Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators that target specific mRNAs for repression and thus play key roles in many biological processes, including insect wing morphogenesis. miR-2 is an invertebrate-specific miRNA family that has been predicted in the fruit fly, Drosophila melanogaster, to be involved in regulating the Notch signaling pathway. We show here that miR-2 plays a critical role in wing morphogenesis in the silkworm, Bombyx mori, a lepidopteran model insect. Transgenic over-expression of a miR-2 cluster using a Gal4/UAS system results in deformed adult wings, supporting the conclusion that miR-2 regulates functions essential for normal wing morphogenesis. Two genes, abnormal wing disc (awd) and fringe (fng), which are positive regulators in Notch signaling, are identified as miR-2 targets and validated by a dual-luciferase reporter assay. The relative abundance of both awd and fng expression products was reduced significantly in transgenic animals, implicating them in the abnormal wing phenotype. Furthermore, somatic mutagenesis analysis of awd and fng using the CRISPR/Cas9 system and knock-out mutants also resulted in deformed wings similar to those observed in the miR-2 overexpression transgenic animals. The critical role of miR-2 in Bombyx wing morphogenesis may provide a potential target in future lepidopteran pest control.
Keywords: CRISPR/Cas9, Gal4/UAS, miR-2 cluster, transgene, wing disc
Introduction
Insects are the only class of winged invertebrates and this is a key factor contributing to their abundance and distribution, and their impact on human welfare.1 An understanding of the molecular basis of wing morphogenesis could provide a starting point for the development of novel pest management strategies. MicroRNAs (miRNAs) are a class of endogenous, non-coding, single-stranded RNAs, ∼22 nucleotides (nt) in length, that regulate gene expression at the post-transcription level by binding to the 3′-end untranslated regions (UTR) of their mRNA targets, leading to their degradation, decreased stability or translational inhibition. We propose that a functional analysis of the roles of miRNAs in wing morphogenesis could provide the basis for developing tissue-specific developmental defects for controlling holometabolous insects of agricultural and medical significance.
Several miRNAs have been implicated in wing development of the fruit fly, Drosophila melanogaster.2 For example let-7 mutant flies have smaller wings,3 and the bantam miRNA can affect the stability of the dorsal-ventral affinity barrier in the wing disc.4 miR-9a-deficient mutants have wing margin defects, whereas their overexpression results in a severe loss of sensory organ precursors.5 miRNA iab-4 regulates a homeotic transformation of halters to wings.6 miR-315 and miR-8 can regulate Wingless signaling,7,8 while miR-1 can regulate Notch signaling.9 However, the role of miRNAs in wing development of in non-drosophilid insects is largely unknown.
Sequence analyses show that some mature miRNAs are conserved phylogenetically in different species and are often clustered in the genome.10,11 miR-2 comprises a large invertebrate-specific family with most members clustered in the genome that is conserved in insects.12 Members of miR-2 in D. melanogaster have been predicted to regulate the Notch signaling pathway.13,14 The Notch pathway in the fruit fly is involved in the determination of wing vein cells and patterning of the wing margins.15,16 Furthermore, miRs-2/13 regulates embryonic development by repressing the proapoptotic factors hid, grim, reaper and sickle.17,18 This linkage of cell death and abnormal wing phenotypes is a key feature of many mutants in D. melanogaster.19,20 It remains to be verified in other holometabolous insects.
Genomic mapping of known miRNAs has enabled identification of orthologs in a number of species, including the silkworm, Bombyx mori.21-24 More than 487 miRNAs have been identified in B. mori (miRBase release 21, June 2014), but their functions are only beginning to be understood. We show here that miR-2 and their target genes, abnormal wing disc (Bmawd) and fringe (Bmfng), mediate wing morphogenesis in this lepidopteron model insect. Silkworms exhibiting severe wing abnormalities were produced by suppressing Bmawd or Bmfng expression products using CRISPR/Cas9-mediated genome engineering. The phenotypes of these mutant silkworms were indistinguishable from those produced by transgenic over-expression of miR-2.
