Abstract
Vertebrates developed immunoglobulin heavy chain (IgH) class switch recombination (CSR) to express different IgH constant regions. Most double-strand breaks for Ig CSR occur within the repetitive portion of the switch regions located upstream of each set of constant domain exons for the Igγ, Igα or Igε heavy chain. Unlike mammalian switch regions, Xenopus switch regions do not have a high G-density on the non-template DNA strand. In previous studies, when Xenopus Sμ DNA was moved to the genome of mice, it is able to support substantial CSR when it is used to replace the murine Sγ1 region. Here, we tested both the 2 kb repetitive portion and the 4.6 kb full-length portions of the Xenopus Sμ in both their natural (forward) orientation relative to the constant domain exons, as well as the opposite (reverse) orientation. Consistent with previous work, we find that the 4.6 kb full-length Sμ mediates similar levels of CSR in both the forward and reverse orientations. Whereas, the forward orientation of the 2 kb portion can restore the majority of the CSR level of the 4.6 kb full-length Sμ, the reverse orientation poorly supports R-looping and no CSR. The forward orientation of the 2 kb repetitive portion has more GG dinucleotides on the non-template strand than the reverse orientation. The correlation of R-loop formation with CSR efficiency, as demonstrated in the 2 kb repetitive fragment of the Xenopus switch region, confirms a role played by R-looping in CSR that appears to be conserved through evolution.
Keywords: Recombination, Activation-induced deaminase, AID, RNA:DNA hybrid, Immunoglobulin, Isotype switch, B Cell, Antibody, Secondary response, Genetic instability, Chromosomal rearrangement, Gene rearrangement, Xenopus, Amphibian, Immune system
1. Introduction
DNA recombination of any of multiple IgH class switch regions located downstream of a VDJ exon has only been documented in tetrapods [1,2]. Other means of IgH isotype switching are employed by fish and sharks [3]. In the most widely studied amphibian system, Xenopus, switching occurs from IgM to IgG or to IgX, a functional analog of IgA. However, these two kinds of events occur under different conditions. Although AID is active at a wide range of temperatures in exothermic vertebrates, in Xenopus the conditions known to produce IgG switching are more restricted. Thymectomized Xenopus express IgX but not IgG, and the absence of T cells does not affect mucosal IgX response [4,5]. In contrast, switching to IgG requires T cell help, and Xenopus T cell function is temperature-dependent. There is little or no IgG produced during an antibody response at 18–19 °C, and skin graft rejection times are slowed. Over the animal’s lifetime, IgM is the prominent serum Ig, contributes a major role in an on-going response that can last for months, and without hyperimmunization is not overtaken by IgG [6–8]. This last observation is in striking contrast to mammals, where most of the Ig of a given specificity is in the switched form (IgG, A or E) [9].
The regions mediating class switch recombination (CSR) first appear in amphibian IgH. In Xenopus the 7.3 kb stretch between the 3′-most JH and Cμ contain multiple repeats, which is the region corresponding to sites of switch junctions between IgM and IgX [10]. One stretch, from 2957 to 5609 bp, consists of 23 non-identical repeats of 150 bp. In mammalian B cells, most double-strand breaks (DSBs) for CSR occur in the repetitive portion, based on the position of the recombination [11–13]. The boundaries of the remaining portion of the switch regions are less certain, with less clearly defined roles, except for the I exon promoter located upstream of the I exon and which is responsible for initiating transcription through the downstream switch region.
In one elegant study,1 a portion of Xenopus Sμ (XSμ) was used in place of the Sγ1 region in the mouse genome [14]. Only the central 2 kb portion of this 4.6 kb region is repetitive (Fig. 1), and the distinctive feature of the repeats is that they are rich in WGCW [10]. The 4.6 kb piece was able to function at about 25–50% of the efficiency as a similar size segment of murine Sγ1 [14]. The Xenopus 4.6 kb portion has a much lower G-density and fewer G-clusters but a higher WGCW density. We have recently shown that G-clusters are important for initiating R-loop formation, and G-density is important for R-loop elongation in vitro and in murine B cells [15–19]. R-loops generated at mammalian switch regions are thought to provide single-stranded DNA regions that allow AID to deaminate cytosines [11,12,20]. Based on the lack of G-density and G-clusters, the 4.6 kb segment did not appear likely to form R-loops in our in vitro biochemical system [21], and so it was not clear what contribution R-loop formation brings to IgH CSR.
