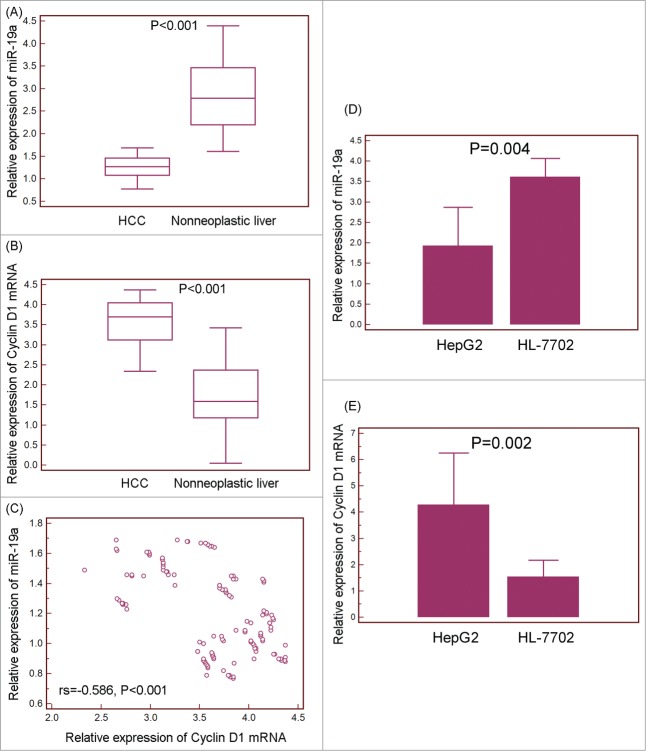
MiR-19a, together with miR-17, miR-18a, miR-20a, miR-19b-1 and miR-92-1, belongs to the miR-17-92 cluster, which is a highly conserved gene cluster located on chromosome 13q31.3.38 Growing evidence shows that the human genomic region encoding the miR-17-92 cluster often plays a crucial role in tumorigenesis. Similar with other members in this cluster, miR-19a has been reported to be dysregulated in various human cancers, such as laryngeal squamous cell carcinoma, breast cancer, non-small cell lung cancer, gastric cancer, colorectal cancer, cervical carcinoma, and bladder cancer.39-42 Especially, Han et al.43 observed the downregulation of miR-19a in primary HCC samples of patients who had developed HCC recurrence compared to those with non-recurrence, implying that this miRNA might function as a tumor suppressor in HCC. Cyclin D1, located within chromosome 11q13, belongs to the G1 cyclin family and plays a role in regulating the transition through the G1 phase of the cell cycle.44 Accumulating studies have reported that Cyclin D1 functions as an oncogene and is overexpressed in a variety of human malignancies, including B-cell lymphomas, parathyroid adenoma, esophageal carcinoma, breast carcinoma, non-small-cell lung carcinoma, HCC, gastric cancer, pancreatic cancer and colorectal cancer.45-53 In this experimental validation, we, at first, performed qRT-PCR to evaluate the relationship between miR-19a and Cyclin D1 mRNA expression in HCC tissues and cells in vitro. As shown in Figure 4A and B, the expression levels of miR-19a in HCC tissues were significantly lower than those in adjacent nonneoplastic liver tissues (HCC vs. Normal: 1.28 ± 0.26 vs. 2.81 ± 0.72, P<0.001, Fig. 4A), while Cyclin D1 mRNA expression was dramatically increased in HCC tissues compared to adjacent nonneoplastic liver tissues (HCC vs. Normal: 3.55 ± 0.56 vs. 1.78 ± 0.86, P<0.001, Fig. 4B). More interestingly, the Spearman Correlation analysis clearly showed negative correlation between miR-19a and Cyclin D1 mRNA expression in HCC tissues (rs = −0.586, P<0.001, Fig. 4C). These findings were consistent with those based on HCC cells in vitro system (Fig. 4D,E).
Figure 4.
Inverse correlation between microRNA (miRNA)-19a and Cyclin D1 expression in human hepatocellular carcinoma (HCC) tissues and cells. (A) Expression levels of miR-19a in 130 pairs of HCC and adjacent nonneoplastic liver tissues were detected by qRT-PCR and normalized to RNU6B. Statistical analysis showed that the expression level of miR-19a in HCC tissues was significantly lower than that in adjacent nonneoplastic liver tissues (HCC vs. Normal: 1.28 ± 0.26 vs. 2.81 ± 0.72, P<0.001). (B) Expression levels of Cyclin D1 in 130 pairs of HCC and adjacent nonneoplastic liver tissues were detected by qRT-PCR and normalized to GAPDH. Statistical analysis showed that Cyclin D1 mRNA expression was dramatically increased in HCC tissues compared to adjacent nonneoplastic liver tissues (HCC vs. Normal: 3.55 ± 0.56 vs. 1.78 ± 0.86, P<0.001). (C) Spearman Correlation analysis clearly showed inverse correlation between miR-19a and Cyclin D1 mRNA expression in HCC tissues. (D) Expression levels of miR-19a in HCC cell line HepG2 and normal human liver cell line HL-7702 were detected by qRT-PCR and normalized to RNU6B; (E) Expression levels of Cyclin D1 in HCC cell line HepG2 and normal human liver cell line HL-7702 were detected by qRT-PCR and normalized to GAPDH. All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
Dysregulation of miR-19a/Cyclin D1 axis associates the advanced tumor progression of human HCC
To evaluate whether miR-19a and/or Cyclin D1 expression was associated with clinicopathological features of patients with HCC, we analyzed the association of miR-19a and/or Cyclin D1 expression with T stage, tumor grade, presence of cirrhosis, underlying liver disease including alcohol abuse, viral hepatitis B and C, sex, and age (Table 1). We chose the median expression values of miR-19a and Cyclin D1 expression as the cutoff points. HCC tissues with an expression value exceeding the cutoff points for miR-19a or Cyclin D1 were deemed to be low expressions of miR-19a or Cyclin D1; all other scores were considered to be high expressions of miR-19a or Cyclin D1. Of 130 HCC patients, 14 (10.77%) were both high expression of miR-19a and Cyclin D1, 16 (12.31%) were both low expression of miR-19a and Cyclin D1, 48 (36.92%) were miR-19a-high and Cyclin D1-low expression, and 52 (40.00%) were miR-19a-low and Cyclin D1-high expression. Statistical analysis showed that the low miR-19a expression (P = 0.01, Table 1), high Cyclin D1 expression (P = 0.01, Table 1) and miR-19a-low/Cyclin D1-high expression (P = 0.006, Table 1) were all more frequently found in HCC tissues with high tumor stage (T3˜4) than those with low tumor stage (T1˜2), suggesting that the dysregulation of miR-19a/Cyclin D1 axis may be implicated into the aggressive progression of HCC.
