Summary
During fetal development nephrons of the metanephric kidney form from a mesenchymal progenitor population that differentiates en masse before or shortly after birth. We explored intrinsic and extrinsic mechanisms controlling progenitor lifespan in a transplantation assay that allowed us to compare engraftment of old and young progenitors into the same young niche. The progenitors displayed an age-dependent decrease in proliferation and concomitant increase in niche exit rates. Single cells transcriptome profiling revealed progressive age-dependent changes with heterogeneity increasing in older populations. Age-dependent elevation in mTor and reduction in Fgf20 could contribute to increased exit rates. Importantly, 30% of old progenitors remained in the niche for up to a week post engraftment, a net gain of 50% to their lifespan, but only if surrounded by young neighbors. We provide evidence in support of a model in which intrinsic age-dependent changes affect inter-progenitor interactions that drive cessation of nephrogenesis.
Keywords: Kidney, Nephron, aging, Stem cells, mTor, Fgf20
Graphical Abstract
Introduction
The mammalian kidney is a vital excretory organ responsible for filtering metabolic wastes from the blood, maintaining pH/electrolyte balance, regulating blood pressure and bone densities, which are accomplished by the functional unit of the kidney, termed the nephron. The adult kidney possesses the capacity to repair existing nephrons but cannot replace nephrons lost with age or disease with new ones (Humphreys et al., 2008; Rinkevich et al., 2014). Instead, the functional lifespan of the metanephric kidneys depends on overabundance of nephrons. Nephron loss during the course of normal aging has been shown to contribute to hypertension, glomerulosclerosis and end-stage renal disease (ESRD) especially in those with low nephron numbers or afflicted with diseases such as diabetes (Barker et al., 2000; Bertram et al., 2011; Hoy et al., 2006; Keller et al., 2003; Lackland et al., 2000; Schreuder, 2008; Schreuder et al., 2008). Therefore, adult health depends on the number of nephrons generated during embryonic development (nephron endowment), a concept extending to many other conditions and known as the Barker Hypothesis. Identifying interventions that can increase nephron endowment will therefore have positive impact on adult health.
How nephron endowment is determined is not fully understood. The metanephric kidney generates nephrons in a reiterative process involving reciprocal interaction between the ureteric bud (UB) tip and the surrounding cap mesenchyme (CM), both derived from the intermediate mesoderm (but see (Taguchi et al., 2014)). The CM secretes GDNF that promotes both growth and branching of the UB tip (Costantini and Kopan, 2010; Hendry et al., 2011). Cell lineage analysis using tamoxifen-controlled Cre driven by the progenitor marker genes Six2, Cited1 or GDNF revealed that the CM represents self-renewing, multipotent nephron progenitors (Boyle et al., 2008; Cebrian et al., 2014; Kobayashi et al., 2008; Mugford et al., 2009). In turn, the UB secrets WNT9b that contributes to CM self-renewal and differentiation of sub-sets of CM cells (Karner et al., 2011). Wnt9b instructs a few progenitors to differentiate in every branching cycle by inducing Wnt4/Fgf8 and possibly, by down-regulating Cited1 (Brown et al., 2013; Karner et al., 2011). Induced cells undergo mesenchymal to epithelial transition (MET) and form a pretubular aggregate (PTA) at the lateral side of the UB, which will polarize to form renal vesicles (RV) and develop further into mature nephrons (Kopan et al., 2007). This entire process is repeated in the mouse ~12 times (Short et al., 2014) and ends in a wave of differentiation generating multiple nephrons per UB tip, reminiscent of arcading in humans embryos (Al-Awqati and Goldberg, 1998; Brunskill et al., 2011; Hartman et al., 2007; Rumballe et al., 2011). CM progenitors, the UB and stromal cells contribute to the maintenance of the progenitor state. It has been shown that FGF9/20 (produced by CM cells), BMP7 (made by stroma and CM cells) and WNT9b (made by the UB) work in concert to maintain the balance of self-renewal and differentiation (reviewed in (Kopan et al., 2014)). In the mouse, the nephron progenitors stop self-renewing and differentiate to form the final nephrons by P3 (Short et al., 2014). The mechanistic basis for the shift in balance from self-renewal to differentiation remains elusive. The leading hypotheses propose that the UB and the stroma regulate the niche environment to control this process. Alternatively, a change in the concentration of critical niche factors brought about by the reduction in CM/UB ratio or a parturition-associated signal determines when nephrogenesis ends by shifting the balance towards differentiations (Costantini, 2010; Hartman et al., 2007; Rumballe et al., 2011; Short et al., 2014). Support for the latter comes from studies inducing prematurity in mice (Stelloh et al., 2012). However, human normally complete nephrogenesis before birth and premature infants continue to generate nephrons for at least 40 days post partum (Rodriguez et al., 2004; Sutherland et al., 2011). At the other end of the spectrum, it has been recently established that a pulse of diphtheria toxin that eliminated 40% of CM cells at the beginning of nephrogenesis resulted in a 40% reduction in nephron numbers, indicating that nephron endowment is determined by the size of the progenitor pool (Cebrian et al., 2014). Interestingly, in this experiment nephrogenesis ended at the same time (P3) as in untreated mice (Cebrian et al., 2014), consistent with a process controlled by the surviving CM cells or their environment but not by the CM/UB ratio. Recent findings showing that CM cells secrete at least two factors (FGF9, 20) required to maintain their niche (Barak et al., 2012) highlights CM as an important contributor to its own niche and suggests that juxtacrine signaling between CM cells could actively regulate the balance of self-renewal vs. differentiation, thus determining when nephrogenesis ends.
Determining which mechanism(s) are at play has important implications for therapeutic interventions aimed at increasing nephron endowment in at risk individuals, but investigations into this mechanism have been hampered due to the lack of definitive progenitor assays as found in other stem cell fields (Hendry et al., 2011; Little and McMahon, 2012). Moreover, detecting an intrinsic change in CM cells with traditional genetic methods cannot be accomplished without simultaneously altering the overall niche environment (Barak et al., 2012). To preserve the niche, an assay akin to competitive repopulation assays (Morrison and Weissman, 1994) is needed to tease apart the relative contribution of intrinsic and extrinsic cues in regulating progenitors cells in solid organs (Barbe and Levitt, 1991; 1995; O'Leary and Stanfield, 1989; Schlaggar and O'Leary, 1991).
Herein we report a kidney progenitor assay based on similar principle to the competitive repopulation assay, and the conclusions derived from its application to probe the niche-engrafting ability of CM cells of different ages and genotypes. Heterochronic transplantation of nephron progenitors isolated from kidneys of different ages revealed that their ability to remain in a young niche is inversely correlated with age. This change precedes the cessation of nephrogenesis and occurs through a combination of slower proliferation and faster exit from the niche. At the single cell level, these changes are manifested on the transcriptional level in a heterogeneous population of nephron progenitors that gradually transition to an “old” signature from mid-gestation to birth. Consistently, single-cell RNA sequencing revealed changes in genes involved in cell cycle/mitosis, adhesion, mTor pathway and biosynthetic machinery. Importantly, when single old cells are immersed in a young environment, they remain in the niche beyond their chronological exit age, suggesting that younger cell engage in juxtacrine signaling that promotes retention in the niche. Supporting this observation, transplantation of young and old wild type cells into an Fgf20−/− young niche revealed that nearly 90% old cells remaining in the niche made contact with a co-injected WT cells whereas in WT niche the old cells are randomly distributed. Together, these data suggest that intrinsic, age-dependent changes in the CM alter the juxtacrine environment. When a critical number of cells acquire the “old” signature, retention signals no longer can prevent mass exit of progenitors.
Results
FACS Sorted Nephron Progenitors Engraft Into The CM Niche
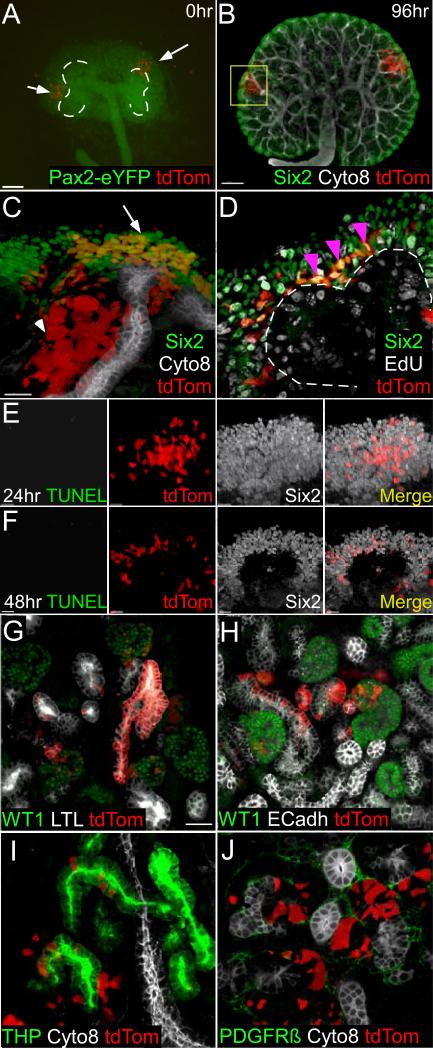
To isolate CM progenitors we purified GFP+tdTomato+ cells from Six2TGC +/tg; Rosa+/tdTomato metanephroi (Kobayashi et al., 2008) using FACS (Figure S1A). GFP expression is extinguished when progenitors differentiate, ensuring that FACS based on GFP expression collected only Six2+ progenitors but not their descendants. Conditional activation of the Rosa+/tdTomato allele by Cre within Six2+ progenitors indelibly labels progenitors and their descendants. The ~105 sorted GFP+tdTomato+ progenitors obtained in each experiment were concentrated to achieve an approximate density of 100-150 cells/nl (see Experimental Procedures). Purified nephron progenitors and the host niche survived when we injected progenitors with a manual microinjector (Narishige IM-6) into the CM of E12.5 Pax2Cre+/tg; Rosa+/eYFP (henceforth Pax2-eYFP) recipient kidney explants under a fluorescent stereoscope (Figure 1A). Injection into E11.5 metanephroi proved difficult as the injection pressure compromised the tissue (data not shown). Each injected niche contained 50-100 injected cells (data not shown); the E12.5 host niche contained ~2000 cells (Short et al., 2014)) ensuring that the incoming cells will not dominate the niche environment. Tunnel staining as well as cleaved caspase-3 staining found negligible level of cell death following the injection (Figures 1E-F and Figure S1D-E).