Fig. 1.
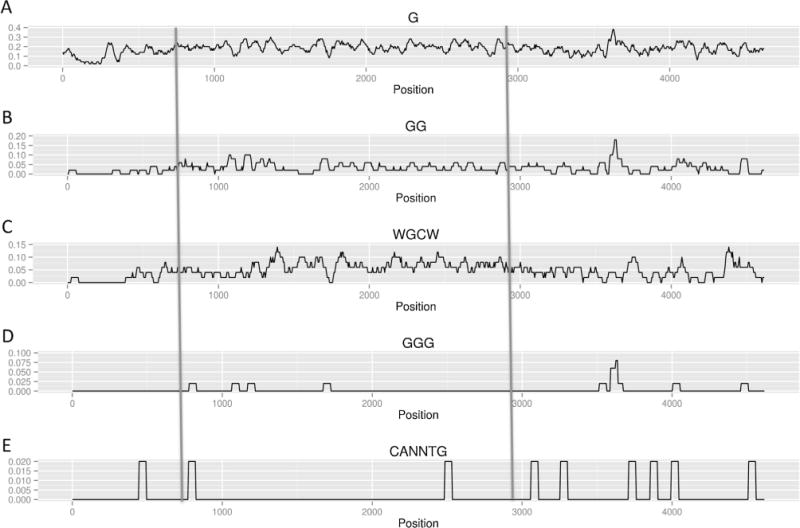
Frequency of G, GG, WGCW and E-box motif in the physiologic orientation of Xenopus IgH Sμ switch region. Different DNA sequence motif frequencies (e.g., GG or WGCW) are displayed across the entire Xenopus IgH Mu switch region (x-axis) in the physiologic (forward) orientation. The 2 kb repetitive portion of the switch region is between two long vertical lines that cut across all of the plots A–E. CANNTG represents naturally-occurring E-box motifs. The y-axis is the frequency of the indicated motif.
Here, we have taken a closer look at the 4.6 kb segment and its central 2 kb repetitive portion. We have used a chromosomal exchange system to position these Xenopus DNA segments in place of the murine Sα region [22]. We find that the physiologic (forward) orientation of the Xenopus 2 kb repetitive portion is much more active for transcription and in driving IgH CSR relative to the reverse orientation of the same fragment (Fig. 2 & Supplementary Fig. S1). In contrast, either orientation of the larger 4.6 kb portion supports a high level of CSR that is similar to that of the 2 kb segment (despite a much lower transcription for either orientation of the 4.6 kb segment than the forward orientation of the 2 kb segment). We also find that the forward orientation of the 2 kb repetitive portion is able to form R-loops efficiently in vivo, whereas, the reverse orientation is not. Based on these results, we discuss possible relationships between G-clustering, R-loop formation, transcription, and IgH CSR for the Xenopus CSR sequences.
Fig. 2.
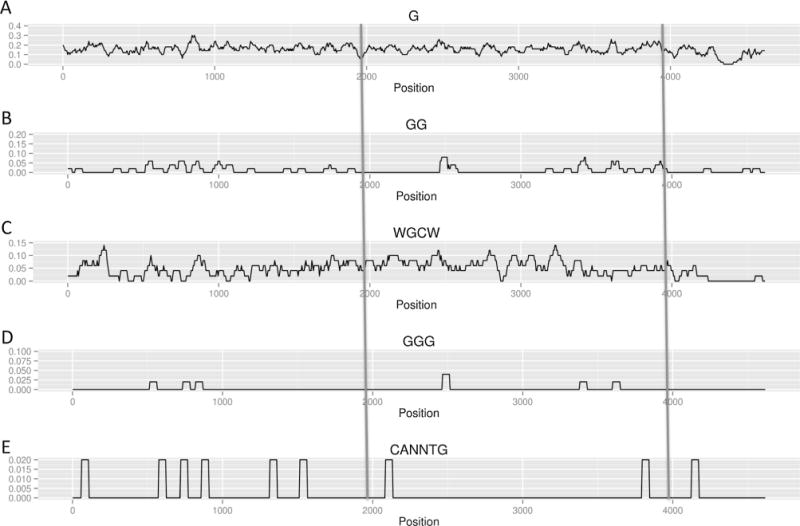
Frequency of G, GG, WGCW and E-box motif in the nonphysiologic (reverse) orientation of Xenopus IgH Sμ switch region. Different motifs frequencies are displayed across the entire Xenopus IgH Sμ switch region in the reverse orientation. The repetitive portion is between the two long vertical lines, as in Fig. 1. CANNTG represents E-box motif. The y-axis is the frequency of the indicated motif.
