Abstract
Early infant diet has significant impacts on the gut microbiota and developing immune system. We previously showed that breast-fed and formula-fed rhesus macaques develop significantly different gut microbial communities, which in turn are associated with different immune systems in infancy. Breast-fed animals manifested greater T cell activation and proliferation and harbored robust pools of T helper 17 (TH17) cells. These differences were sustained throughout the first year of life. Here we examine groups of juvenile macaques (approximately 3 to 5 y old), which were breast-fed or formula-fed in infancy. We demonstrate that juveniles breast-fed in infancy maintain immunologic differences into the fifth year of life, principally in CD8+ memory T cell activation. Additionally, long-term correlation networks show that breast-fed animals maintain persistent relationships between immune subsets that are not seen in formula-fed animals. These findings demonstrate that infant feeding practices have continued influence on immunity for up to 3 to 5 y after birth and also reveal mechanisms for microbial modulation of the immune system.
Keywords: breast-milk, rhesus macaque, gut microbiota, T cell activation, TH17 cells
Introduction
The nutrition an infant receives can have lifelong effects, which are partly mediated by dietary influence on the infant microbiota.1-7 Previous studies have shown that breast- vs formula- fed human infants have significantly different gut microbiotas.8,9 A study in rhesus macaques demonstrated that differences in early infant diet altered the gut microbiota and cytokine production.10 We recently carried out the first detailed study of possible effects of the infant microbiota on a wide range of immune cell phenotypes and functions. We showed that breast-fed and formula-fed rhesus macaque infants have significant differences in the gut microbiota that persist through the first year of life.11 For example, breast-fed infants developed more robust pools of TH17 cells, as well as increased T cell activation and proliferation. These differences in immunologic development may partly explain inter-individual variation in immune responses to infection or vaccination, in infancy or later life.
As a follow-up to our previous study, we have now assessed for how long early infant diet may shape immune cell phenotypes. We investigated the immune systems of previously breast- or formula-fed juvenile rhesus macaques between 3–5 y of age and compared the data to those from our previous infant cohort. The immune systems of juvenile animals were affected by early dietary differences, although the differences between rearing groups were more subtle than was observed in infancy. Striking relationships between immune subsets, shaped by early diet, were documented in network diagrams and these important relationships were sustained into juvenile life. Our findings show that early infant diet has long-standing effects on the immune system, which could result in different responses to immunologic challenges in later life.
Results
Differences in immune subsets shaped by early diet are maintained into juvenile life
To investigate the long-term effects of early infant diet on immune cell subsets, we isolated peripheral blood mononuclear cells (PBMCs) from 12 juvenile rhesus macaques, which were 3–5 y in age (6 breast-fed, 6 formula-fed in infancy). We performed extensive immunophenotyping via flow cytometry, including antibody panels defining more than 115 phenotypes of interest, including antigen-presenting cell and T cell surface, activation, and homing markers. We compared the data obtained to those from 12-month-old infant macaques (6 breast-fed and 6 formula-fed) in our previous study.
We used multivariate regression to determine immune subsets that were significantly associated with rearing status and/or age, recognizing age as an important covariate.11 Some differences in immune subsets found in infancy were maintained in this older cohort. For example, activated T cell subsets were significantly associated with both rearing status and age, indicating that T cell activation remained higher in breast-fed older animals (e.g, CD8+HLA-DR+CD38− memory T cells,% of CD8+; p = 0.004; Fig. 1A). These and other activated subsets had also been seen to be higher in 12-month-old breast-fed macaques, as compared to formula-fed animals. Total CD8+HLA-DR+ memory T cells followed a similar pattern (% of CD8+; p = 0.0008, Fig. 1B), except that age was positively associated with the frequency of these cells.
Figure 1.