Table 1.
Association of microRNA (miR)-19a and/or Cyclin D1 expression with clinicopathologic features of 130 hepatocellular carcinoma patients
| Clinicopathologic Features | Case | miR-19a-low (n, %) | P | Cyclin D1-high (n, %) | P | miR-19a-low/ Cyclin D1-high (n, %) | P |
|---|---|---|---|---|---|---|---|
| Age (years) | |||||||
| ≤50 | 72 | 38 (52.78) | NS | 36 (50.00) | NS | 30 (41.67) | NS |
| >50 | 58 | 30 (51.72) | 30 (51.72) | 22 (37.93) | |||
| Gender | |||||||
| Male | 96 | 50 (52.08) | NS | 48 (50.00) | NS | 40 (41.67) | NS |
| Female | 34 | 18 (52.94) | 18 (52.94) | 12 (35.29) | |||
| Serum AFP | |||||||
| Positive | 72 | 40 (55.56) | NS | 38 (52.78) | NS | 32 (44.44) | NS |
| Negative | 58 | 28 (48.28) | 28 (48.28) | 20 (34.48) | |||
| Tumor stage | |||||||
| T1 | 23 | 4 (17.39) | 0.01 | 4 (17.39) | 0.01 | 1 (4.35) | 0.006 |
| T2 | 40 | 15 (37.50) | 15 (37.50) | 10 (25.00) | |||
| T3 | 52 | 34 (65.38) | 32 (61.58) | 26 (50.00) | |||
| T4 | 15 | 15 (100.00) | 15 (100.00) | 15 (100.00) | |||
| Tumor grade | |||||||
| G1 | 31 | 14 (45.16) | NS | 14 (45.16) | NS | 10 (32.26) | NS |
| G2 | 76 | 39 (51.32) | 37 (48.68) | 30 (39.47) | |||
| G3 | 23 | 15 (65.22) | 15 (65.22) | 12 (52.17) | |||
| Growth pattern | |||||||
| Trabecular | 101 | 53 (52.48) | NS | 51 (50.50) | NS | 40 (39.60) | NS |
| Nontrabecular | 29 | 15 (51.72) | 15 (51.72) | 12 (41.38) | |||
| Cirrhosis | |||||||
| Yes | 86 | 45 (52.33) | NS | 43 (50.00) | NS | 36 (41.86) | NS |
| No | 44 | 23 (52.27) | 23 (52.27) | 16 (36.36) | |||
| Underlying liver disease | |||||||
| Alcoholic | 25 | 13 (52.00) | NS | 13 (52.00) | NS | 10 (40.00) | NS |
| Hepatitis B | 49 | 28 (57.14) | 26 (53.06) | 20 (40.82) | |||
| Hepatitis C | 35 | 17 (48.57) | 17 (48.57) | 14 (40.00) | |||
| Unknown | 21 | 10 (47.62) | 10 (47.62) | 8 (38.10) |
Note: ‘NS’ refers to the differences among groups have no statistical significance.
Dysregulation of miR-19a/cyclin D1 axis predicts poor prognosis of human HCC
Prognosis evaluation is important for making appropriate treatment choices. To determine the prognostic value of miR-19a and/or Cyclin D1 expression in patients with HCC, the Kaplan–Meier method was employed to analyze the correlation of miR-19a and/or Cyclin D1 expression with 5-year disease-free survival and 5-year overall survival of HCC patients. As shown in Figure 5A and B, we observed a trend that 5-year disease-free survival and overall survival of HCC patients with low miR-19a expression were both significantly shorter than those with high miR-19a expression (both P = 0.001, log-rank test). In contrast, the Kaplan-Meier plot of 5-year disease-free survival and overall survival curves stratified by Cyclin D1 expression also showed a significantly negative relationship between Cyclin D1 expression and patients' survival (both P = 0.001, log-rank test, Fig. 5C and D). More interestingly, the HCC patients with miR-19a-low/Cyclin D1-high expression had the shortest 5-year disease-free survival and overall survival compared to other two groups miR-19a-low (high) /Cyclin D1-low (high) and miR-19a-high/Cyclin D1-low, both P<0.001, Figure 5E and F.
Figure 5.