Supplementry material related to this article found, in the online version, at http://dx.doi.org/10.1016/j.molimm.2015.07.039.
2. Materials and methods
2.1. Cell culture and CSR assay
CH12F3.2a and its derivative cells were cultured in RPMI medium supplemented with 10% FCS and 50 μM β-mercaptoethanol. As for CSR assay, healthy cells in log phase were seeded at 5 × 104 cells/ml in medium with 1 μg/ml anti-CD40 (eBioscience #16-0404-86), 5ng/ml IL-4 (R&D #404-ML-010) and 0.5 ng/ml TGF-β1 (R&D #240-B-002), and grown for 72 h [22]. Cells were stained with FITC-conjugated anti-mouse IgA antibody (BD #559354) and analyzed by flow cytometry. CSR efficiency was determined by the percentage of IgA+ cells.
2.1. Plasmid construction
The 2 kb repetitive portion of XSμ was digested with ClaI and NruI from the plasmid pDR128, and cloned into the exchange vector. The 4.6 kb full-length XSμ was amplified by PCR from Xenopus genomic DNA (sequence information at GenBank: AF002166.1) and cloned into the exchange vector. The entire 4.6 kb region was sequenced for confirmation.
2.2. Cellular targeting and screening
Five micrograms exchange vector and one microgram Cre-expression vector were cotransfected into 1F7 cells by electroporation (Lonza) [22]. Transfected cells were serially diluted and seeded in 96-well plates. After 72 h, ganciclovir (Sigma–Aldrich #G2536-100MG) was added at a final concentration of 2 μg/ml. At 7 days after transfection, single clones were picked for puromycin sensitivity test at a final concentration of 1 μg/ml. Puromycin-sensitive clones were screened by PCR around upstream and downstream boundaries of LoxP sites and across the entire switch region. Clones were also examined by Southern blot. At least five clones were used for CSR assay [22].
2.3. Germ-line transcript quantification
Two million healthy cells at a density around 1×106 cells/ml were supplemented with anti-CD40, IL-4 and TGF- β1 for 6 h, and total RNA was extracted with GenElute™ Mammalian Total RNA Kit (Sigma–Aldrich #RTN350). 10% of RNA was reverse transcribed into cDNA with M-MuLV Reverse transcriptase (NEB M0253S), and 10% of the RT products were analyzed with real-time PCR. β-Actin was used as an internal control. Each sample was done in duplicates, and at least three independent cellular clones were analyzed for each construct.
2.4. S9.6 purification
ATCC HB-8730 hybridoma line (generously provided by Bradley Cairns) was cultured in a CELLine 1000 bioreactor (Satorius Biotech, NY) according to manufacturer’s instructions. Harvested antibody (culture supernatant) was purified on a column packed with Protein G Sepharose 4 Fast Flow (GE Healthcare) equilibrated with 1×phosphate buffered saline (PBS).
2.5. S9.6 immunoprecipitation
Healthy cells in log phase were seeded at 3×105 cells/ml in medium with anti-CD40, IL-4 and TGF-β1 and × grown for 24 h. Genomic DNA was prepared by overnight proteinase K digestion, phenol-chloroform extraction and ethanol precipitation. Genomic DNA was digested with EcoRI; importantly, RNase A was added at this step to prevent S9.6 antibody binding to RNA species in subsequent steps [23,24]. Five microgram fragmented genomic DNA was incubated with 5 μg S9.6 antibody in 400 μl IP buffer (10 mM) sodium phosphate (pH 7.0, 140 mM NaCl, 0.1% Tween 20) for 2 h at 4 °C. Ten microliter pre-blocked Dynabeads (Invitrogen 10004D) were added into the mixture and gently rotated at 4 °C. After 2 h, beads were washed with IP buffer three times, and treated with proteinase K overnight. DNA bound to the beads was recovered by phenol-chloroform extraction, and quantified by real-time PCR.