Figure 1. FACS sorted nephron progenitors engraft into the CM niche.
(A) A representative E12.5 Pax2-eYFP kidney injected with Six2 TGC +/tg; Rosa +/tdTomato nephron progenitors at two different sites. Dotted line marks the branching UB. (B). A recipient kidney 96hrs after injection. Six2 antibody (green) labels the progenitor cells in the periphery. Cytokeratine-8 antibody (Cyto8, white) labels the collecting duct. (C). Enlarged image of one injection site (yellow frame in B). Arrow indicating injected progenitors in the CM and arrowheads pointing at differentiated descendants in the nephron epithelia. (D). 3hr pulse labeling of injected kidneys with EdU by the end of 48hrs (white). Magenta arrows point at triple positive (Six2+; tdTom+; EdU+) cells. (E, F) TUNEL staining of a kidney explant 24hrs (E) and 48hrs (F) after transplantation. Six2 antibody stains all CM (white), tdTomato (Red) labels injected cells, and TUNEL (green) labels apoptotic cells. (G). tdTomato+ progenitors differentiated into podocytes precursor [green, Wilm's tumor 1 (Wt1)], proximal tubule [white, Lotus Tetragonolobus Lectin (LTL)], (H) distal tubule (white, ECadLO) and (I) thick ascending limb of Loop of Henle [green, Tamm-Horsfall Protein (THP)] but not UB (white, ECadHI and Cyto8) or (J) stroma (green, PDGFRβ). Scale bar, 1mm in A, 200μm in B, 30 μm in C and D, 20μm in E and F and 30μm in G-I. See also Figure S1.
The explants containing injected cells were cultured for 4 days post injection, fixed, and immunostained to assess the relative retention of injected cells in the CM (exiting cells differentiated into nephron epithelia; Figures 1A-C). We acquired Z-stack images on a confocal microscope to construct 3D images of each injection site and quantified all Six2+ tdTomato+ cells within the niche using ImarisTM software (see Experimental Procedure). The analysis clearly demonstrated that transplanted nephron progenitors engrafted into the CM niche, retained Six2 expression, proliferated (EdU+; Figure 1D) and produced descendants that differentiated into nephron epithelia (Figures 1B and 1C). Extending the culture to 7-days post injection allowed us to identify tdTomato-expressing cells in the CM as well as in every segments of a nephron including podocytes precursors (Wilm's Tumor 1; WT1; Figures 1G-H), proximal tubules (Lotus Tetraglobus Lectin; LTL; Figure 1G)), loop of Henle (Tamm-Horsfall Protein, THP; Figure 1I), and distal tubules (E-Cadherin (ECadLO) positive, Cytokeratin8 (Cyto8) negative; Figure 1H). We did not observe tdTomato cells in the UB (ECadHI, Cyto8+; Figure 1H) or the stromal lineage (PDGFRb; Figure 1I). These results mirror in vivo lineage studies (Kobayashi 2008) and confirmed that the survival and differentiation capacity of progenitors was unaffected by the isolation and injection protocols.
Heterochronic Transplantation Indicates That Young Six2+ Progenitors Contribute A Larger Fraction of Niche Resident Cells Than Old Ones After Four Days
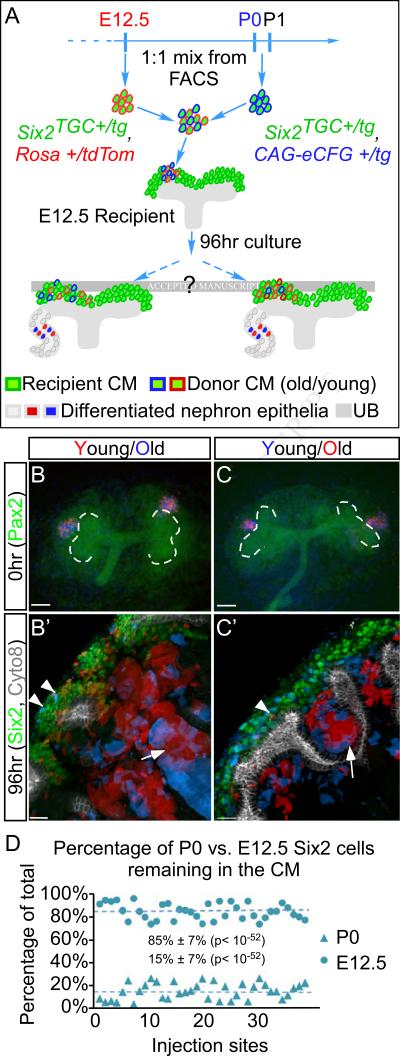
Having established a progenitor engraftment assay, we used it to evaluate renal progenitor engraftment and niche residence (hereafter referred to as engraftment) as a function of age. We FACS purified GFP+tdTom+ young progenitors from E12.5 Six2TGC +/tg; Rosa+/tdTomato embryos (Figure S1A-f; descendants GFP– tdTom+), and GFP+CFP+ old progenitors from P0 Six2TGC +/tg; CAG-eCFP +/tg pups (Figure S1A-e; descendants GFP–CFP+), concentrated and injected them in a 1:1 mixture into the CM of an E12.5 Pax2-eYFP recipient kidney explant. This ensured that the effects of injection site and cell number would impact the old and young equally. After four days the injected metanephroi were fixed, stained and processed for imaging as described in the Experimental Procedures.
If intrinsic changes (i.e., “cellular clock”) controlled exit with no input from the niche, all old progenitors will be expected to exit the CM and differentiate en masse by the objective P4 corresponding to four days in culture, whereas the young progenitors will still reside in the CM. At the other extreme, if old and young Six2+ progenitors were identical and exit from the niche was controlled by the UB and/or stroma, similar numbers of young and old cells will engraft in the young niche (Figure 2A). We quantified the number of Six2+ red and blue cells in Z-stacks acquired by confocal imaging. To normalize the data and plot it across multiple experiments we calculated the percentage of red and blue cells out of the total (100%) injected cells remaining in the niche at day 4 (Figures 2D). The analysis of 37 independent niches injected on 4 different dates revealed that both young and old progenitors produced descendants that differentiated into nephron epithelia (Figures 2B’ and 2C’). However, on average, 85%±7% E12.5 cells remained in the niche after 4 days in culture whereas only 15%±7% of the P0 cells remained (5.7-fold; p<10−23, Figure 2D). Importantly, the same results were obtained when the E12.5 Six2TGC +/tg; CAG-eCFP +/tg (blue, young) cells and P0 Six2TGC +/tg; Rosa +/tdTomato (red, old) were used (compare Figures 2B and B’, 2C and C’). The fact that lineage label has no effect on engraftment was further confirmed by co-injection of progenitors of the same age (young/young or old/old), where cells of different colors behave the same (compare Figures S1 B and B’, C and C’). The only difference we observed was that fewer old cells were detected in the niche by the end of 4 days (Figures S1B-B’, C-C’). Importantly, if the number of cells injected or the location determined the outcome, the results from 30 to 40 independent injection sites would display great variation. Instead, we see strong reproducibility in the outcome indicating independence from the exact location within the CM and initial number injected, validating the quantitative power of the assay.
Figure 2. More Young Six2+ progenitors remain in the CM niche after 4-day culture.
(A) Schematic diagram of the experimental design. Lack of intrinsic differences predicts an even contribution from old and young cells to the CM 4-days after injection (left), whereas variation in intrinsic properties would predict exit of most old cells (right). (B-C) Representative Pax2-eYFP metanephroi injected with a 1:1 mixture of FACS sorted tdTomato or CFP positive Six2+ cells. (B’-C’) Immunostaining of the explants after 4 days in culture. Six2 (green) marks the CM and Cyto8 (white) labels the UB. Both young and old Six2+ cells contributed to the differentiated nephron epithelia (arrows), whereas the majority of cells remaining in the CM are young, regardless of the color. Only a few old cells could be found in each niche (arrowheads). (D) Quantification of injected cells remaining in individual niche after 4 days as the percentage of total injected cells remaining in that niche. On average, 5.7-fold more young cells remained in the niche compared to old cells in a total of 37 niches (stdev= ±8%, p<−52). Scale bars in (B-C) 1mm and (B’-C’) 30μm. See also Figure S1.