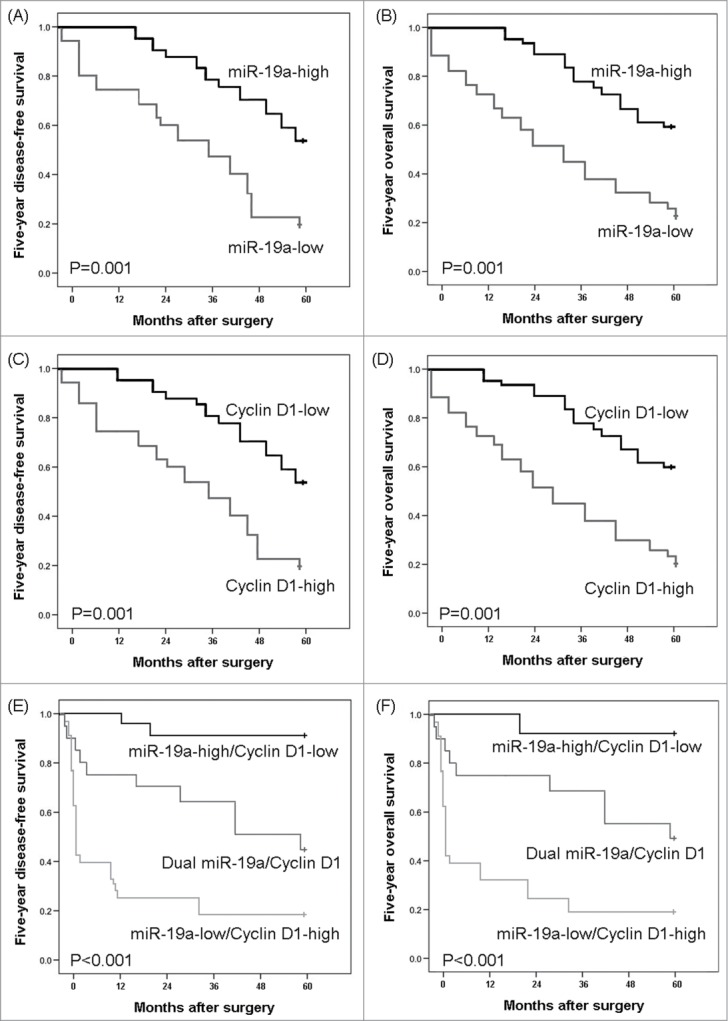
Disease-free survival and overall survival curves for two groups defined by microRNA (miRNA)-19a expression (A and B, respectively), Cyclin D1 expression (C and D, respectively) and miR-19a/Cyclin D1 expression (E and F, respectively) in patients with hepatocellular carcinoma (HCC).
Furthermore, the multivariate analysis found that miR-19a expression, Cyclin D1 expression and combined miR-19a/Cyclin D1 expression were all independent poor prognostic factors for both 5-year disease-free survival (P = 0.01, 0.01 and 0.003, respectively, Table 2) and 5-year overall survival (P = 0.01, 0.01 and 0.002, respectively, Table 2) in HCC. Notably, the prognostic value of combined miR-19a/Cyclin D1 expression was more significant than that of miR-19a and Cyclin D1 expression alone. These clinical evidence highlighted the prognostic potential of miR-19a/Cyclin D1 axis in HCC.
Table 2.
Multivariate survival analysis of five-year overall and disease-free survival in 130 patients with hepatocellular carcinoma
| Five-year overall survival | Five-year disease-free survival | |||||
|---|---|---|---|---|---|---|
| Features |
HR |
95% CI |
P |
HR |
95% CI |
P |
| Serum AFP | 1.931 | 0.685-4.056 | 0.063 | 1.953 | 0.615-4.273 | 0.062 |
| Tumor stage | 2.879 | 1.366-5.196 | 0.009 | 2.686 | 1.386-6.009 | 0.01 |
| Tumor grade | 1.563 | 0.609-4.088 | 0.081 | 1.551 | 0.607-4.466 | 0.086 |
| Presence of cirrhosis | 1.919 | 0.738-4.102 | 0.063 | 1.921 | 0.793-4.219 | 0.062 |
| miR-19a expression | 2.768 | 1.321-6.032 | 0.01 | 2.608 | 1.331-5.863 | 0.01 |
| Cyclin D1 expression | 2.629 | 1.309-5.946 | 0.01 | 2.572 | 1.300-5.626 | 0.01 |
| miR-19a/Cyclin D1 expression | 4.182 | 1.618-8.382 | 0.002 | 4.067 | 1.581-8.139 | 0.003 |
MiRNA-19a directly targets cyclin D1 in HCC cells
Our data mentioned above suggested that the dysregulation of miR-19a/Cyclin D1 axis may contribute to HCC progression and patients' prognosis. Next, we would like to investigate the exact mechanisms by which miR-19a/Cyclin D1 axis acts on hepatocarcinogenesis.
To verify the regulatory effect of miR-19a on Cyclin D1 in HCC cells, we respectively transfected HepG2 cells with miR-19a mimics, miR-19a mimic control (negative control, NC), miR-19a inhibitor, anti-miRNA control and blank control culture medium (mock). After 24 h post-transfection, the expression level of Cyclin D1 protein in HepG2 cells overexpressed miR-19a was significantly lower than those in NC and mock groups (both P<0.05, Fig. 6A˜C), while Cyclin D1 protein expression was dramatically increased in HepG2 cells with the suppression of miR-19a (P = 0.01, Fig. 6B and C).
Figure 6.
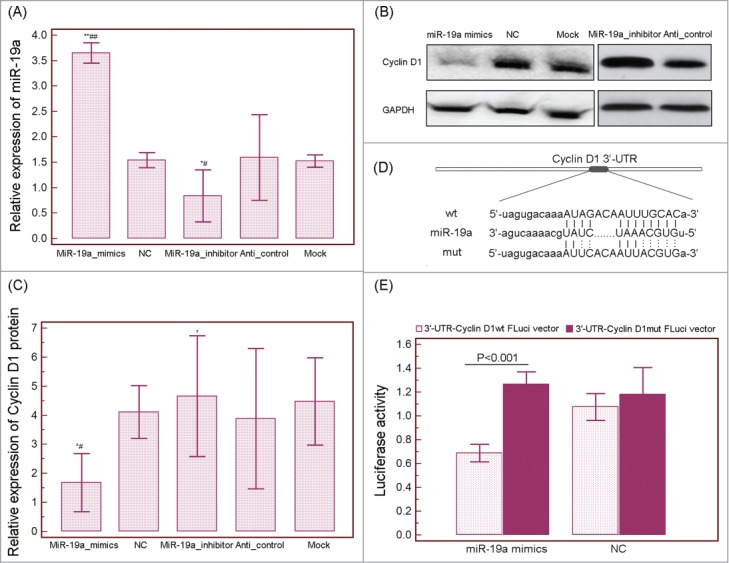
MicroRNA (miRNA)-19a directly targets Cyclin D1 in HCC cells. (A) QRT-PCR analysis showing relative expression of miR-19a in HepG2 cells transfected with miR-19a mimics, miR-19a mimic control (negative control, NC), miR-19a inhibitor, anti-miRNA control (Anti-control) and blank control culture medium (mock). (B and C) Relative expression of Cyclin D1 protein in HepG2 cells transfected with miR-19a mimics, miR-19a mimic control (negative control, NC), miR-19a inhibitor, anti-miRNA control (Anti-control) and blank control culture medium (mock) detected by Western blot analysis. GAPDH was used as an internal loading control. ‘*’ and ‘**’: P<0.05 and P<0.001, respectively, compared with the data of miR-19a mimic control group or anti-miRNA control; ‘#’ and ‘##’: P<0.05 and P<0.001, respectively, compared with the data of mock group. (D) RNA sequence alignment showing the 3'-UTR of Cyclin D1 mRNA contains a complementary site for the seed region of miR-19a. Cyclin D1mut is amutant with substitutions in the complementary region as a negative control. (E) Luciferase report assay was performed to verify whether Cyclin D1 was a direct target of miR-19a. The luciferase activity was detected after transfection of FLuci vector (3'-UTR-Cyclin D1wt FLuci vector or 3'-UTR-Cyclin D1mut FLuci vector) into the miR-19a mimic or miR-19a mimic control (negative control, NC) transfected HepG2 cells.