3. Results
3.1. The repetitive XSμ region mediates orientation-dependent CSR
To examine the ability to drive CSR of the Xenopus Sμ sequence, we used a system that allows us to exchange out the genomic switch region and replace it with any desired sequence in a mouse B-cell line, CH12F3.2a [22]. This cell line is able to specifically and efficiently switch to IgA upon cytokine stimulation. In a CH12F3.2a-derived cell line called 1F7, the endogenous Sα was replaced with a positive–negative selection cassette (PuroΔTK, provides puromycin resistance and ganciclovir sensitivity) flanked by two different loxP sites. The sequence of interest is cloned into an exchange vector with the same loxP sites as 1F7 cells. This exchange vector is cotransfected into 1F7 cells along with a Cre-expressing vector. Cre mediates recombination between the exchange vector and the 1F7 chromosome at the corresponding loxP sites (Fig. 3). The successful replacement of the selection cassette by the sequence of interest gives rise to ganciclovir resistant clones, which are screened with a puromycin sensitivity test and PCR analysis for all clones.
Fig. 3.

Experimental system to exchange the switch regions at the Sa locus of the murine B cell line, CH12F3.2a The 1F7 subclone, which is resistant to puromycin and sensitive to ganciclovir, are transfected with both an exchange vector containing the XSμ and a CRE-expression vector. Cre mediates recombination between the exchange vector and the 1F7 chromosome at the corresponding loxP sites. Cells that have successfully replaced the selection cassette by the sequence of interest would survive during ganciclovir selection, which are further screened with a puromycin sensitivity test and PCR.
We first wanted to verify whether XSμ is able to support efficient CSR at the Sα locus. We replaced the endogenous Sα region with the 4.6 kb Xenopus Sμ region (designated as full-length XSμ hereafter), and found that both orientations result in slightly more than 11% cells that have switched to IgA (Fig. 4). Unlike mammalian switch regions, XSμ does not seem to have an orientation preference during CSR, which agrees with previous findings [14].
Fig. 4.

The Repetitive Portion of XSμ Mediates Orientation-Dependent CSR, but Not the Full-Length XSμ. FACS analysis of CSR is shown. Each data symbol represents an independent clone (diamond: 4.6 kb full-length, square: 2 kb repetitive portion), and the orientation of the Xenopus switch region is shown on the bottom. The bars represent the mean of each group.
Among the 4.6 kb Xenopus Sμ region, most of the repetitive sequence is limited to the 2 kb zone described above [10]. To evaluate the roles of this repetitive zone, we integrated the 2 kb repetitive region (between two blue lines in Figs. 1 and 2) into 1F7 cells in each of the two orientations. We found that the forward 2 kb region is able to support almost 70% CSR of the 4.6 kb full-length XSμ region (~8% switching to IgA). In contrast, the reverse 2 kb region has no CSR higher than background (<1% switching to IgA) (Fig. 4).
Therefore, we conclude that both orientations of the 4.6 kb full-length XSμ and the forward orientation of the 2 kb repetitive portion of XSμ are able to support efficient CSR, whereas, the reverse orientation of the 2 kb repetitive portion of XSμ does not support any significant CSR. Thus, the 2 kb repetitive portion of XSμ is orientation-dependent in directing Ig CSR, but the 4.6 kb full-length XSμ is not.
3.2. R-loop Formation contributes to different levels of CSR
R-loops have been documented at mammalian switch regions [18,25–27], and considered to provide stable single-stranded DNA regions, which are necessary for AID to deaminate C’s in DNA [11,12]. However, whether Xenopus switch regions can support R-loops has been an open question. The S9.6 antibody, which preferentially recognizes RNA:DNA hybrid duplexes in preference to DNA:DNA or RNA:RNA of the same sequence [23,28], was employed here for R-loop detection. Cells with the 2 kb repetitive portion of XSμ were stimulated with cytokines for 24 h before the genomic DNA were harvested and digested with the restriction enzyme, EcoRI. Half of the genomic DNAs were treated with RNase H, which specifically digests RNA in RNA:DNA hybrids, whereas, the other half were not in order to provide a test of whether the S9.6 IP was dependent on the R-loop conformation.
The forward 2 kb repetitive portion of XSμ forms significantly more R-loops than the reverse orientation (Fig. 5A and B). With RNase H treatment, the signal of the fragment containing the 2 kb repetitive portion of XSμ drops significantly, confirming the RNA:DNA component of the R-loop (Fig. 5). Unlike mammalian switch regions [16], neither orientation of the 2 kb portion of XSμ forms R-loops in vitro (Supplementary Fig. S2), raising the possibility that weak R-loops may be stabilized in vivo but not in vitro by factors yet to be defined.