Results
Ectopic overexpression of a miR-2 cluster induces deformed wings
A search of the silkworm database (http://sgp.dna.affrc.go.jp/KAIKObase/) revealed a miR-2 cluster located on chromosome 1 comprising 5 miRNAs, miR-2a-1, miR-13a, miR-13b, miR-2a-2 and miR-2b, with similar seed sequences (Fig. 1A). A Gal4/UAS transgenic system was established to over-express miR-2 to probe its functions in vivo. An A3-Gal4 activator line25-27 was used to drive ubiquitously an effector UAS line that contained a pre-miR-2 cluster 1004 base-pairs (bp) in length flanking genomic DNA sequences (Fig. 1A). The mCherry open reading frame (ORF) was placed at the 5′-end of the pre-miR-2 sequence as a marker.25 Transgenic animals (n = 32) were screened by fluorescence microscopy25 (Fig. S1A–C), and genomic insertion sites identified using inverse-PCR (Fig. S2). The UAS-miR-2 line crossed with A3-Gal4 (Fig. 1A) animals, and their offspring (A3 > miR-2) show expression of both EGFP and mCherry (Fig. S1D–F). Quantitative gene amplification of transcripts (QPCR) of miR-2 in samples derived from the epidermis, fat body, mid gut and wing discs of wandering-stage (3 days prior to pupation) hybrid larvae revealed that miRs-2/13 transcript levels were significantly higher (P < 0.001) throughout when compared to control animals (UAS-miR-2) (Fig. 1B).
Figure 1.
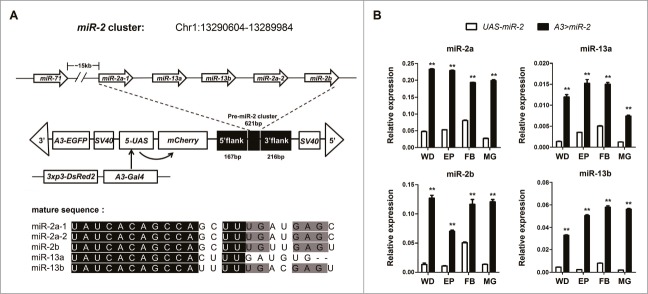
The miR-2 cluster in Bombyx mori, transgene structures and abundance measurements of miRs-2/13. (A) Schematic representations of the genomic organization of the miR-2 cluster, the miR-2 over-expression vector UAS-miR-2 and Gal4 vector pBacA3-Gal4. The members of miR-2 family (miR-2a-1, miR-13a, miR-13b, miR-2a-2 and miR-2b) cluster in B. mori with miR-71, an evolutionarily unrelated miRNA.12 (B) miRs-2/13 expression levels determined by QPCR in the tissues of wandering-stage transgenic and control larvae. mRNA abundance of A3>miR-2 was significantly higher than control UAS-miR-2, miRs-2/13. WD- wing disc, EP- Epidermis, FB- Fat body, MG- Mid gut. The asterisks (* or **) indicate the significant differences (P < 0.05 or P < 0.01, respectively) compared with the relevant control with a 2-tailed t-test. Error bars depict ± SEM.
The silkworm has a pair of meso- and meta-thoracic discs located laterally in larvae.28 These discs evert during the prepupal stage of metamorphosis and the nascent wings form outside the pupal body.29,30 The wing buds expand and inflate during adult eclosion and give rise to the corresponding fore- and hind-wings of the adult.28 All transgenic animals in the hybrid A3 > miR-2 line developed to the adult stage supporting the conclusion that the ubiquitous overexpression of miR-2 did not cause lethality in the subadult stages. However, 80% (n = 80) of A3 > miR-2 heterozygotes moths had abnormal wings characterized by folding, curling and small size (Fig. 2). In addition, wing discs of late 5th instar A3>miR-2 larvae were small and appeared poorly developed with veins that failed to extend from the base of the discs and lacked tracheal invasion (Fig. 2). Adult hybrid animals had disordered and atrophied veins. Furthermore, both larval and adult wing margins were irregular in morphology.
Figure 2.