To verify whether Cyclin D1 was a direct target of miR-19a, the luciferase reporter containing the complimentary seed sequence of miR-19a at the 3′-UTR region of Cyclin D1 mRNA was constructed (Fig. 6D). Luciferase activity was detected at 48 h after the co-transfection of FLuci vector (3′-UTR-Cyclin D1wt FLuci vector or 3′-UTR-Cyclin D1mut FLuci vector), miR-19a mimic or NC mimic, and RLuci vector in HepG2 cells. As shown in Figure 6E, the luciferase activity was significantly decreased in HepG2 cells co-transfected with 3′-UTR-Cyclin D1wt FLuci vector and miR-19a mimic compared with those co-transfected with 3′-UTR-Cyclin D1mut FLuci vector and miR-19a mimic (P<0.001, Fig. 6E), suggesting that the fragment at the 3′-UTR of the Cyclin D1 mRNA was the complementary site for the miRNA-19a seed region.
These findings suggested that Cyclin D1 be a direct target of miR-19a.
MiR-19a inhibits HCC cell proliferation, cell cycle progression and promotes cell apoptosis by targeting cyclin D1
The associations of miR-19a downregulation in HCC tissues with advanced tumor stage and poor patients' prognosis prompted us to verify whether it acted as a tumor suppressor in this malignancy. Since it is of critical to understand the regulation of cell proliferation for developing novel and efficient therapeutic strategies for the treatment of HCC, in vitro cell proliferation assay was performed in this study to investigate the effect of miR-19a on proliferation of HepG2 cells. As shown in Figure 7A, the enforced expression of miR-19a significantly inhibited cell proliferation of HepG2 cells (P = 0.01). Subsequently, we observed that miR-19a overexpression could lead to substantial accumulation of the cell population at the G1 stage of the cell cycle (Fig. 7B) and also promoted apoptosis in HepG2 cells (Fig. 7C).
Figure 7.
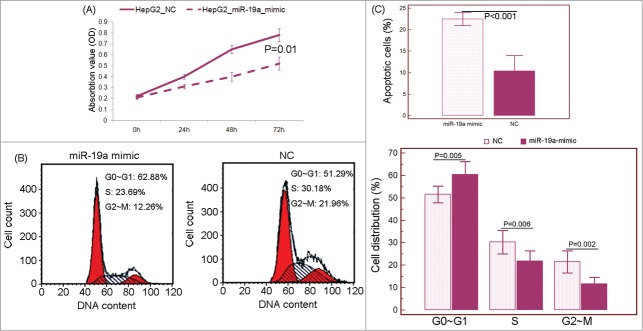
MicroRNA (miRNA)-19a inhibits hepatocellular carcinoma (HCC) cell proliferation, cell cycle progression and promotes cell apoptosis in vitro. (A) MTT assay showed that miR-19a overexpression could inhibit cell proliferation of HepG2 cells. (B) miR-19a overexpression led to substantial accumulation of the cell population at the G1 stage of the cell cycle. (C) miR-19a overexpression promoted apoptosis in HepG2 cells. ‘NC’: miR-19a mimic control (negative control).
To ascertain the roles of Cyclin D1 in miR-19a regulated cell proliferation, cell cycle progression and apoptosis, we further investigated whether knockdown of the endogenous Cyclin D1 was able to mimic the effect of miR-19a restoration. We inhibited the expression of Cyclin D1 via siRNA (Fig. 8A) and confirmed that Cyclin D1 knockdown inhibited cell proliferation (Fig. 8B), cell cycle progression in HepG2 cells, possibly be G1-phase cell cycle arrest (Fig. 8C), and also promoted cell apoptosis (Fig. 8D).
Figure 8.
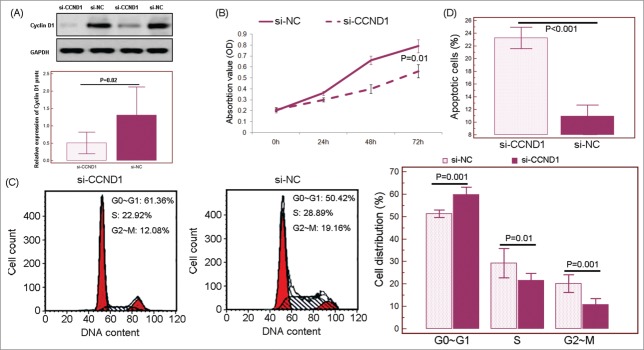
Inhibition of Cyclin D1 was responsible for the tumor suppressive effects of microRNA (miRNA)-19a in hepatocellular carcinoma (HCC) cells in vitro. (B) MTT assay showed that inhibition of Cyclin D1 could inhibit cell proliferation of HepG2 cells. (C) Inhibition of Cyclin D1 led to substantial accumulation of the cell population at the G1 stage of the cell cycle. (D) inhibition of Cyclin D1 promoted apoptosis in HepG2 cells. ‘si-CCND1’: the small interfering RNA targeting human Cyclin D1 transcript; ‘si-NC’: siRNA control.