Fig. 5.
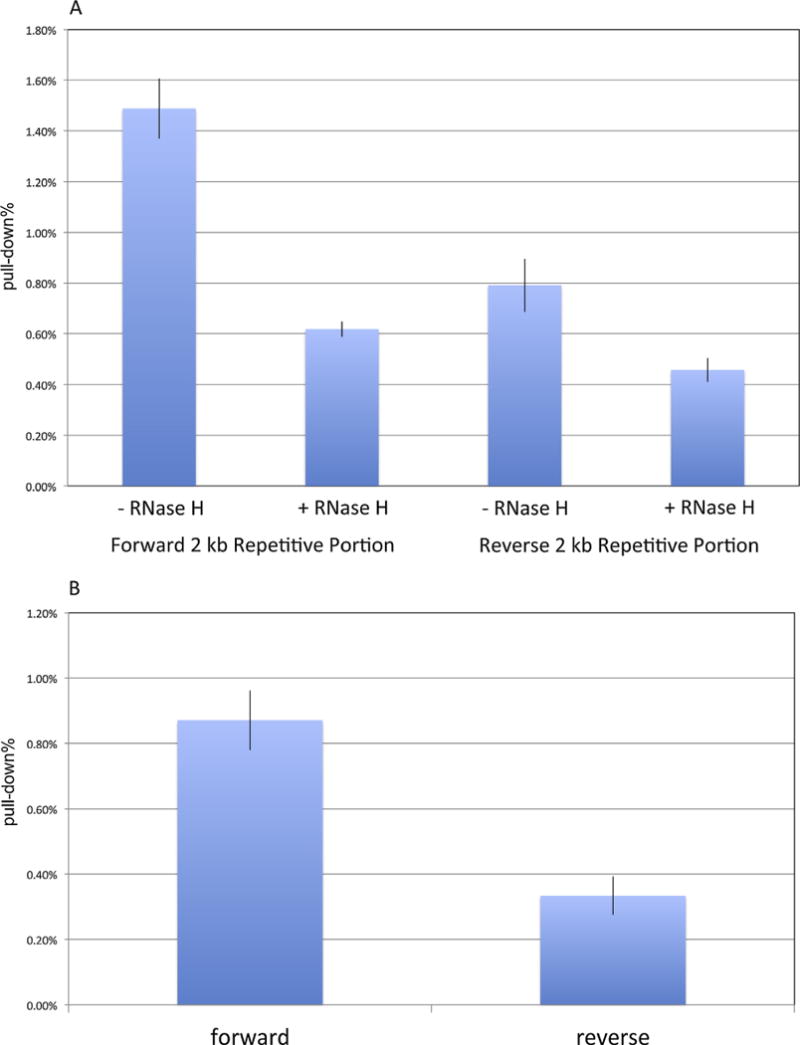
The repetitive portion of XSμ supports levels of R-loop formation in an orientation-dependent manner.
A. Immunoprecipitation with the S9.6 antibody was performed on cellular constructs containing both orientations of the 2 kb repetitive portion of XSμ at the endogenous Sα locus. Half of the genomic DNA were pretreated with RNase H prior to immunoprecipitation, as labeled on the bottom. Background signals from mock samples with no antibody were subtracted. Values were normalized to the total input DNA to calculate the pull-down percentage. Three independent IP experiments were performed for each cell line. Error bars represent SEM.
B. Values of the pull-down percentage of RNase H samples were subtracted in both orientations.
3.3. Transcription may play a role in XSμ during CSR
Transcription through switch regions has been shown to be important for CSR [12,13,29]. For mammalian switch regions, inverting switch regions decreases CSR dramatically [30]. To examine whether the transcription differs between the two orientations of the Xenopus Sμ segments, we randomly picked three cellular clones from either the forward or reverse orientation of the 2 kb repetitive portion of XSμ and the 4.6 kb full-length portion, and then checked their transcription levels. We found that the forward orientation has much more transcription than the reverse orientation, which is quite low (Fig. 6).
Fig. 6.

Orientation of the 2 kb repetitive portion of XSμ Affects transcription but transcription is low for the 4.6 kb full-length portion irrespective of orientation.