These results detected an age-depended difference in engraftment in Six2+ nephron progenitors. However, the presence of both “uninduced” (Six2+Cited1+Wnt4–) and “induced” (Six2+ Cited1– Wnt4+, committed to MET) cells in the Six2 population (Hendry et al., 2011; Mugford et al., 2009; Rumballe et al., 2011) suggests that observed difference could reflect (1) an increase in the fraction of induced progenitors in older Six2 population, (2) cell intrinsic changes in the uninduced progenitors or (3) a combination of both.
Cited1+ Nephron Progenitors engraft better than age-matched Six2+ progenitors, but their engraftment potential also declines with age
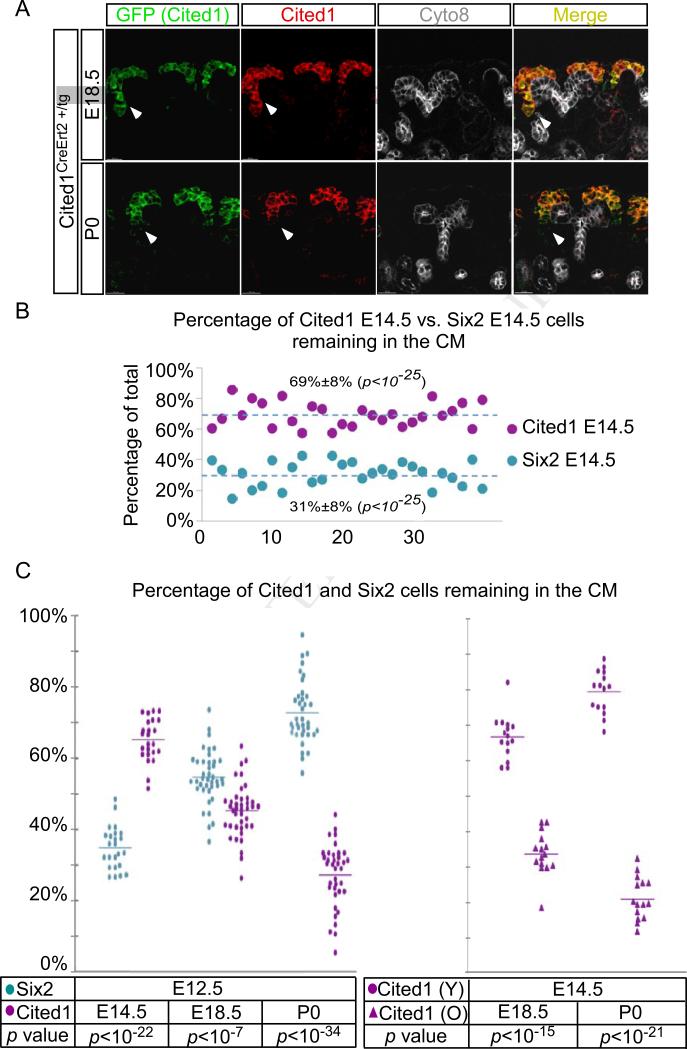
To test if our results reflect age-dependent cell intrinsic changes within the uninduced nephron progenitors, we sorted progenitors from Cited1CreERT2-GFP +/tg mouse in which CreERT2-IRESeGFP expression is directed by Cited1 enhancers/promoter (Boyle et al., 2008), expressed in “uninduced” but not the “induced” progenitors (Brown et al., 2013; Mugford et al., 2009). We first confirmed that GFP expression in the CM of Cited1CreERT2-GFP +/tg mouse recapitulates endogenous Cited1 protein (Figure 3A and Figure S2A and S2B). Next, we isolated the GFP+ tdTomato+ progenitors from E14.5 Cited1CreERT2-GFP +/tg; Rosa +/tdTomato-A mouse and co-injected them with the same number of E14.5 Six2TGC +/tg; CAG-eCFP+/tg progenitors into E12.5 recipients. Note that both lineage reporters used here do not require Cre activity to be turned on: Rosa tdTomato is constitutively expressed from the Rosa+/tdTomato-A allele where the stop cassette has already been floxed out (see Supplemental Experimental Procedures). Similarly, the ubiquitously expressed actin promoter drives the constitutive expression of CFP. As expected, we observed more Cited1+ cells engrafting in the CM (mean 69%±8%) than Six2+ (31%±8%; n=28, p<10−25) consistent with Cited1-GFP marking the uninduced nephron progenitor population (Figure 3B).
Figure 3. The ability of Cited1+ progenitors to engraft in the niche is inversely correlated with age.
(A) Antibody staining of Cited1-driven GFP (green), endogenous Cited1 protein (red) and Cyto8 (white, marking the UB). GFP overlaps with the endogenous protein (arrowhead). (B) Quantification of niche engraftment of E14.5 Cited1+ and Six2+ cells 4 days after co-injection. Cited1+ progenitors engraft better (69% vs. 31%, stdev=8%, p<10-25). (C) Left-Distribution of E12.5 Six2+ cells and E14.5, E18.5 and P0 Cited1+ progenitors in the CM niche 4 days after co-injection (E14.5: 65% vs. 35%, stdev=6%, n=24; E18.5: 45% vs. 55%, stdev=7%, n=40; P0: 27% vs. 73%, n=37). Right- Distribution of E14.5 vs. E18.5 and P0 Cited1+ progenitors in the CM niche 4 days after co-injection (E18.5 vs. E14.5: 67% vs. 33%, stdev=6%, n=15; P0 vs. E14.5: 79% vs. 21%, stdev=6%, n=15). See also Figure S3.
We next asked if Cited1+ cells isolated at different ages during embryonic and postnatal development retained an engrafting advantage relative to E12.5 Six2+ cells. If Cited1 progenitors were refractory to induction until forced to differentiate at P3, then Cited1+ cells of all ages would engraft better than a E12.5 Six2+ population containing a fraction of cells already committed to differentiation. To test if this were the case we co-injected Cited1CreERT2-GFP +/tg; Rosa+/tdTomato cells from E14.5, E18.5 embryos or P0 pups with equal number of E12.5 Six2TGC+/tg; CAG-eCFP+/tg cells into a young niche (n≥24 from two independent experiments with reversed labeling). The results show a continuous decline in the percentage of Cited1+ cells populating in the CM relative to the young, E12.5 Six2+ cells, dropping from 65% (Figure 3; n=24, p<10−22) at E14.5 to 45% (p<10−7 n=40) at E18.5 and 27% (p<10−34; n=37) at P0 (Figure 3C). We confirmed that the Cited1+ E14.5 population had the highest fraction of engrafting progenitors by co-injection of E18.5 or P0 Cited1 progenitors with E14.5 Cited1 progenitors (E14.5 vs. E18.5: 67% vs. 33%; n=15, p<10−15 and E14.5 vs. P0: 79% vs. 21%; n=15, p<10−21; Figure 3E).
The cell number in the niche increases when cells undergo proliferation and decreases when cells undergo cell death or exit. If we assume the proliferation rate (kpr), exit rate (ke) and death rate (kd) are all proportional to the cell number (x), then the differential equation describing the change in cell number (dx) as a function of time (dt) can be written as
As the rate of cell death is negligible (Figure S1D-E), the final cell number remaining depends on the proliferation rates and exit rates of the two co-injected progenitor populations. Although the initial number of cells injected was not captured, we know the ratio of young/old both at the start of the injection (50/50) and at the organ culture endpoint (Fig. 3C). Hence, we performed a series of calculations on the basis of ratio to solve the differential equation and extract information regarding proliferation and exit rates (see equations in Experimental Procedures and full explanation in Supplemental information).