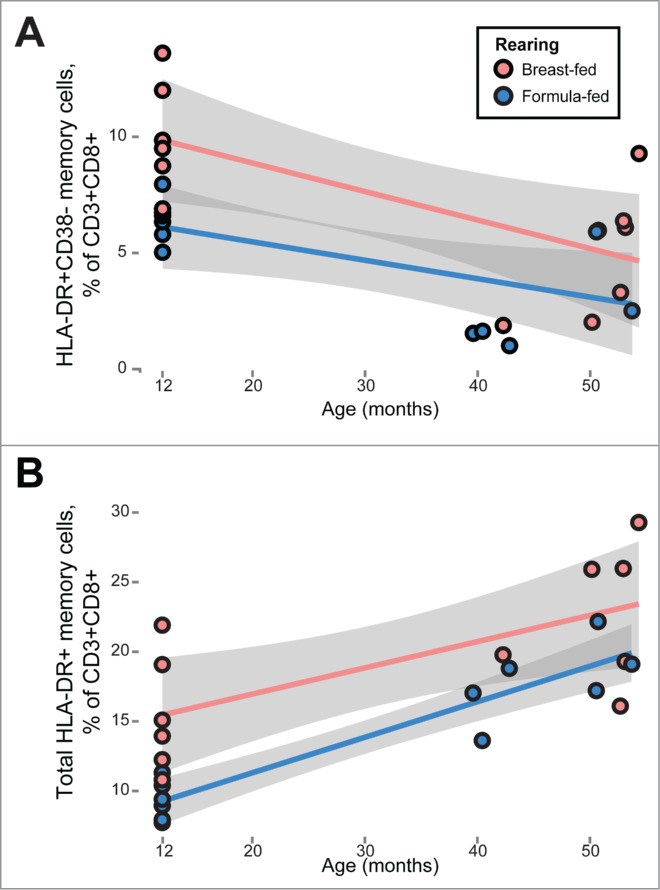
Persistent differences in T cell activation between previously breast-fed and formula-fed rhesus macaque juveniles. Multivariate mixed-effects regression revealed differences in T cell activation between breast- and formula- fed animals (red and blue markers, respectively), while controlling for age and sex. Breast fed animals manifested higher fractions of (A) CD8+HLA-DR+CD38− memory T cells (p = 0.004) and (B) the total HLA-DR+ fraction among CD8+ memory T cells (p = 0.0008). Gray bands represent 95% confidence intervals.
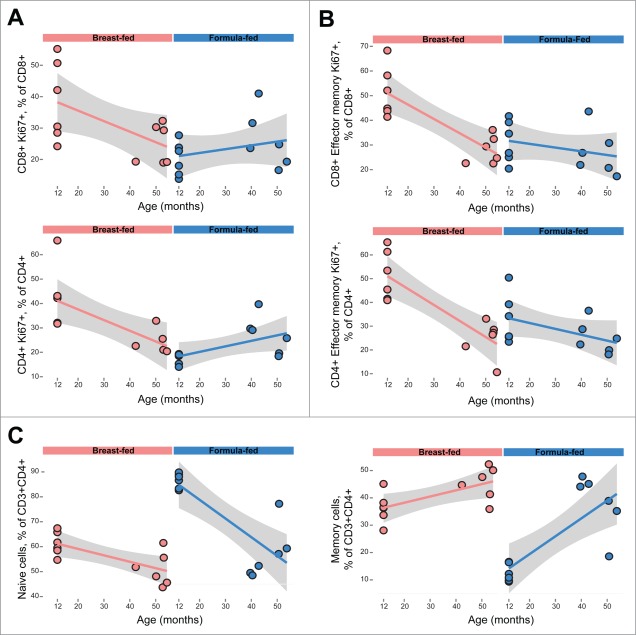
Other subsets found to be different between rearing groups in our 12-month-old infants were no longer different in the juvenile cohort. These populations include proliferative subsets (Ki-67+) and memory and naïve cells within the CD4+ T cell compartment (Fig. 2). In both CD4+ and CD8+ T cells and in the corresponding effector-memory T cells, 12-month-old breast-fed infants manifest higher Ki-67 expression than formula-fed infants. In our juvenile cohort these differences disappear, with the frequency of these cells in previously breast-fed juveniles apparently declining to match that in their previously formula-fed counterparts (Figs. 2A and 2B). In the cases of CD4+ memory and naïve T cells (Fig. 2C), similarly, there are significant differences in these subsets at 12 months: the ratio of memory to naïve cells is much higher in breast-fed infants. In previously formula-fed juveniles, this ratio seems to “catch up” with that seen in breast-fed peers, indicating development of CD4+ memory T cells to an extent eventually matching that in breast-fed animals (Fig. 2C).