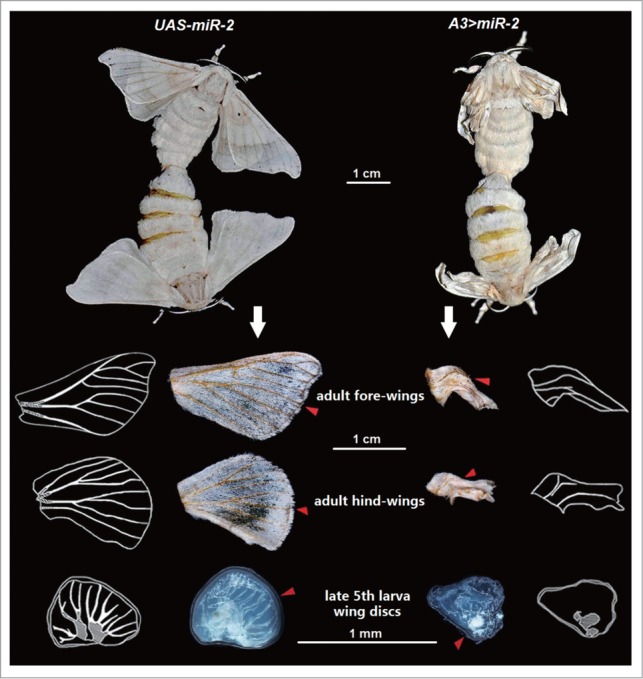
Effect of miR-2 overexpression in Bombyx mori. The A3 > miR-2 silkworms (right column) show abnormal wing phenotypes after miR-2 overexpression, while the control group (left column) UAS-miR-2 are normal. The wing veins of A3 > miR-2 fail to extend at the late 5th larval instar stage (red arrowhead) and the wing margin is irregular. The fore- and hind-wings of A3 > miR-2 moths are curled with disordered and atrophied veins (red arrowhead). Schematic diagrams show the wing margin and wing vein phenotypes.
miR-2 targets Bmawd and Bmfng to regulate wing morphogenesis
Three miRNA target-prediction tools, miRanda, PITA and microTar,25,31,32 were used in combination to identify 113 putative miR-2 family targets in B. mori (Table S1). Since the over-expression of the miR-2 cluster produced the abnormalities in wing disc development, we focused on 2 putative target genes, Bmawd and Bmfng (Table S2), which are well-known regulators in Notch signaling for insect wing development.
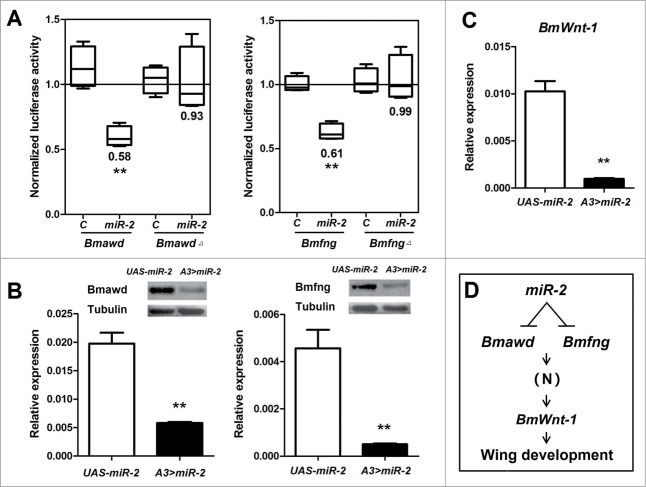
miR-2 reduced reporter activity up to ∼42% and ∼39% compared to controls using the wild-type Bmawd and Bmfng 3′-end UTRs, respectively, in a Dual Luciferase Reporter Assay in HEK293T cells (Fig. 3A). These findings were supported further by observing no effect on reporter activity when using forms of Bmawd and Bmfng mutated in the seed-binding regions of the respective 3′-end UTRs (Fig. 3A; Fig. S3).
Figure 3.
Bmawd and Bmfng are the direct targets of miR-2. (A) miR-2 targets the Bmawd and Bmfng 3′-end UTR in vitro. The firefly luciferase activity was normalized to renilla luciferase activity and then normalized to the median of the control group (the line in the box and the number below the box show the median). Bmawd△ and Bmfng△ defined as forms of Bmawd and Bmfng mutated in the seed-binding regions of the respective 3′-end UTRs. (B) Over-expression of the miR-2 cluster in vivo results in a decrease of the Bmawd (71%) and Bmfng (89%) at transcript levels and this is consistent with protein levels. (C) The transcript level of the downstream effector gene, BmWnt-1, is reduced by 91%. The asterisks (* or **) indicate the significant differences (P < 0.05 or P < 0.01, respectively) compared with the relevant control with a 2-tailed t-test. Error bars depict ± SEM. (D) Model showing regulation of miR-2 in B. mori. miR-2 represses Bmawd and Bmfng, which then affects the expression of BmWnt-1 by Notch signaling (N) leading to the wing development.30,33
QPCR and immuno-blot analyses confirmed that overexpression of miR-2 reduced significantly Bmawd and Bmfng transcript and protein levels in A3 > miR-2 wandering-stage larval wing discs when compared to controls (Fig. 3B). Additionally, the transcript abundance of BmWnt-1, which is a B. mori homolog of wingless whose function is required later in the wing development morphogenetic pathway30,33 was reduced 91% (Fig. 3C, D).