Flow cytometry analysis of propidium iodide stained Cited1+ cells as well as analysis of cyclin/CDK gene expression in our single cell RNA-seq data revealed an increase in G1 and decrease in S/G2/M fraction over time (Figure S2B), consistent with a decrease in measured cell proliferation rate (Short et al. 2014). To test if the change in proliferation rates alone could account for our observations, we assumed that exit rates were unchanged in cells of different ages and compared the calculated proliferation rate to the observed rates (Short et al. 2014) (see Supplemental Experimental Procedures). This model did not reproduce the observed rates (Short et al., 2014) indicating that the assumption “constant exit rates” is wrong. Plugging in the observed proliferation rates in the model (Short et al., 2014) allowed us to conclude that the exit rate of E14.5 Cited1+ cells is slower than the exit rate of E12.5 Six2+ cells, which contain an induced, Six2+Cited1–Wnt4+ population and that P0 Cited1+ cells exit at a faster rate than E18.5 Cited1+ cells. The exit rate of E14.5 cells could not be predicted accurately without a measurement of their exact proliferation rate. If E14.5 cells proliferate faster than once a day, our model predicted that their exit rates would be faster than those of E18.5 Cited1+ cells. Conversely, if E14.5 Cited1+ cells divide at a rate slower than once a day, then the change in exit rates will increase monotonously from E14.5 to E18.5 to P0. Consistent with this prediction, the extrapolated age-dependent change in exit rate among Six2+ cells is non-monotonous with the lowest rates displayed at E17.5 (Figure 6 in (Short et al., 2014))
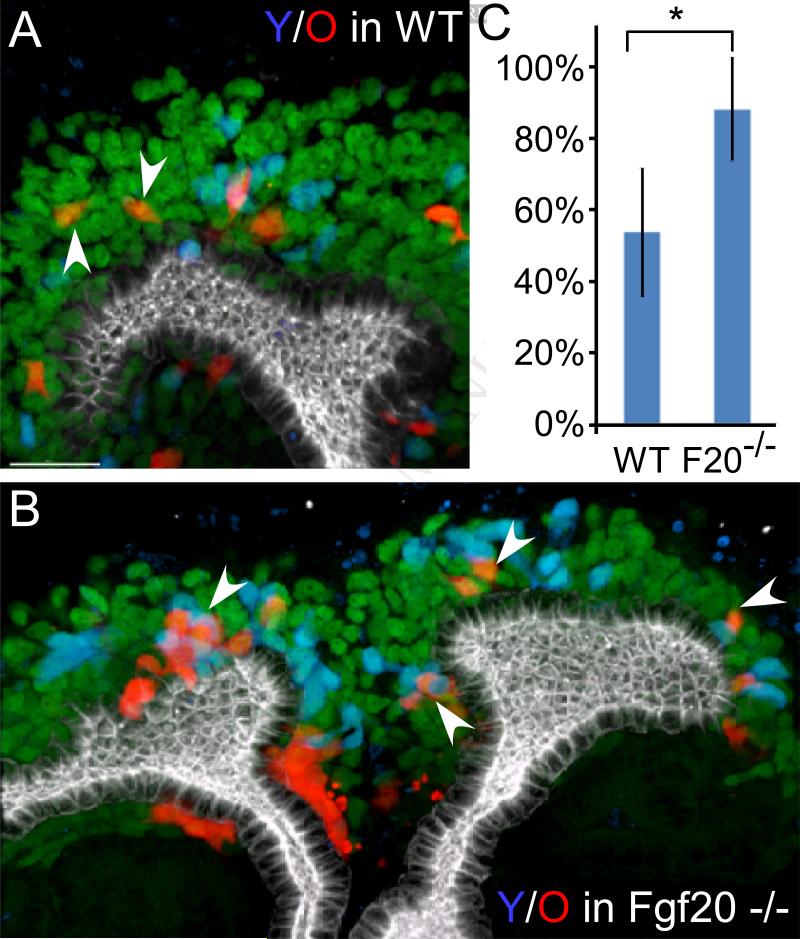
Figure 6. FGF20 signaling helps old cells to remain in the niche.
(A) Representative staining of a wild type niche showing only a few old cells in direct contact with co-injected young cells (arrowheads). (B) Representative staining of Fgf20−/− niche injected with wild type Six2+ P0 (red) and E12.5 (blue) progenitors. Old (red) Six2+ cells in contact with young (blue) co-injected cells marked with arrowheads. Scale bars, 30um. (C) Quantification of percentage (y-axis) of old cells in contact with the wild type co-injected cells in the wild type or FGF20−/− niches (x-axis) (p<0.001).
We conclude that most Cited1+ cells progressively lose their ability to remain in a young niche as they aged through a combination of slower self-renewal and faster exit rates (which could also be achieved by an increase in asymmetric cell division; (Itzkovitz et al., 2012)). Importantly, these results established that age-dependent changes within the “uninduced”, Cited1+ population precede the cessation of nephrogenesis. Interestingly, a third of the cells were still resident in the niche at least two days past the time they would have differentiated in situ, indicating that the decision of each progenitor to remain or exit the niche cannot depend only on its intrinsic properties.
Old Progenitors Can Engraft If They Associate With Young Cells
Our experimental paradigm combines old cells with young cells, whereas in situ they are surrounded with cells of their own age. Retention of old progenitors in the CM could reflect long-range stroma or UB signals, or the change in the composition of their immediate cellular environment. Whereas long-range signals will impact cells at any distribution, a neighborhood or “community effect” (Gurdon, 1988; Gurdon et al., 1993; Stüttem and Campos-Ortega, 1991) predicts that old cell within clusters that are insulated from the young environment will preferentially exit the niche. By contrast, single old cells that make contact with the young neighbors will likely behave like their neighbors and would be retained in the CM. To distinguish between these possibilities we examined the distribution of old cells remaining in the CM.
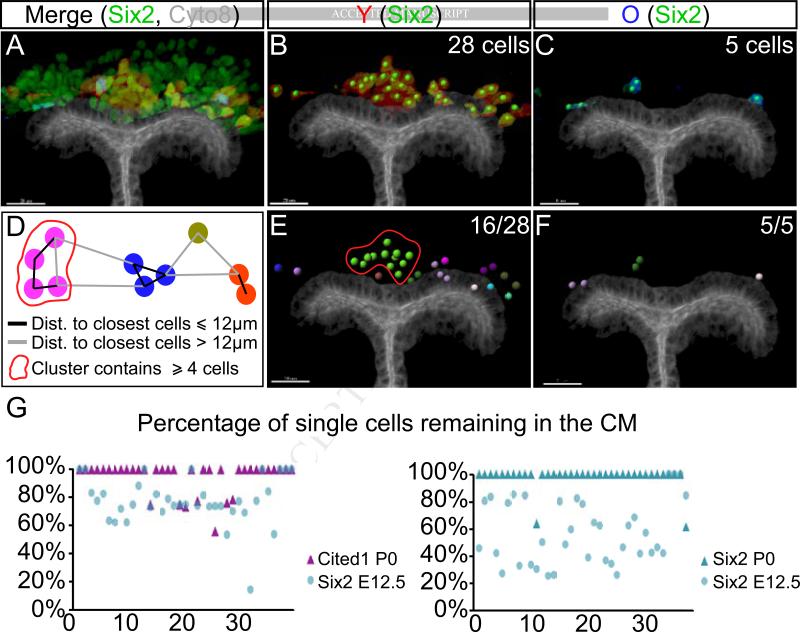
To analyze the neighborhood of the old cells remaining within the niche at the end of 4-day culture we calculated the distance between the nuclei of progenitors labeled with the same color. We binned cells into two groups: those within less than one cell diameter (~12μm) from at least one neighbor, and those with no neighbor within 12μm distance (Figure 4A-F). To account for cells undergoing mitosis, we arbitrarily decided that “groups” must contain 4 or more cells, all the rest were classified as “single” (Figure 4D). This analysis showed that in young and old Six2 cell co-injections, ~50% of injected young cells were found in clusters in 34 of 37 injected niches (Figure 4G). Strikingly, in 35 of 37 niches 100% of old Six2+ cells were categorized as a “single” (Figure 4G). Similarly, 100% of old Cited1 cells were classified as “single” in 30 out of 37 niches when co-injected with young Six2 cells. To determine if this represented random assortment of the injected cells, we performed logistic regression with two random effects (date and injection nested within date). The analysis established that the observed differences in distribution were non-random (p<0.002, Odds ratio 48.30 with 95% confidence interval of 12.048/166.667), and that the probability for an “all single” distribution for P0 and E12.5 cells was 86.86% and 13.11%, respectively (Least square means (LSM); Table S1). Increasing the cell diameter parameter to 15μM did not change the conclusions (Table S1).
Figure 4. Old cells remaining in the niche are surrounded by young neighbors.
(A-C) A representative example of the Imaris software analysis pipeline. Antibody staining labels CM (Six2, green) and UB (Cyto8, white); injected E12.5 tdTomato+ cells are red and P0 CFP+ cells are blue. Six2+ nuclei that are positive for either tdTomato (B) or CFP (C) are marked with bright green dots. (D) Criteria for segregating clusters; see text for detail. (E-F) Counting the number of “single” cells in this niche. 16 out of 28 young cells were single or in small (>4 cell) clusters, whereas 5 out of 5 old cells were single. (G) Quantification of the percentage of “single” Six2+ and Cited1+ cells to total cells by the end of 4-day culture in 37 injected niches (distance to closest cells ≤12μm). See also Figure S3 and Table S1.