Figure 2.
Relative equalization of some immune parameters between rearing groups in juveniles, as compared to infant animals. While infant animals show significant differences between breast and formula fed animals, previously breast- and formula-fed juvenile rhesus macaques show little difference in (A) T cell proliferation (CD4+Ki-67+ and CD8+Ki-67+) (B) Effector memory T cell proliferation (CD8+ and CD4+ shown above and below, respectively) or (C) CD4+ naïve and memory T cells. Gray bands represent 95% confidence intervals.
Similar “catch up” seemed to occur with TH17 cells, in that no significant differences in TH17 cell pools remained between 3- to 5-year-old macaques that were previously breast- or formula-fed (p = 0.53).
Relationships between immune subsets shaped by early diet are sustained into juvenile life
We then assessed the relationships between immune subsets in early and later life. We calculated Spearman correlations between immune variables separately in breast- and formula-fed infant and juvenile animals, that is, in 4 groups separately. We then used Cytoscape (“Merge Networks” tool) to identify correlations that were persistent over time within each rearing cohort. Finally, we looked at the differences between these 2 merged networks (breast-fed vs formula-fed; Fig. 3).
Figure 3.
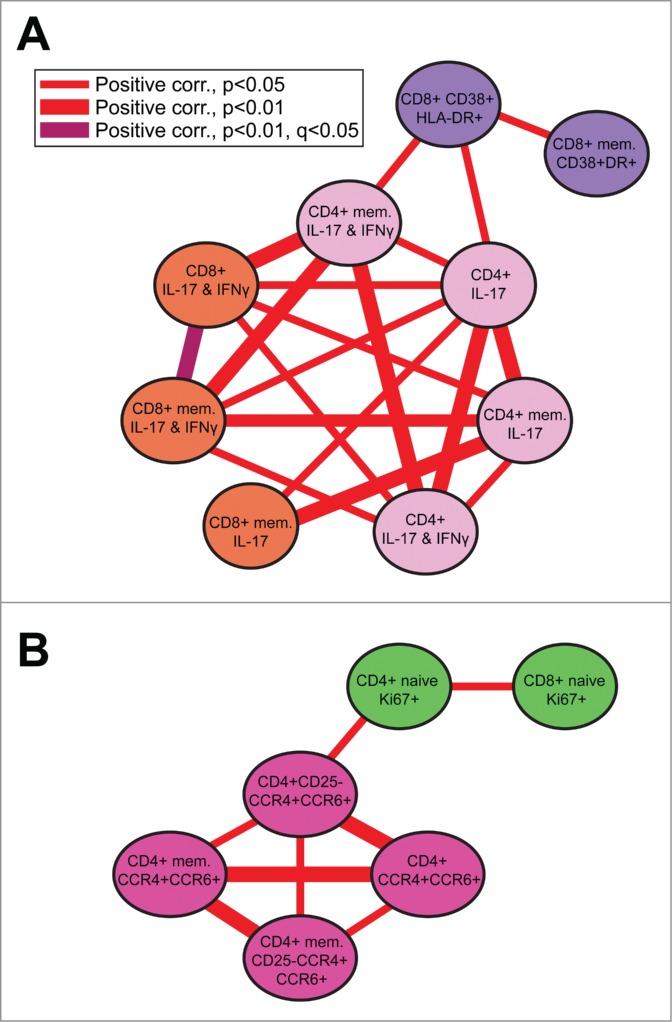
Breast-fed animals maintain correlation networks that formula-fed animals do not. Correlations between immune subsets seen in our breast-fed cohorts (infant and juvenile) were not observed in formula-fed animals. (A) Activated T cells (purple) correlate positively with TH17 cells and memory TH17 cells producing IFN-γ (light pink), as well as with TC17 cells (orange). (B) Proliferating T cells (green) correlated positively with extracellular markers of gut-homing TH17 cells (dark pink). Formula-fed animals do not maintain either network.