Loss-of-function of Bmawd or Bmfng causes wing defect phenotypes
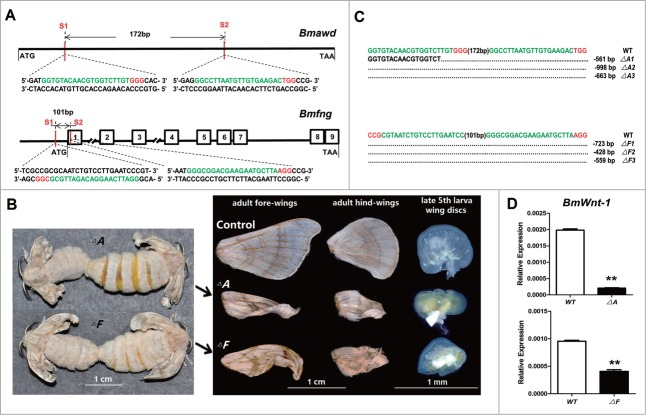
The CRISPR/Cas9 system was used to produce separate loss-of-function mutations in the Bmawd (△A) and Bmfng (△F) loci.34 Two 23-bp sgRNA-targeting sites (S1 and S2) were identified by screening the Bmawd or Bmfng ORF followed the 5′-GG-N18-NGG-3′ rule34-36 (Fig. 4A). Bmawd lacks an intron and the interspace fragment between S1 and S2 was 172 bp in length (Fig. 4A). The Bmfng gene has 9 exons, and its S1 site is localized to the antisense strand of the 5′-end UTR and S2 is localized in exon-1 with a 101 bp interspace fragment between them (Fig. 4A). SgRNA-1 and SgRNA-2 for each gene were co-injected separately with Cas9 mRNA into 480 preblastoderm embryos as described previously.34 Of these, 262 (55%, △A) and 235 (49%, △F) eggs hatched, and 201 △A and 197 △F larvae survived to the adult stage. Knockout of the same genes in Drosophila caused lethality,37,38 but less lethal mutations in Bombyx. A total of 61% △A (n = 122) and 40% △F (n = 78) moths showed abnormal wings identical to those observed in the A3 > miR-2 line (Fig. 4B). PCR-based mutagenesis analysis revealed that targeting sites and their interspace fragments were deleted (Fig. 4C). BmWnt-1 transcripts also were reduced in mRNA extracted from these wings (Fig. 4D).
Figure 4.
Abnormal wing phenotypes caused by Bmawd or Bmfng mutations in Bombyx mori. (A) schematic diagram of the sgRNA-targeting sites (red vertical line). The target site sequence is in green, and the protospacer adjacent motif sequence is in red. The Bmawd gene lacks introns and Bmfng gene has 9 exons (box). (B) △A or △F mutant moths have abnormal wings. (C) Primary structures of deletion mutations in the Bmawd or Bmfng loci. Different types of deletion events are shown with the number of deleted nucleotide. The target site sequence is in green, and the protospacer adjacent motif sequence is in red. (D) The transcript level of BmWnt-1 is down-regulated significantly in △A (89%) and △F (57%). The asterisks (* or **) indicate the significant differences (P < 0.05 or P < 0.01, respectively) compared with the relevant control with a 2-tailed t-test. Error bars depict ± SEM.
Discussion
We provide molecular and genetic support for the conclusion that mir-2 function is essential for wing development in B. mori. miR-2 over-expression in transgenic A3>miR-2 results in mutant moths with abnormally small wing discs observable as early as the 5th instar larvae, and these result in aberrant adult wings.