A. Three independent cellular clones with the 2 kb repetitive portion of XSμ in both orientations were picked for germ-line transcription (GLT) quantification. Healthy cells grown to 106/ml were treated with 1 μg/ml anti-CD40, 5 ng/ml IL-4 and 0.5 ng/ml TGF-β1 (CIT) for 6 h. Total RNA was extracted, reverse-transcribed into cDNA and quantified by real-time PCR. β-Actin was used as an internal control. In parallel, the same cellular clones without CIT treatment were used to measure the background of GLT. The error bar represents the SEM.
B. Three independent cellular clones with the 4.6 kb full-length of XSμ in both orientations were picked for germ-line transcription (GLT) quantification. The experiment was otherwise identical to that in panel A.
In contrast, the transcription through the 4.6 kb full-length XSμ is very low in either orientation (Fig. 6B) and is similar to the low level seen for the reverse orientation of the 2 kb repetitive portion (Fig. 6A). The average transcription levels of 4.6 kb XSμ are about 30% (forward) or 40% (reverse) of that of the reverse 2 kb repetitive region. These levels of transcription are all very low relative to the level of transcription when the forward orientation of the 2 kb repetitive region is at the Sα locus (200-fold higher than the forward orientation of the 4.6 kb full-length region and 60-fold higher than the reverse orientation of the 2 kb repetitive region). The fact that the 4.6 kb full-length portion has higher CSR in either orientation than the more highly transcribed 2 kb forward orientation suggests that the lower level of transcription seen for the 4.6 kb full-length portion is adequate to support CSR (i.e., provides the necessary minimal level of transcription) [31], though we discuss other possibilities below.
3.4. E-box motifs are not able to rescue CSR or transcription of the reverse repetitive portion of XS
We wondered why the reverse orientation of the 4.6 kb full-length XSμ has a significantly higher CSR than the reverse orientation of the 2 kb repetitive portion of XSμ. We noted that there are several potential E-box motifs upstream when the 4.6 kb fragment is in the reverse orientation (Fig. 2E). E-box binding proteins are critical for regulating Ig transcription. To investigate whether these E-box motifs account for the IgH CSR difference between the 4.6 kb full-length versus the 2 kb repetitive portion of XSμ, we inserted 2 E-box motifs immediately upstream of the reverse 2 kb repetitive portion of XSμ. Neither CSR (Fig. 7) nor transcription (data not shown) changes significantly. To further test for the possible stimulatory role of the reverse XSμ, we inserted an additional 5 E-box motifs upstream of the reverse orientation of the 2 kb repetitive portion of XSμ for a total of 7 E-box motifs. We found that CSR remains unchanged (Fig. 7). Therefore, we conclude that E-box motifs are not responsible for the orientation dependence of the 2 kb repetitive portion versus the lack of such dependence in the 4.6 kb full-length region of XSμ.
Fig. 7.
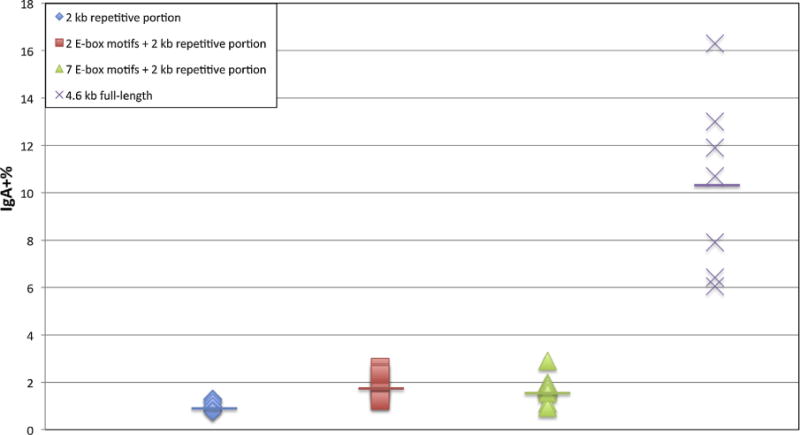
Effect of E-box Motifs on CSR in the reverse orientation of the repetitive portion of XSμ. Two (square) or seven (triangles) E-box motifs were inserted upstream of the reverse orientation of the 2 kb repetitive portion of XSμ, and at least 6 cellular clones were picked for FACS. The FACS data of the 2 kb repetitive portion (diamond) and 4.6 kb full-length (X symbols) of XSμ in the reverse orientation were included as controls.