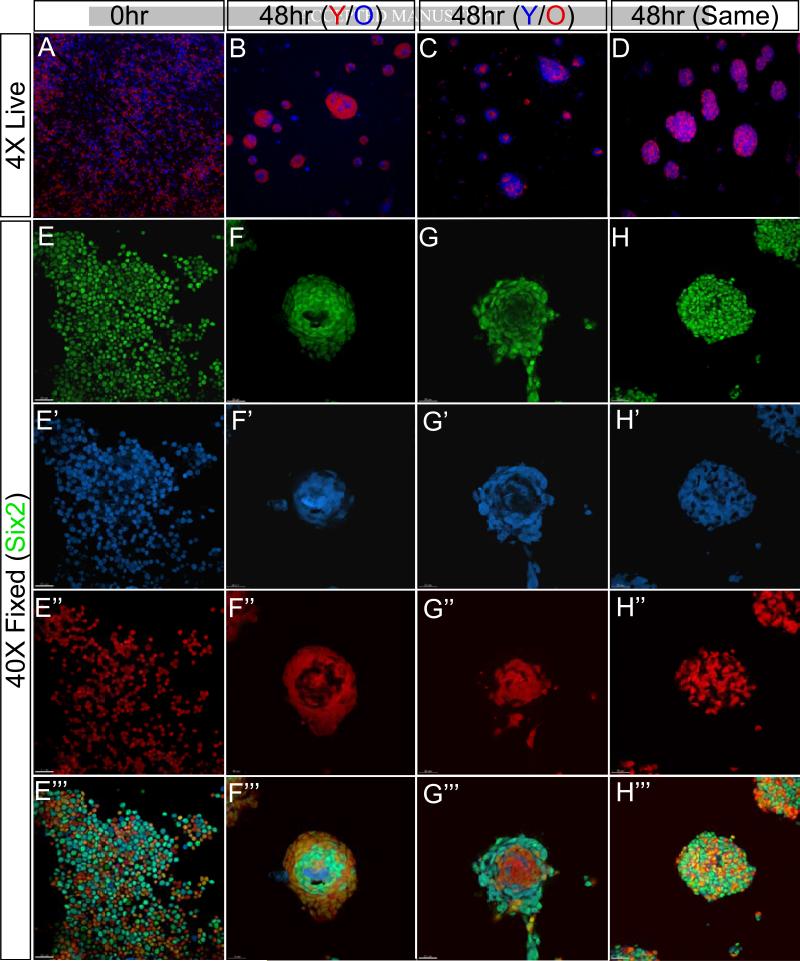
We next examined if differences in adhesion prevented old-old aggregation but allowed young-young and old-young association. We cultured progenitor cell mixtures (old-old, old-young and young-young progenitors at 1:1 ratio) in the presence of heparin, Fgf9 and BMP7 (Barak et al., 2012). After 48hr in vitro, the cultures were analyzed using anti-Six2 immunofluorescence (Figure 5). We observed that a well-mixed initial suspension of cells formed many small aggregates by the end of 48hrs. When the aggregates contained cells of the same age, the colored cells were mixed randomly (Figures 5D, H-H’”); aggregate size did not vary with age. In contrast, a mixture of young and old Cited1+ cells sorted young cells to the periphery around a cluster of old cells (Figures 5B, F-F’” and 5C, G-G’”), demonstrating a clear difference in age-dependent adhesion properties. It is important to note that a few young cells were located in the center and a few old cells at the periphery, indicating heterogeneity in adhesive properties. This result reinforced the idea that a community effect influenced the outcome, not the failure of old cells to associate with their own kind (Gurdon, 1988; Gurdon et al., 1993; Stüttem and Campos-Ortega, 1991).
Figure 5. Young and old progenitors display differential adhesion properties when cultured in vitro.
(A-D) A 1:1 mixture of FACS sorted E12.5 and P0 Six2+ cells imaged live with a fluorescent stereoscope immediately after plating on top of a filter (A) and 48hr culture with heparin, FGF9 and BMP7 (B-D). (E-H) High magnification image of fixed heterochronic co-cultures. Antibody staining of Six2 marked in green. Note that the initial culture was a monolayer of evenly distributed cells (E-E’”). By the end of 48hrs, all cells formed aggregates (F-F’” to H-H’”). Scale bar, 30μm.
The “long-lived” old cells may no longer proliferate, failing to produce any epithelial descendants. To address this we examined metanephroi co-injected with young and old cells after seven days in culture. We could detect old cells in the CM and their decedents in an S-Shaped body as well as more mature epithelia along the cortical-medullar axis (Figure S3). Thus, old progenitors in the CM are actively proliferating and producing differentiated descendants long after their original exit time.
We have previously demonstrated that Fgf20/9 are juxtacrine CM factors required to maintain nephron progenitors (Barak et al., 2012). We noticed that the number of cells containing Fgf20 transcripts decreased dramatically at P0. To determine if Fgf20 contributes to the niche retention signal, we co-injected wild type young and old nephron progenitors into Fgf20-null recipients. The mutant host niche environment did not change the fraction of old progenitors remaining in the CM but remarkably, 90% of the remaining progenitors maintained contact with at least one co-injected wild type young progenitor (Figure 6B), a behavior that is significantly less frequent in wild type host niche (Figure 6A and 6C, p<0.001). This experiment suggests that Fgf20 and/or additional factors produced by co-injected young wild type cells (but not Fgf20−/− host cells) are important for determining progenitor lifespan in the CM.
Single cell RNA sequencing reveals distinct, age-dependent pattern of gene expression
The age-dependent change in engraftment could reflect a decrease in the fraction of true “stem” cells in the old population or a gradual change in the self-renewal/differentiation capacity of many cells in the entire Cited1 population over time. To differentiate between these possibilities, we performed single-cell RNA-seq (scRNA-seq) analysis on E14.5, E18.5 and P0 Cited1 cells to reveal cellular heterogeneity that is often masked at the population level. To extend our analysis to E12.5 nephron progenitors we incorporated single-cell RNA-seq data derived from E12.5 Cited1+Six2+ nephron progenitors from whole kidneys (Brunskill et al., 2014) (see Supplemental Experimental Procedures for detail).
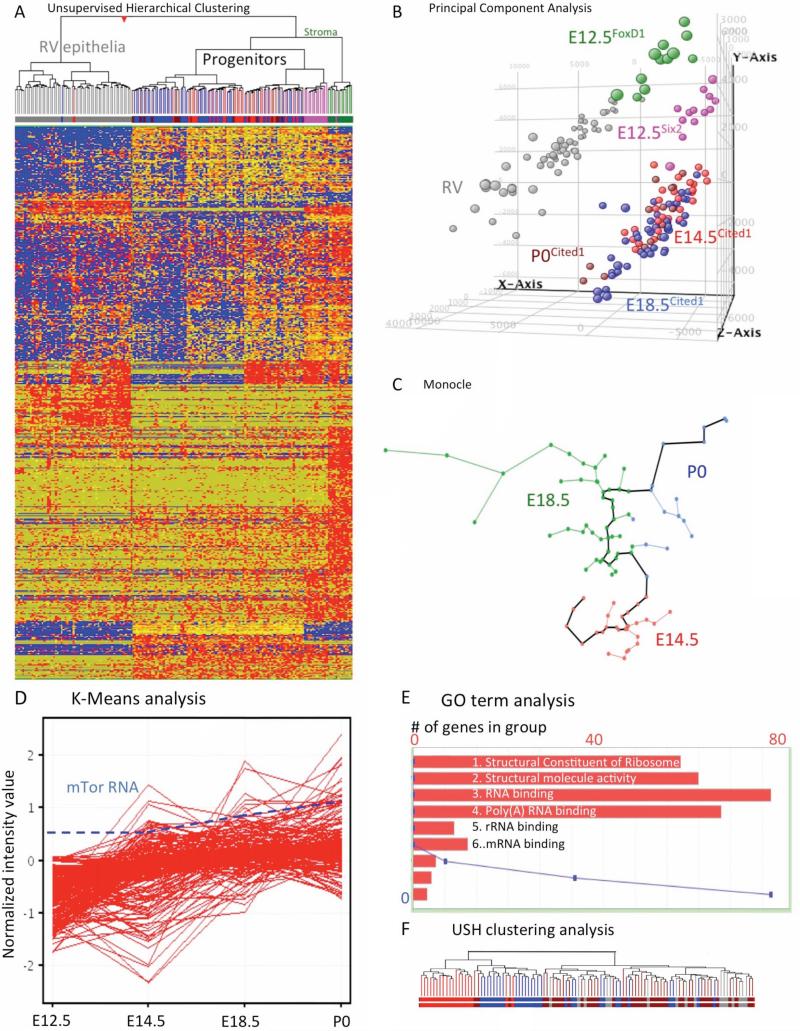
To identify the molecular changes that unfold during the aging of nephron progenitors, we used unsupervised clustering tool (Tamayo et al., 1999) to analyze the single cell expression profiles. The data did not support the presence of a true “stem” cell population that is found in all age groups, but instead an heterogeneous population with age-dependent progression towards an “old” signature” (Figure S4A). Interestingly, whereas there were no E12.5 progenitors and only a few E14.5 progenitor cells that resembled older progenitors, no P0 progenitors possessed a “young” transcriptional signature (Figure S4A). Next we performed a similar analysis with a larger dataset including E12.5 metanephroi scRNA-seq data (Brunskill et al., 2014). As shown in Figure 7A, all Cited1+ progenitors are distinguished from stroma (green) or RV epithelia (gray). E12.5 progenitors formed a distinct group, with the 91 Cited1+ progenitors displaying progressive change with age (Figure 7A, Figure S7A and Table S2 and S3). Principal component analysis (PCA, Figure 7B and Figure S7B), which uses an unsupervised algorithm to reduce the dimensionality of the original data by finding highly ranked components contributing to formation of distinct data groups (Hornquist et al., 2002; Liu et al., 2002; Peterson, 2002), distinguished Cited1 nephron progenitors from all other cells within the population. This suggests that the progenitor transcriptome is progressively evolving over time towards an “old” or epithelial-like signature, a process that occurs at different rates in individual cells. Collectively, this analysis not only identified age-dependent changes in gene expression among Cited1 nephron progenitors, it found heterogeneity within all age groups with some young Cited1+ progenitor cells already displaying an “old” signature, perhaps prior to exit from the niche.