The combined results also support the conclusion that Bmawd and Bmfng are major miR-2 targets and are involved in normal wing development. The awd gene encodes a protein with nucleoside diphosphate kinase (NDP kinase) activity and is a homolog of human metastasis suppressor gene, Nm23.39,40 awd in D. melanogaster is required for proper Notch signaling41 and mutations in the gene results in adults with wing margin and vein-thickening defects similar to the Notch phenotype.41 Loss of awd function also represses wingless (wg) expression.41 dsRNA-mediated ablation of awd function in the lepidoptera, Spodoptera litura42 and Diaphorina citri,43 also results in defects in wing formation. The fng gene in D. melanogaster encodes a glycosyltransferase that can modify the Notch receptor and thereby modulate the signaling pathway during wing development.44,45 Loss of Bmfng function in B. mori also inhibits the activation of the Notch signaling pathway and results in an abnormal wing phenotype and reduced BmWnt-1 transcript level observed in the flügellos mutants.30 Although the seeds of miR-2 family were conserved in insects,12 their predicted targets changed in different species because of the diverse 3′-end UTRs. For example, the Drosophila dme-miR-2 had target binding sites in the 3′-end UTRs of Dmefng but not in Dmeawd (Table S3).
dsRNA-mediated RNAi by using directly injection of dsRNA was seldom effective in lepidoptera, especially during larval stage. An alternative approach is to apply Cas9 system in Bombyx. The Cas9 sgRNA-mediated mutagenesis of the Bmawd and Bmfng loci in this study produced abnormal wing phenotypes similar to those in the A3 > miR-2 overexpression line and a reduction of the transcript level of BmWnt-1.
These data are consistent with the conclusion that miR-2 has a major role in wing morphogenesis in B. mori by regulating the expression of Bmawd and Bmfng to a proper level required for normal wing formation. Manipulating miR-2 may be useful for developing novel methods to control lepidopteron pests by establishing a sex-specific flightless strain to permit genetic sexing.46
Materials and methods
Silkworm strains and plasmids
A multivoltine, nondiapausing silkworm strain, Nistari, was used for germ-line transformation and subsequent experiments. Larvae were reared on fresh mulberry leaves under standard conditions.
The pre-miR-2 cluster and genomic 5′- and 3′-end flanking sequences were amplified by PCR. Complete products 1004 bp in length were sub-cloned into a piggyBac plasmid pmR-IE1-mCherry (Clontech). DNA sequences of mCherry-5′F-premiR2-3′F were PCR-amplified25 using primers HRF and HRR (Table S4), and at the 3′-end of 5×UAS in the XhoI site of pBacA3EGFP-UAS-SV40 to generate the final plasmid UAS-miR-2.
miRNA target predictions
Three miRNA prediction software packages, PITA, miRanda and microTar,25,31,32 were used jointly to predict the miR-2 targets. Thresholds were set to a score of ≥ 140 for miRanda (default), ddG ≤ 0 for PITA, and energy ≥ 0.5 for microTar.25,31,32 The 3′-end UTRs sequences of B. mori genes were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/).
Dual luciferase reporter (DLR) assay
For high transfection efficiency and low background expression of targets, the mammalian HEK293T cell line was used for the DLR assay. A 1004 bp genomic fragment containing the pre-miR-2 cluster was cloned into the pmR-mCherry (Clontech) multiple cloning site (MCS) for the expression of miR-2 cluster (miR-2-F & R, Table S4). Mutated sequences (Fig. S3) were introduced into the 3′-end UTRs of Bmawd and Bmfng by PCR. The wild-type or mutant 3′-end UTRs were cloned and inserted into the pGL3-promoter plasmid (Promega, Madison, WI) between the firefly luciferase ORF and SV40 poly (A) DNA fragments. HEK293T cells were transfected with 5 ng of the pGL3 reporter plasmid, 5 ng of the pRL-TK control plasmid and 100 ng of the pmR-mCherry-miR-2 expression plasmid mixed with 0.5 μl Lipofectamine® 2000 Transfection Reagent (Invitrogen) in 5 μl Opti-MEM® I Reduced Serum Medium (Gibco) in each well of a 96-well plate. The Dual-Luciferase® Reporter Assay (Promega) was performed 24 h after the transfection according to the manufacturer's protocol. Experiments were performed triplicate, each with 4 technical repeats. The mean of the relative luciferase expression ratio (firefly luciferase/renilla luciferase) of the control was set to 1.