Animals that were previously breast-fed sustained long-term correlation networks not seen in formula-fed animals (Fig. 3). Activated CD8+ T cells (CD38+HLA-DR+; purple nodes in Fig. 3A) were positively correlated with IL-17-producing CD4+ T cells (TH17 cells) and memory TH17 cells producing interferon gamma (IFNγ; light pink nodes; Fig. 3A). Frequency of these TH17 cells, in turn was correlated with that of IL-17-producing CD8+ T cells (TC17 cells, orange nodes). In addition, in this same breast-fed group, we identified sustained correlations between proliferative T cells (Ki-67+, green nodes) and extracellular markers of gut-homing TH17 cells (CCR4+ and CCR6+ cells in Figure 3B, darker pink nodes). None of these networks were found in the previously formula-fed group, indicating that the relationships involved were not as dominant or consistent after formula feeding.
Discussion
In this follow-up to our early infant diet cohort study, we found that some immunologic differences seen in infancy are sustained into juvenile life. Markers of T cell activation remained associated with the rearing status and age of an animal, especially those marking activation of CD8+ memory T cells. Other immunologic features became more uniform between the infant and juvenile periods, suggesting that the effects of other exposures came to dominate those that were earlier attributable to infant feeding practices. Markers of T cell proliferation and relative proportions of memory and naïve T cells had equalized between rearing groups by the juvenile stage. The exact timing of and impetus for these changes are unknown but will be revealed by careful longitudinal studies.
When we investigated the relationships between immune subsets over time, we found that breast-fed animals maintained correlation networks that formula-fed animals did not. Breast-fed animals maintained positive correlations between T cell activation and proliferation and TH17 cells. These sustained relationships are likely driven by the influence of breast-feeding in infancy. Possibly, for example, TH17 cell development is driven in breast-fed animals by characteristics of the early infant microbiota, via effects on T cell activation, while in formula-fed animals later influences such as viral infection are more important. We did not analyze the gut microbiota for the juvenile cohort. Studies in humans have shown that the gut microbiota differences imposed by different early diets dissipate over time.12-15 Possibly, homogenization of the gut microbiota between rearing groups (all animals in this study received identical diets) could explain homogenization of many immune cell subsets.
Possible consequences of early infant feeding for infection and inflammation in later life
These new data suggest that effects of early infant diet may be different at various subsequent life stages.
Infancy
The largest immunologic effects of early infant diet were observed at 9–12 months of age, a time period that in humans is considered to be part of infancy. Our studies demonstrate a link between breast feeding, the associated breast feeding-specific microbiota, and T cell activation and proliferation, as well as increases in TH17 cell pools. TH17 cells are important modulators of intestinal immunity whose development is induced by gut commensal bacteria.16 Larger preexisting TH17 cell pools have been associated with lower viral loads when rhesus macaques are infected with SIV.17 The increased T cell activation and proliferation set points may be caused by stimulation of the T cell compartment by a more diverse gut microbiota or may be associated with specific bacterial colonizers or their metabolites.11
Juvenile to early adulthood
By 3–5 y of age, previously breast- vs. formula-fed macaques demonstrated no detectable differences in T cell proliferation, or in levels of memory and naïve T cells. However, differences in T cell activation were sustained, suggesting that infant feeding could be determinative of the quality of certain immune responses even after a long period of time. Hosts with increases in these cells may manifest more rapid responses against infection.18,19 Alternatively or in addition, the altered T cell activation may be reflective of changed antigen presenting cell function, perhaps due to durable changes in the microbiota.20
Later adult life