These data suggest that the tumor suppressive roles of miR-19a may be mediated by inhibiting its target gene Cyclin D1.
Conclusion
This systematic investigation provides a framework to identify featured miRNAs and their target genes which are potent effectors in the occurrence and development of HCC. Our data imply that miR-19a may play important inhibitory roles in HCC malignancy via regulating Cyclin D1. More importantly, miR-19a/Cyclin D1 axis might have promising applications as a therapeutic target and a prognostic marker for patients with HCC.
Materials and Methods
miRNA expression profiles
Five publicly available datasets of miRNA expression profiles were chosen in this study, including Wei_HCC (110 non-cancerous liver samples vs. 110 HCC samples),15 Murakami_HCC (25 non-cancerous liver samples vs. 25 HCC samples),16 URA_HCC (9 non-cancerous liver samples vs. 26 HCC samples),17 Wang_HCC (18 non-cancerous liver samples vs. 18 HCC samples)18 and Qi_HCC_serum/urine (Healthy controls vs. HCC_serum/urine).19 The detailed information about these datasets was provided in Supplementary Table 3. Normalized miRNA data were downloaded directly. After deleting redundant data, a total of 117 HCC differentially expressed miRNAs, including 64 upregulated and 53 downregulated miRNAs, were identified for further analysis. The detailed information of the HCC differentially expressed miRNA list was shown in Supplementary Table 4.
miRNA target prediction
Validated targets for above HCC differentially expressed miRNAs were collected from miRTarBase (Release 4.5: Nov. 1, 2013; http://mirtarbase.mbc.nctu.edu.tw/), which has accumulated more than fifty thousand miRNA-target interactions (MTIs) which are all validated experimentally by reporter assay, western blot, qPCR, microarray and next-generation sequencing experiments.54 Here, we only collected the MTIs which are validated experimentally by reporter assay, western blot and qPCR. In order to facilitate data analysis, the different ID types for validated target genes were converted to gene symbol from NCBI-Gene.
Gene ontology (GO) and pathway enrichment analysis for validated genes of HCC differentially expressed miRNAs
Database for Annotation, Visualization and Integrated Discovery55 (DAVID, http://david.abcc.ncifcrf.gov/home.jsp,version 6.7) was used for GO enrichment analysis. Then, the pathway enrichment analysis based on data from the FTP service of KEGG56 (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/, Last updated: Oct 16, 2012) was also performed.
Network analysis
Interaction data collection
Interaction data used in the network construction were imported from eight existing databases including Human Annotated and Predicted Protein Interaction Database (HAPPI),57 Reactome,58 Online Predicted Human Interaction Database (OPHID),59 InAct,60 Human Protein Reference Database (HPRD),61 Molecular interaction Database (MINT),62 Database of Interacting Proteins (DIP),63 and PDZBase.64 The detailed information on these databases is described in Supplementary Table 5. In order to facilitate data analysis, the different ID types for proteins or genes were all converted to gene symbol from NCBI-Gene.
Network construction
Interactions between validated target genes of HCC differentially expressed miRNAs were used to construct miRNA-mediated gene regulatory network. The interaction data were obtained from eight existing databases as mentioned above. Then, we applied Navigator software (Version 2.2.1) and Cytoscape (Version 2.8.1) to visualize the networks.
Defining network topological feature set
We defined four measures including ‘Degree’, ‘Node betweenness’, ‘Closeness’ and ‘K coreness’ for evaluating the topological property of each node i in the interaction network. Among them, ‘Degree’ is defined as the number of links to node i; ‘Node betweenness’ is defined as the number of shortest paths between pairs of nodes that run through node i; ‘Closeness’ is defined as the inverse of the farness which is the sum of node i distances to all other nodes; ‘K coreness’ is used to measure the centrality of node i.65
Then, we used Markov clustering algorithm to divide all nodes in miRNA-mediated gene regulatory network into different functional modules. Functional modularity analysis can identify nodes which are highly interconnected within the network and usually involved in the same biological modules or pathways.
Experimental validation
Patients and tissue samples
This retrospective study was approved by the Research Ethics Committee of 302nd Hospital of PLA, Beijing, China. Written informed consent was obtained from all patients. All tissues specimens were handled and made anonymous according to the ethical and legal standards.
We collected 130 self-pairs of HCC specimens and adjacent nonneoplastic liver tissues from 130 patients with primary HCC who underwent a curative liver resection at the 302nd Hospital of PLA, Beijing, China. All the tissue specimens were snap-frozen in liquid nitrogen and stored at −80°C following surgery for qRT-PCR assay. These patients were diagnosed as HCC between 2001 and 2008, and none had chemotherapy or radiotherapy before the surgery. World Health Organization (WHO) criteria was used for the patients' diagnosis. Based on the Edmondson grading system and the sixth edition of the tumor-node-metastasis (TNM) classification of the International Union against Cancer, tumor differentiation and tumor stage of each patient were respectively determined. Table 1 summarized the clinicopathological features of all patients with HCC enrolled in the current study.
All 130 patients with HCC were performed follow-up (median follow-up period: 8.6 years). Postoperative surveillance included routine clinical and laboratory examinations every third month, computed tomography scans of the abdomen, and radiographs of the chest every third month. After 5 years, the examination interval was extended to 12 months.
Cell culture
Human HCC cell line HepG2 was obtained from the American Type Culture Collection (Manassas, VA, USA) and was cultured in DMEM (Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibico, USA), 2 mM L-glutamine and antibiotics. Normal human liver cell line HL-7702 was obtained from the American Type Culture Collection (Manassas, VA, USA) and was maintained in RPMI 1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibico, USA). Two cell lines were both maintained at 37°C in a humidified chamber supplemented with 5% CO2.