Figure 7. Singe cell RNA-seq analysis of Cited1+ progenitors.
(A) A heat map depicting the expression of genes in Cited1+ progenitors (E12.5, E14.5, E18.5 and P0), stromal cells (FoxD1+, E12.5) and RV cells. Unsupervised clustering visualize the expression patterns of genes (E12.5 magenta; E14.5 red; E18.5 blue; P0 brown; Foxd1+ stroma green; RV grey). Red, blue and yellow intensities indicate high, low and intermediate expression levels, respectively. (B) Analysis of single-cell RNA-Seq data using principle component analysis (PCA) (C) The algorithm Monocle was utilized to organize the cells using “pseudo-time analysis” of E14.5, E18.5 and P0 Cited1+ cells. (D) Gene expression profile from K-means cluster #1 showing the increase of ribosomal biosynthesis genes during progenitors aging. A dashed line represents the concomitant increase in expression of mTOR mRNA. (E) Graphic illustration showing the top five GO terms from K-means cluster 1. The X-axis displays the number of genes in the specified GO term, the Y-axis plots the p-value as a blue line. (F) Hierarchical clustering of cells based on genes in K-means cluster #1 shows the segregation of progenitors into three major clusters: one containing only E12.5 progenitors, a cluster containing E12.5, E14.5 and a few E18.5 progenitors and a large cluster containing only E18.5 and P0 progenitors. See also Table S2-7.
Since the E12.5 progenitors were run on a different plate and formed a clearly distinct group, we performed age-dependent clustering of the single-cell data from E14.5, E18.5 and P0 nephron progenitors using Monocle, an algorithm that uses an independent component analysis (ICA) to predict temporal processes by ordering single-cell expression data into what is referred to as ‘pseudotime’ (Trapnell et al., 2014; Trapnell et al., 2013). As shown in Figure 7C and Table S4) Monocle ordered progenitors into five distinct groups based on gene expression and created a ‘pseudotime’ beginning at E14 and ending at P0. Collectively, these analyses confirmed that age-dependent gene expression trends are detected with multiple algorithms.
We utilized K-means cluster analysis to identify enriched biological pathways and processes in the E12.5 to P0 progenitors. A total of twelve different gene expression patterns were identified and each of these clusters was analyzed using gene set enhancement analysis tool (ToppGene.cchmc.org) with false discovery rate (FDR) cutoff of p<0.05 (Table S5, S6 and S7). One of the most statistically significant clusters was enriched in genes related to ribosome biogenesis (p<10−103), RNA binding (p<10−48), biological processes related to poly(A) RNA binding (p<10−44) and rRNA binding (p<10−14; Figure 7D and E, Table S5, S6 and S7). Accordingly, GO term analysis of the 328 organizing genes that Monocle used identified structural constituent of ribosome (p<10−9) and RNA binding (p<10−6) as the most statistically significant terms (Table S4).
Several computational methods identified the coordinated production of a significant number of ribosomal proteins and ribosomal RNAs (rRNA) as one possible contributing factor to aging in nephron progenitors. Increased ribosome biogenesis is reminiscent of elevated mTOR activity (Iadevaia et al., 2014). Indeed, the expression profile of mTor increased 1.5-fold as progenitors aged and positively correlated with increased ribosomal biosynthesis gene expression. To determine the degree of heterogeneity of ribosome biosynthesis present within the progenitors, we examined the ribosomal gene expression profile using unsupervised hierarchical clustering in E12.5, E14.5, E18.5 and P0 Cited1+ progenitors. This analysis segregated the progenitors into three major clusters: one consisting of only E12.5 progenitors, a second containing E12.5 and E14.5 progenitors, and a third containing E14.5, E18.5 and P0 progenitors (Figure 7F). Again, we observed young progenitors that cluster with old progenitors but not the converse. Thus, inclusion of E12.5 progenitors polarized the age-dependent gene expression trends and highlighted the level of heterogeneity present within nephron progenitors.
In addition to identifying gene clusters whose expression increases as progenitors age, we sought to identify those clusters in which there was progressive decline in gene expression (Table S5, S6 and S7). Four of the 12 clusters in our analysis met these parameters and the GO terms associated with these clusters were dominated genes related to mitotic cell cycle (p<10−18) and cell division (p<10−18). We examined each of these clusters further for GO terms related to extracellular matrix, given the significant differences we observed in adhesion between young and old progenitors. One of these clusters contained GO terms related to, extracellular matrix (p<10−3, not shown). Inspection of this cluster revealed several genes such as fibronectin leucine rich transmembrane protein 3 (flrt3), which has been implicated in modulating FGF signaling and homotypric adhesion (Bottcher et al., 2004; Karaulanov et al., 2006), and the secreted proteins fibronectin (Fn1) and thrombospondin-1 (Thbs1), an adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions. Fn1 and Thbs1 are progressively diminished as progenitors age. Conversely, Tnc (tenascin C) levels increase in aging progenitors. These and other ECM molecules may explain the differential sorting we reported (see Figure 5).
Discussion
Our understanding of the molecular and cellular mechanisms that control nephrogenesis is growing rapidly in recent years fields (Costantini and Kopan, 2010; Hendry et al., 2011; Kopan et al., 2014; Little and McMahon, 2012). Using a competitive transplantation assay and single cell RNA-seq analysis we discovered that intrinsic, age-dependent changes within the CM contribute to regulating CM lifespan. Young and old Cited1+ progenitors display different proliferation and exit rates; elevated mTor signaling, reduced Fgf20 signaling, and altered ECM could be among the drivers of age related change.
How do cell intrinsic changes affect a progenitor's choice to stay or exit the niche? Nephron progenitors could contain an internal “clock” that determined when they would exit the niche. Such a “clock” could be the manifestation of uncharacterized gene networks uncovered in our transcriptional analysis, or mechanism that counts cell cycle divisions, or dilution of inherited chromatin modifiers. Our data indicates that if such a clock exists, it can be easily reset: as many as 30% of the P0 cells remain in the niche and produce differentiating descendants for long as 7 days, or P7 in their original age. Investigators creating synthetic niches reached similar conclusion (Barak et al., 2012; Brown et al., 2015; Taguchi et al., 2014). Younger cells surrounded those old cells that remained in the niche past the subjective age of P3. It is important to keep in mind that unlike more typical stem/progenitor niches established by a separate population of support cells, the nephron progenitors are an integral part of their own niche (Barak et al., 2012). Cell intrinsic changes modulate both the environment and how a progenitor responds to the environment established in collaboration with other CM residents. Indeed, transplanting P0 and E12.5 progenitors cells into a host niche deficient in Fgf20 showed that the company of Fgf20 expressing cells improved retention in the niche.
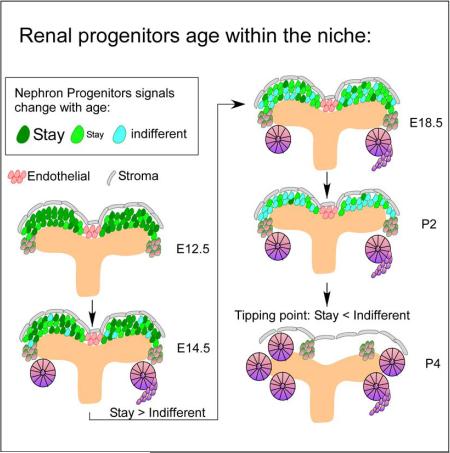
Combined with the detailed analysis of niche dynamics (Short et al., 2014) and the observation that randomly ablating nearly half the progenitors preserved the timing for secession of nephrogenesis (Cebrian et al., 2014), our observations led us to propose a testable model for how intrinsic changes and juxtacrine signaling could determine the timing of secession from nephrogenesis without evoking a clock (Figure 8). First, because not all cells change at the same rate, we assume that age-dependent intrinsic changes alter the ratio between niche-supportive “young” cells and niche-indifferent or disruptive “old” cells. The change from “young” to “old” occur progressively through development, increasing the fraction of “old” cells with time. Further, we assume that a critical number of cell-cell contacts with young neighbors (involving juxtacrine FGF signaling) are required to maintain niche integrity. Lastly, we assume that after P1, the CM reaches a tipping point where enough “old” cells have formed, diluting the retention signals (or producing a critical amount of the exit signal) creating an exit-reinforcing environment that results in arcading-like, en mass differentiation of all remaining progenitors to nephrons. This model could explain why randomly killing half the cells does not change the timing, why some but not all Fgf9+/–; Fgf20−/− progenitors have accelerated niche exit, why old cells can be retained in a young niche as long as they are in contact with FGF20 expressing neighbors, and why old cell aggregates cannot be found after 4-5 days in culture. The model predicts that the choice of a nephron progenitor to stay or exit the niche requires reinforcement by the local environment. Whether the young niche “resets” key features of the “old” signature (perhaps by modulation of SMAD signaling, (Brown et al., 2015)) or simply delays their exit remains to be determined.