We have not yet assessed immunologic differences in adult macaques that may be ascribed to infant feeding. Given the data reported here on juvenile animals, which are suggestive of waning effects of infant feeding and possibly greater effects of more proximal exposures, a large study may be required to understand lifelong effects of infant diet. However, the long-term connections between immune subsets, reflected in correlation networks, may indicate that early diet has a long-term impact that is sustained by long-term effects on the microbiota. Different microbiotas and associated drivers of adaptive immune cell subsets would suggest the likelihood of different responses to interventions targeting the immune system. Among previously breast-fed animals, but not formula-fed animals, T cell activation (HLA-DR+) and proliferation (Ki-67+) were positively associated with TH17 cells. Thus, future therapies that might induce TH17 cells through T cell activation and proliferation may be more effective in breast-fed individuals.21
Breastfeeding provides important protection against infectious diseases, which account for over two-thirds of the 12 million annual deaths in children younger than 5 y in less developed countries.22-27 Nonetheless, we know little about the specific immunophenotypic changes established by breast-feeding that confer such advantages, or the mechanisms by which those differences shape an infant's risk of disease. We can infer specific risks or protective effects where mechanisms are well understood.28 For example, animals with increased T cell activation (as a result of previous breast-feeding) may have more rapid responses to infection or quantitatively greater responses to vaccination.18,19 Such T cell activation, however, might be detrimental in the setting of chronic HIV infection.29 Setting aside such specific cases, we still know relatively little about how natural immune variation in healthy individuals contributes to disease risk overall. To understand the impact of such inter-individual variability, more longitudinal and interventional experiments must be performed.
Our data show that early infant diet has a profound and long-lasting effect on the immune system. The initial divergence is powered by differing gut microbiotas developing in breast- and formula- fed infants. While the effects do wane over time, various immune subsets seem to equalize at different rates. Factors other than the infant microbiota, such as cytomegalovirus infection, also contribute important variability to the immune system.30 Thus, although work remains to be done in order to clarify the interactions between early and later exposures, large differences attributable to infant feeding doubtless explain some of the immune variation seen between individuals, particularly between infants.31
Materials and Methods
Ethics statement
This study was performed under strict compliance with the NIH Guide for the Care and Use of Laboratory Animals. Established policies of the Institutional Animal Care and Use Committee (IACUC) of the University of California, Davis were followed for sample collections, housing, and medical care. The study was performed at the CNPRC, which is one of 8 centers supported by the National Institutes of Health, Office of the Director (NIH/OD), and is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The CNPRC houses more than 5,000 non-human primates, most of which are rhesus macaques (Macaca mulatta), in outdoor corrals or indoor cages.
Study design
Twelve infant and 12 juvenile rhesus macaques were used for study of immune cell phenotypes. The infants were the same used and described in ref. 11. Juvenile animals were housed in similar outdoor environments. Three of the previously formula-fed animals were specific-pathogen free (SPF) and were therefore seronegative for Herpes B virus, simian immunodeficiency virus (SIV), Type D retrovirus and simian T-lymphotropic virus.32 These 3 were not significantly different from non-SPF formula-fed juveniles in immune variables, as tested by Wilcoxon rank sum test. Our low sample size of previously formula-fed juveniles limits that result, and may warrant further study. Other than SPF status, juvenile animals had no environmental differences between rearing groups besides their early feeding practices.
Sample collection
Blood samples were taken from each rhesus macaque infant 12 months after birth as well as juvenile macaques aged 3–5 years, as cited in ref. 11. Peripheral blood mononuclear cells (PBMCs) were isolated by gradient density purification using Lymphocyte Separation Medium (LSM; MP Biomedicals, LLC, Solon, OH), then washed in medium containing FBS (fetal bovine serum) and 10% DMSO, then cryopreserved in liquid nitrogen.
Immune cell phenotyping by flow cytometry
Frozen rhesus macaque PBMCs were thawed and stained for flow cytometry (including intracellular staining) as previously described in ref. 11.