Molecular biology
Inverse PCR was carried out as described previously25,47 to investigate genomic insertion loci of transgenes. Both lines crossed with pBacA3-Gal4 animals respectively, and their transformed G2 offspring showed the identical phenotype thus we used the G2 offspring of B1 line in subsequent experiments. Real-time quantification of miRNAs48 was performed to analyze the expression level of miR-2 in transgenic and control individuals. Total RNA was extracted from the larval wing discs (n = 80) and other tissues (n = 3) from wandering-stage larvae using TRIzol Reagent (Invitrogen). cDNA was prepared using the RevertAid First-Strand cDNA synthesis kit (Thermo). The small nuclear RNA U6 of B. mori (BmU6) was used as an internal control. The stem-loop primers for cDNA synthesis from miRNA and the primers used for QPCR are listed (Table S4). QPCR conditions were performed as described previously.25 Bmrp49F and Bmrp49R (Table S4) amplifies a 136 bp fragment from the B. mori ribosomal protein 49 (Bmrp49) as an internal control.
Wandering stage wing discs were collected, homogenized in lysis buffer (Beyotime), and samples resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel. The gel was blotted onto a PVDF membrane and this was incubated with rb-anti-Bmawd or rb-anti-Bmfng at 1:500 and HRP-anti-rabbit at 1:5000. The signal was detected using a Pierce ECL immuno-blotting substrate kit (Thermo). The detection of tubulin as a control was performed as described above using mouse-anti-tubulin as the primary antibody.
Cas9/sgRNA preparation and microinjection
The Cas9-sgRNA system was used to target Bmawd or Bmfng locus. S1 and S2 were identified by screening the Bmawd/Bmfng ORF and following the 5′-GG-N18-NGG-3′ rule.34 The sgRNAs were prepared using the MAXIscript® T7 kit (Ambion) according to the manufacturer's instruction. The Cas9 mRNA was synthesized with mMESSAGE mMACHINE® T7 kit (Ambion) from the linearized vector pTD1-T7-Cas9 as previously described.34
Fertilized eggs were collected within 1 hour after oviposition and microinjected within 3 hours. sgRNA-1 (150 ng/μl) and sgRNA-2 (150 ng/μl) of either Bmawd or Bmfng combined with Cas9 mRNA (300 ng/μl) were mixed and co-injected into 480 embryos each. Injected eggs were incubated at 25°C in a humidified chamber for 10–12 days until larval hatching. Larvae were reared with fresh mulberry leaves under standard conditions.
Statistical Analysis
Data were analyzed with GraphPad using 2-tailed t-test. Probability values of less than 0.05 are considered significant. Data are presented as mean with SEM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from National Basic Research Program of China (2012CB114101), National Science Foundation of China (31420103918, 31272037 and 31372257). AAJ was supported in part by NIH NIAID AI29746.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Engel MS, Grimaldi DA. New light shed on the oldest insect. Nature 2004; 427:627-30; PMID:14961119; http://dx.doi.org/ 10.1038/nature02291 [DOI] [PubMed] [Google Scholar]
- 2.Waldron JA, Newbury SF. The roles of miRNAs in wing imaginal disc development in Drosophila. Biochem Soc Trans 2012; 40:891-5; PMID:22817754; http://dx.doi.org/ 10.1042/BST20120035 [DOI] [PubMed] [Google Scholar]
- 3.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 2008; 18:943-50; PMID:18571409; http://dx.doi.org/ 10.1016/j.cub.2008.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becam I, Rafel N, Hong X, Cohen SM, Milan M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 2011; 138:3781-9; PMID:21795284; http://dx.doi.org/ 10.1242/dev.064774 [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev 2006; 20:2793-805; PMID:17015424; http://dx.doi.org/ 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev 2005; 19:2947-52; PMID:16357215; http://dx.doi.org/ 10.1101/gad.1372505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver SJ, Hagen JW, Okamura K, Perrimon N, Lai EC. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc Natl Acad Sci U S A 2007; 104:18151-6; PMID:17989227; http://dx.doi.org/ 10.1073/pnas.0706673104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci U S A 2008; 105:15417-22; PMID:18824696; http://dx.doi.org/ 10.1073/pnas.0807763105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 2005; 102:18986-91; PMID:16357195; http://dx.doi.org/ 10.1073/pnas.0509535102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006; 34:D140-4; PMID:16381832; http://dx.doi.org/ 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marco A, Hooks K, Griffiths-Jones S. Evolution and function of the extended miR-2 microRNA family. RNA Biol 2012; 9:242-8; PMID:22336713; http://dx.doi.org/ 10.4161/rna.19160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 2005; 1:e13; PMID:16103902; http://dx.doi.org/ 10.1371/journal.pcbi.0010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 2002; 30:363-4; PMID:11896390; http://dx.doi.org/ 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- 15.deCelis JF, Bray S, GarciaBellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development 1997; 124:1919-28; PMID:9169839 [DOI] [PubMed] [Google Scholar]
- 16.Irvine KD, Vogt TF. Dorsal-ventral signaling in limb development. Curr Opin Cell Biol 1997; 9:867-76; PMID:9425353 [DOI] [PubMed] [Google Scholar]
- 17.Boutla A, Delidakis C, Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res 2003; 31:4973-80; PMID:12930946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 2005; 121:1097-108; PMID:15989958; http://dx.doi.org/ 10.1016/j.cell.2005.04.016 [DOI] [PubMed] [Google Scholar]
- 19.James AA, Bryant PJ. Mutations Causing Pattern Deficiencies and Duplications in the Imaginal Wing Disk of Drosophila-Melanogaster. Dev Biol 1981; 85:39-54; PMID:6788628 [DOI] [PubMed] [Google Scholar]
- 20.Rusconi JC, Hays R, Cagan RL. Programmed cell death and patterning in Drosophila. Cell Death Differ 2000; 7:1063-70; PMID:11139279 [DOI] [PubMed] [Google Scholar]
- 21.Tong CZ, Jin YF, Zhang YZ. Computational prediction of microRNA genes in silkworm genome. J Zhejiang Univ Sci B 2006; 7:806-16; PMID:16972323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He PA, Nie Z, Chen J, Lv Z, Sheng Q, Zhou S, Gao X, Kong L, Wu X, Jin Y, et al.. Identification and characteristics of microRNAs from Bombyx mori. BMC Genomics 2008; 9:248; PMID:18507836; http://dx.doi.org/ 10.1186/1471-2164-9-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Zou Q, Tang SM, Wang LG, Shen XJ. Computational identification and characteristics of novel microRNAs from the silkworm (Bombyx mori L.). Mol Biol Rep 2010; 37:3171-6; PMID:19823945 [DOI] [PubMed] [Google Scholar]
- 24.Liu SP, Li D, Li QB, Zhao P, Xiang ZH, Xia QY. MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genomics 2010; 11:148; PMID:20199675; http://dx.doi.org/ 10.1186/1471-2164-11-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling L, Ge X, Li ZQ, Zeng BS, Xu J, Aslam AF, Song Q, Shang P, Huang YP, Tan AJ. MicroRNA Let-7 regulates molting and metamorphosis in the silkworm, Bombyx mori. Insect Biochem Mol Biol 2014; 53:13-21; PMID:25016132 [DOI] [PubMed] [Google Scholar]
- 26.Li ZQ, Ge X, Ling L, Zeng BS, Xu J, Aslam AF, You L, Palli SR, Huang YP, Tan AJ. CYP18A1 regulates tissue-specific steroid hormone inactivation in Bombyx mori. Insect Biochem Mol Biol 2014; 54C:33-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan A, Tanaka H, Tamura T, Shiotsuki T. Precocious metamorphosis in transgenic silkworms overexpressing juvenile hormone esterase. Proc Natl Acad Sci U S A 2005; 102:11751-6; PMID:16087883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kango-Singh M, Singh A, Gopinathan KP. The wings of Bombyx mori develop from larval discs exhibiting an early differentiated state: a preliminary report. J Biosci 2001; 26:167-77; PMID:11426053 [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga TM, Fujiwara H. Identification and characterization of genes abnormally expressed in wing-deficient mutant (flügellos) of the silkworm, Bombyx mori. Insect Biochem Mol Biol 2002; 32:691-9; PMID:12020843 [DOI] [PubMed] [Google Scholar]