Unbiased transcriptional profiling of Cited1+ nephron progenitors at the single cells level further supported this model. When Cited1+ cells were analyzed, the 63 older Cited1+ cells did not seem to harbor a sub-population of “young” stem cells: no P0 cell displayed a signature resembling the E12.5/E14.5 signature (Figure S4A). Conversely, young Cited1+ populations contain some cells already resembling E18.5 or P0 cells. Importantly, the genes/pathways differentiating young from old include pathways with known effects on organismal and stem cell aging, including mTor and the translational machinery (Chen et al., 2009; Lamming et al., 2013; Signer et al., 2014). These data establish that age-dependent intrinsic transcriptional changes occur within the Cited1+ progenitor population, and that these changes precede the cessation of nephrogenesis. Additional finding revealed other potential regulators of the progenitor aging process including RNA binding proteins, adhesion and ECM proteins and chromatin modifiers, suggesting possible roles for translation and epigenetic control in modulating these decisions (Chen et al., 2011; Rosenberg et al., 2011).
Our results offers important insights to a long-standing question in the kidney field: are nephron progenitors true stem cells, or do they more closely resemble a transient amplifying population? The finding that most nephron progenitors only have a finite lifespan even when placed in a supportive environment, and that no age-resistant “stem” signature was present in all age groups, suggests that the majority of CM cells are transient amplifying cells and argues against a model where nephron stem cells are forced to differentiate by the end of nephrogenesis. However, survival of 30% “old” cells in a young environment or in a synthetic niche (Brown et al., 2015) is reminiscent of the emerging concept that “stem” states are niche-induced and cells can retain this state as long as they are within the niche (Tetteh et al., 2014).
We note that our experimental results and model do not rule out possible roles of extrinsic triggers, such as changes in niche topology and birth, which could work in parallel to or even trigger CM-intrinsic changes in regulating progenitor lifespan. An important question for future studies will be to identify which interventions can delay the tipping point, permitting the niche to produce more nephrons. The ultimate test will be translating the insight stemming from analyzing the mouse metanephros to the human kidney, which creates the majority of its nephrons in a post-arcading nephron production period not present in mice.
In conclusion, this study characterized the Cited1+ sub-population of nephron progenitor cells. We described previously unknown age-dependent cell intrinsic changes and proposed a model based on inter-CM cell-cell contacts to explain nephron progenitor lifespan in situ. The functional study and unbiased transcriptome analysis both support the conclusions and provide important insights into the temporal control of he niche lifespan during nephrogenesis. Some of the players uncovered in this initial characterization effort may have important functions in regulating progenitor lifespan and several promising candidate molecules and pathways discussed herein remain to be functionally tested in future studies. The new protocol established here and the data within will accelerate this process of discovery by the community interested in deciphering the role of various candidates in individual CM cells while preserving an intact niche.
Experimental Procedures
Animals
All mice were maintained in the Cincinnati Children's Hospital Medical Center (CCHMC) animal facility according to animal care regulations; Animals Studies Committee of CCHMC approved the experimental protocols (protocol 3D05041). See Supplemental Information for detailed strain information.
Kidney organ culture and FACS sorting of nephron progenitors
Mouse metanephric organ cultures were performed as described by Barak et al. (Barak and Boyle, 2011). For FACS sorting, E12.5, E14.5, E18.5 and P0 kidneys were dissected from control (no transgene) or Six2TGC+/tg or Cited1CreERT2+/tg mice containing either Rosa tdTomato or Actin-CFP reporter. Sorting was performed on BD FACSdiva flow cytometer. Gating was implemented based on negative control profiles to select for live, single cell that was GFP+tdTomato+ or GFP+CFP+ double positive. Sorted cells were collected in kidney media with 5% FBS and, depending on the downstream application were either (a) placed in ice cold kidney media and transported to the injection rig (b) used for RNA-seq, or (c) placed onto trans-well filters in organ culture dishes containing 37°C kidney media supplemented with Heparin (1mg/ml; Sigma), 8.6nM FGF9 (PeproTech), 50ng/ml BMP7 (R&D Systems) for in vitro culture
Transplantation of nephron progenitors
FACS sorted cells plated on the trans-well filter are loaded and concentrated into the capillary needle of the microinjector (see Supplemental Experimental Procedures). The injection set includes a fluorescent stereomicroscope (Leica, FM-10), a manual microinjector (IM-6, Narishige) and a micromanipulator (Narishige) for holding and adjusting the position of the microinjector. Each metanephroi were injected at 2-4 different niches. Injected kidneys were then cultured for another 4-7 days with media changed once every 24hrs.
Immunostaining and confocal imaging
Recipient kidneys together with the trans-well filter were fixed in 4% PFA for 6hrs, washed with PBS and blocked in PBS-BB (PBS containing 1% BSA, 0.2% powdered skim milk and 0.3% Triton-100) overnight at 4°C. After blocking, kidney cultures were incubated with primary antibodies (see Supplemental information for antibodies and dilutions used) in the PBS-BB for 48hrs to allow sufficient penetration. Primary antibodies were washed off for 48hrs at 4°C followed by overnight secondary antibody incubation and another 48hr of wash at 4°C. Stained kidneys together with the trans-well filter were mounted with ProLong® Gold Antifade Reagent (Life Technologies) and allowed to cure overnight before imaging. Confocal imaging was performed on a Nikon A1Rsi inverted confocal microscope with fixed and stained kidney cultures on the glass slide. For each injection site, samples were imaged on a 40X water-immersion lens using a pinhole of 1.2μm on all channels at the resolution of 1024X1024. The z-stacks were taken with an interval of 1.5μm over 50-80μm to produce a stack of consecutive images with 1/3 overlaps.
Imaris analysis
To count the number of injected progenitors remaining in the cap mesenchyme, z-stack images were visualized with Imaris (version 7.2, Bitplane AG). For each injection point, the volume of entire Six2+ CM region containing injected cells and the UB tip is manually captured in the software. Next, Six2+ nuclei, tdTomato and CFP cells in this region are masked in two independent volumes (smoothing 0.62 M, background subtraction with an assumed object diameter of 2.33 M, automatic quality threshold, filter objects by number of voxels 200-500) The number of Six2+; tdTomato+ and Six2+; CFP+ cells is determined by the Imaris™ ‘spots function’ algorithm with the following parameters: XY diameter 6um, background subtraction with automatic threshold applied. The fraction of red and blue cells remaining in the niche of the total (red+blue) is presented as percentage.
Statistical analysis
The percentage of red or blue cells out of total co-injected in multiple sites on different days were combined to derive the average percentage and standard deviation (SD). A two-tailed student t-test was used to calculate the p value. Error estimates of all pooled data were calculated as standard deviation (SD). The percentage of single cells vs groups was calculated separately for cells of each color in individual injected niche. To determine if the observed distribution was due to random assortment of injected cells we performed logistic regression with two random effects (date and injection nested within date). See Table S1 for detailed statistical analysis.
Single cell RNA sequencing and data analysis
Single nephron progenitors obtained from FACS sorting were loaded onto a Fluidigm C1 Chip by captured using a 10uM-17uM filter using the single-cell AutoPrep system and visually inspected using a fluorescent microscope to confirm the presence of a single cell in each well, and to note the location of tdTomato cells in the microfluidic device. cDNA libraries were generated on the same platform using Clontech UltraLow Smarter Amplification chemistry. The cDNA was converted into sequencing libraries using the llumina NexteraXT DNA Sample Preparation kit. A total of 91 single cells were processed by the Fluidigm C1 and single-end 50 sequencing was carried out with Hiseq2500 to an average depth of ~4 million reads per cell. scRNA-seq data was processed using Illumina sequence analysis software and the CCHMC barcode deconvolution pipeline and aligned to the mouse genome (MM10) using cufflinks (Trapnell et al., 2012), normalized, and filtered using a stringent cutoff of >10RPKM in at least 5 of 91 cells to focus on abundant transcripts. A list was filtered by one-way ANOVA with a p>0.5 cut-off. The age of each cell was determined by the unique sequences of the reporter (see supplemental information). Unsupervised clustering was performed using the self-organizing (SOM) map on the GeneSpring platform. Supervised clustering with one-way ANOVA was also used to find genes expressed differently across age groups. The data Accession number for this data is GSE66202.
Mathematical modeling of the nephron progenitor cells
We assume the cell number (x) is proportional to the proliferation; exit and death rates. Thus, the differential equation describing the cell number can be written as
We can solve the differential equation
If x is used for E12.5 Six2 cells and y for Cited1 cells,
Because the same number of Six2 and Cited1 cells were injected (x0=y0):
We solved this equation first assuming that exit rates were fixed in cells of different ages (supplemental information). We next let k=kpr-ke (assuming negligible death rate) to derive a new equation
to predict the exit rate difference (key-kex) based on known proliferation rates and endpoint ratio at day 4. Two possible results can exist depending on whether average cell cycle times at E14.5 are longer or shorter than 1 day (supplemental information).