Statistical analysis
Immunophenotyping data from 12-month-old rhesus macaques (obtained in our previous study) and from our juvenile cohort were used for analysis. All analysis was performed in R, and all plots were created using ggplot2. We used multivariate mixed-effects regression (using lmer in the lme4 package) to assess variables significantly associated with age and rearing status, comparing to a null model testing age only. Significance of random effects was assessed based on the likelihood ratio-test between nested models. Differences between rearing groups at individual time points were calculated using Wilcoxon rank sum test.
To assess long-term correlations between immune subsets, we first separated data into 4 groups based on age cohort (infant vs juvenile) and rearing status. We calculated Spearman correlations between variables. We then used Cytoscape to merge networks based on rearing status and display the resulting networks using the “Merge Networks” tool.33 Final networks were drawn in Illustrator based on Cytoscape results.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under award number K23AI081540 to DJHOC, by the Bill and Melinda Gates Foundation under a “Grand Challenges Exploration” award (#52094) to DJHOC, and by the National Center for Research Resources (P51RR000169) and is currently supported by the Office of Research Infrastructure Programs/OD (P51OD011107). NRN was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number UL1 TR000002 and linked award TL1 TR000133. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Joyce Lee and Paul-Michael Sosa for assistance with sample collection.
Author Contributions
AA and DHO designed the study. AA, NN, and DHO performed the statistical analysis. NN, AA, GML, and DHO wrote the manuscript. GML performed the flow cytometry assay and analysis. DL and KKAVR collected the samples.
References
- 1.Siggers J, Sangild PT, Jensen TK, Siggers RH, Skovgaard K, Stoy AC, Jensen BB, Thymann T, Bering SB, Boye M. Transition from parenteral to enteral nutrition induces immediate diet-dependent gut histological and immunological responses in preterm neonates. Am J Physiol Gastrointest Liver Physiol 2011; 301:G435-45; PMID:21700903; http://dx.doi.org/ 10.1152/ajpgi.00400.2010 [DOI] [PubMed] [Google Scholar]
- 2.Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 2010; 23:23-36; PMID:20450531; http://dx.doi.org/ 10.1017/S0954422410000065 [DOI] [PubMed] [Google Scholar]
- 3.Zetterstrom R, Bennet R, Nord KE. Early infant feeding and micro-ecology of the gut. Acta Paediatr Japon 1994; 36:562-71; PMID:7825464; http://dx.doi.org/ 10.1111/j.1442-200X.1994.tb03247.x [DOI] [PubMed] [Google Scholar]
- 4.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol 2010; 21:149-56; PMID:20434324; http://dx.doi.org/ 10.1016/j.copbio.2010.03.020 [DOI] [PubMed] [Google Scholar]
- 5.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet 2003; 361:512-9; PMID:12583961; http://dx.doi.org/ 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489:220-30; PMID:22972295; http://dx.doi.org/ 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. J Dev Orig Health Dis 2013; 4:203-14; PMID:24353893; http://dx.doi.org/ 10.1017/S2040174412000712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol 2012; 13:r32; PMID:22546241; http://dx.doi.org/ 10.1186/gb-2012-13-4-r32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hascoet JM, Hubert C, Rochat F, Legagneur H, Gaga S, Emady-Azar S, Steenhout PG. Effect of formula composition on the development of infant gut microbiota. J Pediatr Gastroenterol Nutr 2011; 52:756-62; PMID:21593648; http://dx.doi.org/ 10.1097/MPG.0b013e3182105850 [DOI] [PubMed] [Google Scholar]
- 10.O'Sullivan A, He X, McNiven EM, Haggarty NW, Lonnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res 2013; 12:2833-45; PMID:23651394; http://dx.doi.org/ 10.1021/pr4001702 [DOI] [PubMed] [Google Scholar]
- 11.Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, Hartigan-O'Connor DJ. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med 2014; 6:252ra120; PMID:25186175; http://dx.doi.org/ 10.1126/scitranslmed.3008791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr 2009; 98:229-38; PMID:19143664; http://dx.doi.org/ 10.1111/j.1651-2227.2008.01060.x [DOI] [PubMed] [Google Scholar]