- 30.Sato K, Matsunaga TM, Futahashi R, Kojima T, Mita K, Banno Y, Fujiwara H. Positional cloning of a Bombyx wingless locus flugellos (fl) reveals a crucial role for fringe that is specific for wing morphogenesis. Genetics 2008; 179:875-85; PMID:18505883; http://dx.doi.org/ 10.1534/genetics.107.082784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang JH, Ge X, Li ZQ, Wang YQ, Song QS, Stanley DW, Tan AJ, Huang YP. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem Mol Biol 2013; 43:692-700; PMID:23707601; http://dx.doi.org/ 10.1016/j.ibmb.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 32.Ge X, Zhang Y, Jiang JH, Zhong Y, Yang XN, Li ZQ, Huang YP, Tan AJ. Identification of microRNAs in Helicoverpa armigera and Spodoptera litura based on deep sequencing and homology analysis. Int J Biol Sci 2013; 9:1-15; PMID:23289012; http://dx.doi.org/ 10.7150/ijbs.5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiazBenjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 1995; 121:4215-25; PMID:8575321 [DOI] [PubMed] [Google Scholar]
- 34.Wang YQ, Li ZQ, Xu J, Zeng BS, Ling L, You L, Chen YZ, Huang YP, Tan AJ. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res 2013; 23:1414-6; PMID:24165890; http://dx.doi.org/ 10.1038/cr.2013.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al.. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013; 339:819-23; PMID:23287718; http://dx.doi.org/ 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang WY, Fu YF, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JRJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013; 31:227-9; PMID:23360964; http://dx.doi.org/ 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dearolf CR, Hersperger E, Shearn A. Developmental consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Dev Biol 1988; 129:159-68; PMID:3137111; http://dx.doi.org/ 10.1016/0012-1606(88)90170-4 [DOI] [PubMed] [Google Scholar]
- 38.Correia T, Papayannopoulos V, Panin V, Woronoff P, Jiang J, Vogt TF, Irvine KD. Molecular genetic analysis of the glycosyltransferase Fringe in Drosophila. Proc Natl Acad Sci U S A 2003; 100:6404-9; PMID:12743367; http://dx.doi.org/ 10.1073/pnas.1131007100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IM, King CR, Liotta LA, Steeg PS. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature 1989; 342:177-80; PMID:2509941; http://dx.doi.org/ 10.1038/342177a0 [DOI] [PubMed] [Google Scholar]
- 40.Zhao X, Tong F, Jiang C, Zhang Y. Expression and purification of abnormal wing disc gene from silkworm, Bombyx mori. Bulletin of Sericulture 2009; 40:13-6 [Google Scholar]
- 41.Ignesti M, Barraco M, Nallamothu G, Woolworth JA, Duchi S, Gargiulo G, Cavaliere V, Hsu T. Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. BMC Biol 2014; 12:12; PMID:24528630; http://dx.doi.org/ 10.1186/1741-7007-12-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng X, Hu J, Xu X, Wang Z, Hu Q, Jin F, Ren S. Toxic effect of destruxin A on abnormal wing disc-like (SLAWD) in Spodoptera litura fabricius (Lepidoptera: Noctuidae). PLoS One 2013; 8:e57213; PMID:23468937; http://dx.doi.org/ 10.1371/journal.pone.0057213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Shesheny I, Hajeri S, El-Hawary I, Gowda S, Killiny N. Silencing abnormal wing disc gene of the Asian citrus psyllid, Diaphorina citri disrupts adult wing development and increases nymph mortality. PLoS One 2013; 8:e65392; PMID:23734251; http://dx.doi.org/ 10.1371/journal.pone.0065392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature 1997; 387:908-12; PMID:9202123; http://dx.doi.org/ 10.1038/43191 [DOI] [PubMed] [Google Scholar]
- 45.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol 2003; 4:786-97; PMID:14570055; http://dx.doi.org/ 10.1038/nrm1228 [DOI] [PubMed] [Google Scholar]
- 46.Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, Berendonk TU, White-Cooper H, Scaife S, Kim Phuc H, et al.. Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A 2010; 107:4550-4; PMID:20176967; http://dx.doi.org/ 10.1073/pnas.1000251107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan AJ, Fu GL, Jin L, Guo QH, Li ZQ, Niu BL, Meng ZQ, Morrison NI, Alphey L, Huang YP. Transgene-based, female-specific lethality system for genetic sexing of the silkworm, Bombyx mori. Proc Natl Acad Sci U S A 2013; 110:6766-70; PMID:23569267; http://dx.doi.org/ 10.1073/pnas.1221700110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CF, Ridzon DA, Broomer AJ, Zhou ZH, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al.. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005; 33:e179; PMID:16314309; http://dx.doi.org/ 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.