4. Discussion
The parameters of mammalian switch regions that control switching efficiency have been defined as requiring high density of WGCW, G-density, and G clusters. These characteristics permit a high frequency of targeting by AID that generates DNA lesions, the first steps leading to DNA breakage and recombination. The density of G nucleotides predicts the ability to form R-loops, deemed necessary for AID targeting, and in non-mammalian species, such as Xenopus, the switch region had not appeared to support R-looping. Since the ability of Xenopus to undergo H chain class switching has been well established, and in almost all respects is similar to that of mammals, its switch region merited a closer examination as regards the significance of R-looping in the switching mechanism.
Here we report that the 2 kb repetitive portion of the Xenopus IgH Sμ sequence can support sufficient class switch recombination in an orientation-dependent manner in murine B cells. This 2 kb region contains most of the repetitive portion of XSμ (indicated between two blue lines in Figs. 1 and 2), and nearly all of the natural breakpoints between Sμ and other switch regions in Xenopus occur within this 2 kb region [10]. The forward orientation of the 2 kb repetitive portion of XSμ is able to support almost 70% CSR of the 4.6 kb full-length XSμ, whereas the reverse orientation of the 2 kb repetitive portion can barely support any significant CSR level above the assay background. This orientation-dependent difference is likely due to either R-loop formation or transcription, or both. The forward orientation of the 2 kb repetitive switch region has more R-loop formation (Fig. 5). The forward orientation of this segment also has more transcription (Fig. 6A). The orientation-dependence of Ig CSR for the Xenopus 2 kb repetitive Sμ is consistent with similar behavior in mammalian switch regions [22].
An open question is why the 2 kb repetitive region, but not the 4.6 kb full-length, of XSμ show orientation-dependence. The Xenopus IgH Sμ switch region contains 23 repeats, and each repeat is approximately 150 bp [10]. Though its WGCW frequency is similar to that of mammalian switch regions, it is AT-rich [2] and does not form significant R-loops in our in vitro biochemical system (Supplementary Fig. S2). We plotted the frequency of different nucleotide motifs (Figs. 1 and 2) and found that only the frequency of the GG dinucleotides is different between the two orientations of the 2 kb repetitive portion of XSμ (Supplementary Fig. S1). In vitro data have shown that GG can also support a low level of R-loop formation [15]. This may explain why we detect more R-loops in the forward orientation of the 2 kb repetitive portion than the reverse of that same 2 kb segment when assayed in the cellular assay (Fig. 5). The cellular assay appears to be more sensitive than the in vitro assay, perhaps due to stabilizing factors within cells. Therefore, the 2 kb repetitive portion of the XSμ functions similarly to mammalian switch regions, which is consistent with the fact that the repetitive feature is conserved from Xenopus switch regions to mammalian switch regions.
Supplementry material related to this article found, in the online version, at http://dx.doi.org/10.1016/j.molimm.2015.07.039.
Another question is why the reverse orientation of the 4.6 kb full-length XSμ is able to support a high level of CSR, even though the 2 kb repetitive portion in the reverse orientation is not able to do so. Considering the dramatically different transcription levels of cells with different orientations of the 2 kb repetitive portion of XSμ, initially we suspected that the few E-box motifs upstream (outside) of the 2 kb repetitive portion in the reverse orientation may promote transcription and CSR to levels that match that of the forward 4.6 kb portion. However, seven artificial E-box motifs upstream of the 2 kb repetitive portion of XSμ in the reverse orientation did not increase either transcription or CSR relative to only two E-boxes (Fig. 7 and data not shown).
Another possible explanation for the 4.6 kb full-length XSμ being high for CSR (when the 2 kb reverse orientation is not) is the relatively larger number of WGCW sites at the beginning of the 4.6 kb full-length XSμ in the reverse orientation (these are not present in the 2 kb segment). These could be sites of AID-induced nicks and which therefore may promote R-loop formation [17]. Important in this regard, we have previously shown that a nick on the nontemplate DNA strand between a promoter and a switch sequence causes increased R-loop formation in a biochemical system [17].
More GG dinucleotides in the repetitive portion of XSμ in the forward orientation may cause an increase in both transcription and R-loop formation over what is observed for the reverse orientation. We note that the higher transcription for the 2 kb forward segment, which may be due to factors other than GG dinucleotides, is possibly enough to promote R-loop formation, regardless of the GG dinucleotide number. However, the 4.6 kb segment has a high level of CSR in either orientation, despite a low level of transcription in either orientation. Thus, the lower level of transcription seen for both orientations of the 4.6 kb segment and the reverse orientation of the 2 kb segment may be adequate to support a high CSR. Hence, transcription may not be the dominant factor responsible for the orientation-dependence of the 2 kb segment [31].