Supplementary Material
Highlights.
Temporal characterization of Cited1+ progenitor mRNA at the single cell level
Stem cell transplantation assay for metanephric nephron progenitors
Engraftment potential of progenitors declines with age, reflects a community effect
Young cells can extend the lifespan of individual old progenitors in the niche
Acknowledgements
We thank Drs. Andrew McMahon, David Ornitz, Frank Costantini, Lilianna Solnica-Krezel and Timothy Holy for sharing reagents, equipment and animals; Scott Boyle and Hila Barak for expert advice and discussion. Tao Shen, Ahu Turkoz, Nicholas Fahey with animal husbandry. The Siteman Flow Cytometry Core at Alvin J. Siteman Cancer Center at Washington University School of Medicine, and the Research Flow Cytometry Core at CCHMC , which provided cell-sorting service. We thank Dr. Matthew Koffron and Michael Muntifering (Confocal Imaging Core) and Matthew Batie (Clinical Engineering) for training and discussion. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant P30 CA91842. Grant from the March of Dimes (DMO) and NIH support grants, P30DC04665, P30DK052574 and P30AR057235 supported the production of mice. RK, SC and were supported in part by a grant from the National Institutes of Diabetes and Digestive and Kidney Disease (DK066408), by the Alan A. and Edith L. Wolff Chair for Developmental biology (to RK), and by the William K. Schubert Chair for Pediatric Research (to RK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
SC developed the assay, performed most experiments, contributed to their design and co-wrote the manuscript. EWB contributed to several figures and helped write the manuscript. EB, SSP, PJD, NS and BJS performed different aspects of the single cell analysis. CIH and TZ performed the modeling and analysis of the single cell data to extract Exit rates. RK contributed to the concept, to experiential design, data interpretation, and co-wrote of the manuscript.
References
- Al-Awqati Q, Goldberg MR. Architectural patterns in branching morphogenesis in the kidmey Kidney International. 1998;54:1832–1842. doi: 10.1046/j.1523-1755.1998.00196.x. [DOI] [PubMed] [Google Scholar]
- Barak H, Huh S-H, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschké P, Salomon R, Antignac C, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Developmental Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Shiell AW, Barker ME, Law CM. Growth in utero and blood pressure levels in the next generation. Journal of hypertension. 2000;18:843–846. doi: 10.1097/00004872-200018070-00004. [DOI] [PubMed] [Google Scholar]
- Bertram JF, Douglas-Denton RN, Diouf B, Hughson MD, Hoy WE. Human nephron number: implications for health and disease. Pediatr Nephrol. 2011;26:1529–1533. doi: 10.1007/s00467-011-1843-8. [DOI] [PubMed] [Google Scholar]
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Muthukrishnan SD, Guay JA, Adams DC, Schafer DA, Fetting JL, Oxburgh L. Role for compartmentalization in nephron progenitor differentiation. Proc Natl Acad Sci U S A. 2013;110:4640–4645. doi: 10.1073/pnas.1213971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AC, Muthukrishnan SD, Oxburgh L. A Synthetic Niche for Nephron Progenitor Cells. Dev Cell. 2015;34:229–241. doi: 10.1016/j.devcel.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Lai HL, Jamison DC, Potter SS, Patterson LT. Microarrays and RNA-Seq identify molecular mechanisms driving the end of nephron production. BMC Dev Biol. 2011;11:15. doi: 10.1186/1471-213X-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Park JS, Chung E, Chen F, Magella B, Potter SS. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141:3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian C, Asai N, D'Agati V, Costantini F. The number of fetal nephron progenitor cells limits ureteric branching and adult nephron endowment. Cell Rep. 2014;7:127–137. doi: 10.1016/j.celrep.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bellew C, Yao X, Stefkova J, Dipp S, Saifudeen Z, Bachvarov D, El-Dahr SS. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J Biol Chem. 2011;286:32775–32789. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman HA, Lai HL, Patterson LT. Cessation of renal morphogenesis in mice. Dev Biol. 2007;310:379–387. doi: 10.1016/j.ydbio.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry C, Rumballe B, Moritz K, Little MH. Defining and redefining the nephron progenitor population. Pediatr Nephrol. 2011 doi: 10.1007/s00467-010-1750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornquist M, Hertz J, Wahde M. Effective dimensionality of large-scale expression data using principal component analysis. Bio Systems. 2002;65:147–156. doi: 10.1016/s0303-2647(02)00011-4. [DOI] [PubMed] [Google Scholar]
- Hoy WE, D Hughson M, Singh GR, Douglas-Denton R, Bertram JF. Reduced nephron number and glomerulomegaly in Australian Aborigines: A group at high risk for renal disease and hypertension. Kidney Int. 2006;70:104–110. doi: 10.1038/sj.ki.5000397. [DOI] [PubMed] [Google Scholar]
- Iadevaia V, Liu R, Proud CG. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol. 2014;36:113–120. doi: 10.1016/j.semcdb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Itzkovitz S, Blat, Irene C, Jacks T, Clevers H, van Oudenaarden A. Optimality in the Development of Intestinal Crypts. Cell. 2012;148:608–619. doi: 10.1016/j.cell.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaulanov EE, Bottcher RT, Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006;7:283–290. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 2011;138:1247–1257. doi: 10.1242/dev.057646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G, Zimmer G, Mall G, Ritz E, Amann K. Nephron Number in Patients with Primary Hypertension. N Engl J Med. 2003;348:101–108. doi: 10.1056/NEJMoa020549. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Chen S, Little M. Nephron Progenitor Cells: Shifting the Balance of Self-Renewal and Differentiation. Curr Top Dev Biol. 2014;107C:293–331. doi: 10.1016/B978-0-12-416022-4.00011-1. [DOI] [PubMed] [Google Scholar]
- Kopan R, Cheng HT, Surendran K. Molecular Insights into Segmentation along the Proximal-Distal Axis of the Nephron. J Am Soc Nephrol. 2007;18:2014–2020. doi: 10.1681/ASN.2007040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackland DT, Bendall HE, Osmond C, Egan BM, Barker DJ. Low birth weights contribute to high rates of early-onset chronic renal failure in the Southeastern United States. Archives of internal medicine. 2000;160:1472–1476. doi: 10.1001/archinte.160.10.1472. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Sabatini DM, Baur JA. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Invest. 2013;123:980–989. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, McMahon AP. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Zhang Y, Gehan E, Clarke R. Block principal component analysis with application to gene microarray data classification. Statistics in medicine. 2002;21:3465–3474. doi: 10.1002/sim.1263. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Yu J, Kobayashi A, McMahon AP. High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population. Dev Biol. 2009;333:312–323. doi: 10.1016/j.ydbio.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LE. CLUSFAVOR 5.0: hierarchical cluster and principal-component analysis of microarray-based transcriptional profiles. Genome Biol. 2002;3:SOFTWARE0002. doi: 10.1186/gb-2002-3-7-software0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MM, Gomez AH, Abitbol CL, Chandar JJ, Duara S, Zilleruelo GE. Histomorphometric analysis of postnatal glomerulogenesis in extremely preterm infants. Pediatric and developmental pathology : the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society. 2004;7:17–25. doi: 10.1007/s10024-003-3029-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg SL, Chen S, McLaughlin N, El-Dahr SS. Regulation of kidney development by histone deacetylases. Pediatr Nephrol. 2011;26:1445–1452. doi: 10.1007/s00467-011-1796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumballe BA, Georgas KM, Combes AN, Ju AL, Gilbert T, Little MH. Nephron formation adopts a novel spatial topology at cessation of nephrogenesis. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder MF. Interpretation of birth weight data: a note of caution. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2008;52:807. doi: 10.1053/j.ajkd.2008.06.030. author reply 807-808. [DOI] [PubMed] [Google Scholar]
- Schreuder MF, Langemeijer ME, Bokenkamp A, Delemarre-Van de Waal HA, Van Wijk JA. Hypertension and microalbuminuria in children with congenital solitary kidneys. Journal of paediatrics and child health. 2008;44:363–368. doi: 10.1111/j.1440-1754.2008.01315.x. [DOI] [PubMed] [Google Scholar]
- Short KM, Combes AN, Lefevre J, Ju AL, Georgas KM, Lamberton T, Cairncross O, Rumballe BA, McMahon AP, Hamilton NA, et al. Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell. 2014;29:188–202. doi: 10.1016/j.devcel.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelloh C, Allen KP, Mattson DL, Lerch-Gaggl A, Reddy S, El-Meanawy A. Prematurity in mice leads to reduction in nephron number, hypertension, and proteinuria. Translational research : the journal of laboratory and clinical medicine. 2012;159:80–89. doi: 10.1016/j.trsl.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MR, Gubhaju L, Moore L, Kent AL, Dahlstrom JE, Horne RS, Hoy WE, Bertram JF, Black MJ. Accelerated maturation and abnormal morphology in the preterm neonatal kidney. J Am Soc Nephrol. 2011;22:1365–1374. doi: 10.1681/ASN.2010121266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Farin HF, Clevers H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.