Construction of miR-19a expression vectors and cellular transfection
Commercial miR-19a mimics, miR-19a mimic control, miR-19a inhibitor, and nonspecific anti-miRNA control expression vectors were purchased from Ambion (Austin, TX, USA). HepG2 cells were transfected with miR-19a mimics, miR-19a mimic control, miR-19a inhibitor, and nonspecific anti-miRNA control expression vectors with Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol.
Quantitative reverse transcription-PCR (qRT-PCR)
The qRT-PCR analysis for miRNA and mRNA was performed according to the similar protocol of our previous studies.65-66 U6 small RNA and GAPDH were respectively used as internal controls for normalization and quantification miR-19a and Cyclin D1 expression. The primer sequences were listed in Supplementary Table 6. Relative quantification of miRNA and mRNA expression was evaluated using the comparative cycle threshold (CT) method. All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
RNA interference and cellular transfection
To knockdown the expression of the human Cyclin D1 gene, the small interfering RNA (siRNA) targeting human Cyclin D1 transcript (si-CCND1) and siRNA control (si-NC) were constructed (Shanghai GeneChem Co, Ltd., Shanghai, China). Sequence of these siRNAs were: Cyclin D1 siRNA, sense, 5′-CAAGCU CAA GUG GAA CCU GTT-3′, antisense, 5′-CAG GUU CCA CUU GAG CUU GTT-3′; siRNA control, sense, 5′-UUC UCC GAA CGU GUC ACG UTT-3′, antisense, 5′-ACG UGA CAC GUU CGG AGA ATT-3′. HepG2 cells were seeded into 6-well plates and incubated overnight, and then transfected using LipofectamineTM 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. The expression levels of Cyclin D1 in HepG2 cells transfected with si-CCND1 and si-NC were detected by Western blot analysis as described in the next section.
Western blot
The Western blot protocol and semiquantitative analysis were carried out following the protocol of our previous studies.65-66 The specific antibodies were as following: Cyclin D1 antibody (#sc-753, rabbit polyclonal antibody, dilution 1:100, Santa Cruz Biotechnology, Inc. USA) and GAPDH antibody (CW0266, dilution 1:1000 CoWin Biotech). All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
Luciferase reporter assay
The regulatory effect of miR-19a to Cyclin D1 was evaluated by a luciferase reporter assay in HepG2 cells following the protocol of our previous studies.65-66 Briefly, the human Cyclin D1 3′-UTR luciferase reporter construct was generated by cloning Cyclin D1 3′-UTR sequence containing the predicted miR-19a binding site into the 3′-UTR region of the pGL3 luciferase reporter vector (Promega Corporation, Madison WI, USA). The miR-19a binding site-deleted Cyclin D1 3′-UTR luciferase reporter construct was generated by PCR fragments of Cyclin D1 3′-UTR luciferase reporter construct lacking the target site and ligated. HepG2 cells were cultivated in 24-well plates and co-transfected using Fugene (Roche) with 100 ng of pGL3-Cyclin D1-miR-19a constructs, 10 ng miR-19a mimic or NC mimic, and 2 ng pRL-SV40 RLuci vector (Promega). The luciferase activity assay was performed 24 h after transfection using the dual-luciferase reporter assay system (Promega Corporation, Madison WI) according to the manufacturer's instructions. All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
In vitro cell proliferation assay
The in vitro cell proliferation of HepG2 cells transfected with miR-19a mimic, miR-19a mimic control, si-CCND1 and si-NC vectors, respectively, were measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method following the protocol of our previous studies 33-34. All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
In vitro cell cycle and apoptosis analysis
At 48 h post-transfection of miR-19a mimic, miR-19a mimic control, si-CCND1 and si-NC vectors, HepG2 cells were harvested by trypsinization and washed with phosphate-buffered saline (PBS). The effects of miR-19a/Cyclin D1 axis on cell cycle progression were determined by fluorescence-activated cell sorter (FACS) analysis using the Cell Cycle and Apoptosis Analysis Kit (Beyotime Institution of Biotechnology, Shanghai, China) according to the manufacturer's instructions. Ten thousand cells were counted per sample. All experiments were done in triplicate. Mean normalized gene expression ± SE was calculated from independent experiments.