The H chain switching pathway, which evolved in the primitive amphibians, was already established before divergence of lissamphibians that gave rise to modern amphibians and of amniotes. We suggest that even in the tetrapod ancestor, an extended stretch of single-stranded DNA, generated by R-looping, helped to initiate recombination events. Here we have documented some of the factors involved in determining Xenopus switch sequence efficiency to drive CSR, but we acknowledge that the interplay and importance of the factors involved will require further work to dissect.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to MRL and KY. Use of cores for the project was supported in part by a general center grant, P30CA014089 from the National Cancer Institute.
Abbreviations
- IgH
immunoglobulin heavy chain
- Ig CSR
immunoglobulin class switch recombination
- XSm
Xenopus IgH mu switch region
- CIT
anti-CD40,iL4,TGF-β1
Footnotes
Note that this 4.1 kb region corresponds to a 4.6 kb in the NCBI database and is 4.6 kb when we amplified it from the Xenopus genome. Thus, we will refer to it as 4.6 kb hereafter.
References
- 1.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, Zarrin AA. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv Immunol. 2009;101:163–189. doi: 10.1016/S0065-2776(08)01005-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E. Origin of immunoglobulin isotype switching. Curr Biol. 2012;22:872–880. doi: 10.1016/j.cub.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu E, Flajnik MF, Du Pasquier L. A third immunoglobulin class in amphibians. J Immunol. 1985;135:1998–2004. [PubMed] [Google Scholar]
- 5.Mashoof S, Goodroe A, Du CC, Eubanks JO, Jacobs N, Steiner JM, Tizard I, Suchodolski JS, Criscitiello MF. Ancient T-independence of mucosal IgX/A: gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal immunol. 2013;6:358–368. doi: 10.1038/mi.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Pasquier L, Schwager J, Flajnik MF. The immune system of Xenopus. Annu Rev Immunol. 1989;7:251–275. doi: 10.1146/annurev.iy.07.040189.001343. [DOI] [PubMed] [Google Scholar]
- 7.Du Pasquier L, Robert J, Courtet M, Mussmann R. B-Cell development in the amphibian Xenopus. Immunol Rev. 2000;175:201–213. doi: 10.1111/j.1600-065x.2000.imr017501.x. [DOI] [PubMed] [Google Scholar]
- 8.Hadji-Azimi I. Studies on Xenopus laevis immunoglobulins. Immunology. 1971;21:463–473. [PMC free article] [PubMed] [Google Scholar]
- 9.Paul W. Fundamental Immunology. 4th. Raven Press; New York: 1999. [Google Scholar]
- 10.Mussmann R, Courtet M, Schwager J, Pasquier LD. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur J Immunol. 1997;27:2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- 10.Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 12.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 13.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, DuPasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 14.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas, high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol. 2010;30:146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang ZZ, Pannunzio NR, Han L, Hsieh CL, Yu K, Lieber MR. The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites. Cell Rep. 2014;8:557–569. doi: 10.1016/j.celrep.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K, Lieber MR. The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res. 2014;42:13186–13193. doi: 10.1093/nar/gku1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels GA, Lieber MR. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han L, Masani S, Yu K. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2011;108:11584–11589. doi: 10.1073/pnas.1018726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, Leysath CE. The sub-nanomolar binding of DNA–RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit. 2013;26:376–381. doi: 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K, Lieber MR. Complexities due to single-stranded RNA during antibody detection of genomic RNA:DNA hybrids. BMC Res Notes. 2015;8(8):127. doi: 10.1186/s13104-015-1092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 25.Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence-dependence of chromosomal R-loops at the immunoglobulin heavy chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang FT, Yu K, Hsieh CL, Lieber MR. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita K, Harigai M, Fagarasan S, Muramatsu M, Honjo T. A hallmark of active class switch recombination: transcripts directed by I promoters on looped-out circular DNAs. Proc Natl Acad Sci U S A. 2001;98:12620–12623. doi: 10.1073/pnas.221454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinkura R, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 30.Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: effect of a mutation in a STAT6 binding site in the gamma 1 promoter. J Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