- 13.Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2012; 2:94; PMID:23087909; http://dx.doi.org/ 10.3389/fcimb.2012.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 1999; 69:1035S-45S; PMID:10232646 [DOI] [PubMed] [Google Scholar]
- 15.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microb 1982; 15:189-203; PMID:7143428; http://dx.doi.org/ 10.1099/00222615-15-2-189 [DOI] [PubMed] [Google Scholar]
- 16.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al.. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485-98; PMID:19836068; http://dx.doi.org/ 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartigan-O'Connor DJ, Abel K, Van Rompay KK, Kanwar B, McCune JM. SIV replication in the infected rhesus macaque is limited by the size of the preexisting TH17 cell compartment. Sci Transl Med 2012; 4:136ra69; PMID:22649090; http://dx.doi.org/ 10.1126/scitranslmed.3003941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber DL, Wherry EJ, Ahmed R. Cutting edge: rapid in vivo killing by memory CD8 T cells. J Immunol 2003; 171:27-31; http://dx.doi.org/ 10.4049/jimmunol.171.1.27 [DOI] [PubMed] [Google Scholar]
- 19.Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 1998; 8:89-95; PMID:9462514; http://dx.doi.org/ 10.1016/S1074-7613(00)80461-6 [DOI] [PubMed] [Google Scholar]
- 20.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell 2006; 124:849-63; PMID:16497593; http://dx.doi.org/ 10.1016/j.cell.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 21.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Ann Rev Immunol 2009; 27:485-517; PMID:19132915; http://dx.doi.org/ 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- 22.Victoria CG. Infection and disease: the impact of early weaning. Food Nutr Bull 1996; 17:390-96; [Google Scholar]
- 23.Jelliffe DB, Jelliffe EFP. Human Milk in the Modern World. Oxford and New York: Oxford University Press, 1978 [Google Scholar]
- 24.Feachem R, Koblinski M. Interventions for the control of diarrhoeal diseases among young children: promotion of breastfeeding. Bull World Health Organ 1984; 62:271-91; PMID:6610496 [PMC free article] [PubMed] [Google Scholar]
- 25.Jason JM, Nieburg P, Marks JS. Mortality and infectious disease associated with infant-feeding practices in developing countries. Pediatrics 1984; 74:702-27; PMID:6435089 [PubMed] [Google Scholar]
- 26.Cunningham AS, Jelliffe DB, Jelliffe EFP. Breast-feeding and health in the 1980s: a global epidemiologic review. J Pediatr 1991; 118:659-66; PMID:2019919; http://dx.doi.org/ 10.1016/S0022-3476(05)80023-X [DOI] [PubMed] [Google Scholar]
- 27.Murray CJL, Lopez AD. The global burden of disease: a comprehensive assessment of mortality and disability from diseases, injuries and risk factors in 1990 and projected to 2020. Cambridge: Harvard School of Public Health, 1996 [Google Scholar]
- 28.Hartigan-O'Connor DJ, Abel K, McCune JM. Suppression of SIV-specific CD4+ T cells by infant but not adult macaque regulatory T cells: implications for SIV disease progression. J Exp Med 2007; 204:2679-92; PMID:17954571; http://dx.doi.org/ 10.1084/jem.20071068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartigan-O'Connor DJ, Deeks SG. Immunology In: Young B, Hardy D, eds. Fundamentals of HIV Medicine 2012. Edition. Washington: American Academy of HIV Medicine [Google Scholar]
- 30.Brodin P, Jojic V, Gao TX, Bhattacharya S, Angel CJL, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, et al.. Variation in the Human Immune System Is Largely Driven by Non-Heritable Influences. Cell 2015; 160:37-47; PMID:25594173; http://dx.doi.org/ 10.1016/j.cell.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, et al.. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011; 473:523-7; PMID:21562493; http://dx.doi.org/ 10.1038/nature10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanthaswamy S, Kou A, Satkoski J, Penedo MC, Ward T, Ng J, Gill L, Lerche NW, Erickson BJ, Smith DG. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC). Am J Primatol 2010; 72:587-99; PMID:20162538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498-504; PMID:14597658; http://dx.doi.org/ 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]