Statistical analysis
The software of SPSS version13.0 for Windows (SPSS Inc, IL, USA) and SAS 9.1 (SAS Institute, Cary, NC) was used for statistical analysis. The chi-squared test was used to show differences in categorical variables. Patient survival and the differences in patient survival were determined by the Kaplan-Meier method and the log-rank test, respectively. A Cox regression analysis (proportional hazard model) was performed for the multivariate analyses of prognostic factors. Differences were considered statistically significant when P was less than 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81303153, 30901795), State Project for Essential Drug Research and Development (grant number 2013ZX09301307) & Beijing Joint Project Specific Funds and the Fundamental Research Funds for the Central public welfare research institutes (No. ZZ2014036).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin 2005;55:74-108; PMID:15761078; http://dx.doi.org/ 10.3322/canjclin.55.2.74 [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55; PMID:22353262; http://dx.doi.org/ 10.1016/S0140-6736(11)61347-0 [DOI] [PubMed] [Google Scholar]
- 3.Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med 2008;359 2045-7; PMID:18923166; http://dx.doi.org/ 10.1056/NEJMe0807581 [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002;31:339-46; PMID:12149612; http://dx.doi.org/ 10.1038/ng0802-339 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM, Wu JY, Chen HY, Wang ZW, Qiu GQ, Peng ZH. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol 2012;6:445-57; PMID:22552153; http://dx.doi.org/ 10.1016/j.molonc.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 8.Huang JT, Wang J, Srivastava V, Sen S, Liu SM. MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol 2014;4:113; PMID:24904827; http://dx.doi.org/ 10.3389/fonc.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69; PMID:16557279; http://dx.doi.org/ 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 10.Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M . MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ 2015; 22: 46-57; PMID:25190143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer 2012;12:227; PMID:22681717; http://dx.doi.org/ 10.1186/1471-2407-12-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YH, Lin KH, Chen HC, Chang ML, Hsu CW, Lai MW, Chen TC, Lee WC, Tseng YH, Yeh CT. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PLoS One. 2012;7:e37188; PMID:22629365; http://dx.doi.org/ 10.1371/journal.pone.0037188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20; PMID:15652477; http://dx.doi.org/ 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 14.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol 2004;2:e363; PMID:15502875; http://dx.doi.org/ 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami Y1, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006;25:2537-45; PMID:16331254; http://dx.doi.org/ 10.1038/sj.onc.1209283 [DOI] [PubMed] [Google Scholar]
- 16.Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma 2013;60:135-42; PMID:23259781; http://dx.doi.org/ 10.4149/neo_2013_018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 2009;49:1098-112; PMID:19173277; http://dx.doi.org/ 10.1002/hep.22749 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 2012;18:5442-53; PMID:23082062; http://dx.doi.org/ 10.3748/wjg.v18.i38.5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, Chen XQ, Huang L, Zhou QM, Jia WH, et al.. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res 2013;19:4780-91; PMID:23812667; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-2728 [DOI] [PubMed] [Google Scholar]
- 20.Ker CG, Chen HY, Chen KS, Jeng IJ, Yang MY, Juan CC, Chen PH, Lo HY, Chai IC, Shih DS, et al.. Clinical significance of cell differentiation in hepatocellular carcinoma. Hepatogastroenterology 2003;50:475-9; PMID:12749251 [PubMed] [Google Scholar]
- 21.Valente TW, Fujimoto K: Bridging: locating critical connectors in a network. Social Networks 2010, 32:212-20; PMID:20582157; http://dx.doi.org/ 10.1016/j.socnet.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayanan T, Gersten M, Subramaniam S, Grama A. Modularity detection in protein-protein interaction networks. BMC Res Notes 2011;4:569; PMID:22206604; http://dx.doi.org/ 10.1186/1756-0500-4-569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 2007;3:e59; PMID:17447836; http://dx.doi.org/ 10.1371/journal.pcbi.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen G, Jia H, Tai Q, Li Y, Chen D. miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis 2013;34:211-9; PMID:23087084; http://dx.doi.org/ 10.1093/carcin/bgs320 [DOI] [PubMed] [Google Scholar]
- 25.Chung GE, Yoon JH, Myung SJ, Lee JH, Lee SH, Lee SM, Kim SJ, Hwang SY, Lee HS, Kim CY. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep 2010;23:113-9; PMID:19956871; http://dx.doi.org/ 10.3892/or_00000682 [DOI] [PubMed] [Google Scholar]
- 26.Sun G, Shi L, Yan S, Wan Z, Jiang N, Fu L, Li M, Guo J. MiR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed Res Int 2014;2014:687826; PMID:24995320; http://dx.doi.org/ 10.1155/2014/687826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge W, Yu DC, Li QG, Chen X, Zhang CY, Ding YT. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin Lab 2014;60:427-34; PMID:24697119 [DOI] [PubMed] [Google Scholar]
- 28.Jiang QQ, Liu B, Yuan T. MicroRNA-16 inhibits bladder cancer proliferation by targeting Cyclin D1. Asian Pac J Cancer Prev 2013;14:4127-30; PMID:23991964; http://dx.doi.org/ 10.7314/APJCP.2013.14.7.4127 [DOI] [PubMed] [Google Scholar]
- 29.Ma Q, Wang X, Li Z, Li B, Ma F, Peng L, Zhang Y, Xu A, Jiang B. microRNA-16 represses colorectal cancer cell growth in vitro by regulating the p53/survivin signaling pathway. Oncol Rep 2013;29:1652-8; PMID:23380758; http://dx.doi.org/ 10.3892/or.2013.2262 [DOI] [PubMed] [Google Scholar]
- 30.Cai CK, Zhao GY, Tian LY, Liu L, Yan K, Ma YL, Ji ZW, Li XX, Han K, Gao J, et al.. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep 2012;28:1764-70; PMID:22922827; http://dx.doi.org/ 10.3892/or.2012.1995 [DOI] [PubMed] [Google Scholar]
- 31.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al.. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008;14:1271-7; PMID:18931683; http://dx.doi.org/ 10.1038/nm.1880 [DOI] [PubMed] [Google Scholar]
- 32.Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A 2010;107:8231-6; PMID:20406904; http://dx.doi.org/ 10.1073/pnas.1002080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, et al.. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 2008;182:509-17; PMID:18695042; http://dx.doi.org/ 10.1083/jcb.200801079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009;50:113-21; PMID:19441017; http://dx.doi.org/ 10.1002/hep.22919 [DOI] [PubMed] [Google Scholar]
- 35.Qin X, Wang X, Wang Y, Tang Z, Cui Q, Xi J, Li YS, Chien S, Wang N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci U S A 2010;107:3240-4; PMID:20133739; http://dx.doi.org/ 10.1073/pnas.0914882107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res 2013;32:21; PMID:23594563; http://dx.doi.org/ 10.1186/1756-9966-32-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, et al.. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res 2009;39:786-94; PMID:19473441; http://dx.doi.org/ 10.1111/j.1872-034X.2009.00502.x [DOI] [PubMed] [Google Scholar]
- 38.Lin Q, Chen T, Lin Q, Lin G, Lin J, Chen G, Guo L. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J Surg Oncol 2013;107:767-71; PMID:23609137; http://dx.doi.org/ 10.1002/jso.23312 [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, Liu J, Kang Y, He Y, Liang B, Yang P, Yu Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res 2014;33:67; PMID:25107371; http://dx.doi.org/ 10.1186/s13046-014-0067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014;14:448; PMID:24938880; http://dx.doi.org/ 10.1186/1471-2407-14-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P, Nie Y, Wu K, Shi Y, Fan D. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis 2014;5:e1144; PMID:24675462; http://dx.doi.org/ 10.1038/cddis.2014.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Xia HW, Ge XJ, Zhang YC, Tang QL, Bi F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac J Cancer Prev 2013;14:7421-6; PMID:24460313; http://dx.doi.org/ 10.7314/APJCP.2013.14.12.7421 [DOI] [PubMed] [Google Scholar]
- 43.Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM, Wu JY, Chen HY, Wang ZW, Qiu GQ, Peng ZH. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol 2012;6:445-57; PMID:22552153; http://dx.doi.org/ 10.1016/j.molonc.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter T, Pines J. Cyclins and cancer. Cell 1991;66:1071-4; PMID:1833062; http://dx.doi.org/ 10.1016/0092-8674(91)90028-W [DOI] [PubMed] [Google Scholar]
- 45.Motokura T, Arnold A. Cyclin D1 and oncogenesis. Curr Opin Genet Dev 1993;3:5-10; PMID:8453274; http://dx.doi.org/ 10.1016/S0959-437X(05)80334-X [DOI] [PubMed] [Google Scholar]
- 46.Ok CY, Xu-Monette ZY, Tzankov A, O'Malley DP, Montes-Moreno S, Visco C, M?ller MB, Dybkaer K, Orazi A, Zu Y, et al.. Prevalence and clinical implications of cyclin D1 expression in diffuse large B-cell lymphoma (DLBCL) treated with immunochemotherapy: a report from the International DLBCL Rituximab-CHOP consortium program. Cancer 2014;120:1818-29; PMID:24648050; http://dx.doi.org/ 10.1002/cncr.28664 [DOI] [PubMed] [Google Scholar]
- 47.Mallya SM, Arnold A. Cyclin D1 in parathyroid disease. Front Biosci 2000;5:D367-71; PMID:10704427; http://dx.doi.org/ 10.2741/Mallya [DOI] [PubMed] [Google Scholar]
- 48.Iizuka S, Oridate N, Nashimoto M, Fukuda S, Tamura M. Growth inhibition of head and neck squamous cell carcinoma cells by sgRNA targeting the cyclin D1 mRNA based on TRUE gene silencing. PLoS One 2014;9:e114121; PMID:25437003; http://dx.doi.org/ 10.1371/journal.pone.0114121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravikumar G, Ananthamurthy A. Cyclin D1 expression in ductal carcinoma of the breast and its correlation with other prognostic parameters. J Cancer Res Ther 2014;10:671-5; PMID:25313758; http://dx.doi.org/ 10.4103/0973-1482.138135 [DOI] [PubMed] [Google Scholar]
- 50.Lu JW, Lin YM, Chang JG, Yeh KT, Chen RM, Tsai JJ, Su WW, Hu RM. Clinical implications of deregulated CDK4 and Cyclin D1 expression in patients with human hepatocellular carcinoma. Med Oncol 2013;30:379; PMID:23292829; http://dx.doi.org/ 10.1007/s12032-012-0379-5 [DOI] [PubMed] [Google Scholar]
- 51.Tong WW, Tong GH, Chen XX, Zheng HC, Wang YZ. HIF2α is associated with poor prognosis and affects the expression levels of survivin and cyclin D1 in gastric carcinoma. Int J Oncol 2015;46:233-42; PMID:25338835; http://dx.doi.org/ 10.3892/ijo.2014.2719 [DOI] [PubMed] [Google Scholar]
- 52.Bachmann K, Neumann A, Hinsch A, Nentwich MF, El Gammal AT, Vashist Y, Perez D, Bockhorn M, Izbicki JR, Mann O Cyclin D1 is a strong prognostic factor for survival in pancreatic cancer: analysis of CD G870A polymorphism, FISH and immunohistochemistry. J Surg Oncol 2015; 111: 316-23. [DOI] [PubMed] [Google Scholar]
- 53.Ripple MJ, Parker Struckhoff A, Trillo-Tinoco J, Li L, Margolin DA, McGoey R, Del Valle L. Activation of c-Myc and Cyclin D1 by JCV T-antigen and β-catenin in colon cancer. PLoS One 2014;9:e106257; PMID:25229241; http://dx.doi.org/ 10.1371/journal.pone.0106257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al.. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014;42(Database issue):D78-85; PMID:24304892; http://dx.doi.org/ 10.1093/nar/gkt1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 2003;4:P3; PMID:12734009; http://dx.doi.org/ 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 56.Wixon J, Kell D. The kyoto encyclopedia of genes and genomes–KEGG. Yeast 2000;17:48-55; PMID:10928937; http://dx.doi.org/ 10.1002/1097-0061(20000930)17:3%3c225::AID-YEA34%3e3.3.CO;2-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen JY, Mamidipalli S, Huan T. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics 2009;10 1: S16; PMID:19594875; http://dx.doi.org/ 10.1186/1471-2164-10-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 2009;37:D619-22; PMID:18981052; http://dx.doi.org/ 10.1093/nar/gkn863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics 2005;21:2076-82; PMID:15657099; http://dx.doi.org/ 10.1093/bioinformatics/bti273 [DOI] [PubMed] [Google Scholar]
- 60.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A. The IntAct molecular interaction database in 2010. Nucleic Acids Res 2010;38:D525-31; PMID:19850723; http://dx.doi.org/ 10.1093/nar/gkp878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S. Human protein reference database – 2009 update. Nucleic Acids Res 2009;37:D767-72; PMID:18988627; http://dx.doi.org/ 10.1093/nar/gkn892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res 2010;38:D532-9; PMID:19897547; http://dx.doi.org/ 10.1093/nar/gkp983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Hum Genomics 2009;3:291-7; PMID:19403463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 2005;21:827-8; PMID:15513994; http://dx.doi.org/ 10.1093/bioinformatics/bti098 [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q, Li Z, Li B1, Lin N. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer 2014;13:253; PMID:25424347; http://dx.doi.org/ 10.1186/1476-4598-13-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC, Ling XH, Fu X, Dai QS, Cai C, Chen JH, et al.. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int J Cancer 2014;135:541-50; PMID:24382668; http://dx.doi.org/ 10.1002/ijc.28707 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.