Abstract
The W chromosome of the silkworm Bombyx mori has been known to determine femaleness for more than 80 years. However, the feminizing gene has not been molecularly identified, because the B. mori W chromosome is almost fully occupied by a large number of transposable elements. The W chromosome-derived feminizing factor of B. mori was recently shown to be a female-specific PIWI-interacting RNA (piRNA). piRNAs are small RNAs that potentially repress invading “non-self” elements (e.g., transposons and virus-like elements) by associating with PIWI proteins. Our results revealed that female-specific piRNA precursors, which we named Fem, are transcribed from the sex-determining region of the W chromosome at the early embryonic stage and are processed into a single mature piRNA (Fem piRNA). Fem piRNA forms a complex with Siwi (silkworm Piwi), which cleaves a protein-coding mRNA transcribed from the Z chromosome. RNA interference of this Z-linked gene, which we named Masc, revealed that this gene encodes a protein required for masculinization and dosage compensation. Fem and Masc both participate in the ping-pong cycle of the piRNA amplification loop by associating with the 2 B. mori PIWI proteins Siwi and BmAgo3 (silkworm Ago3), respectively, indicating that the piRNA-mediated interaction between the 2 sex chromosomes is the primary signal for the B. mori sex determination cascade. Fem is a non-transposable repetitive sequence on the W chromosome, whereas Masc is a single-copy protein-coding gene. It is of great interest how the piRNA system recognizes “self ”Masc mRNA as “non-self” RNA.
Keywords: amplification cycles, ping-pong, piRNA, PIWI proteins, silkworm, sex determination
The ZW sex determination system is found in a diverse range of animals including moths. In the mulberry silkworm Bombyx mori, females are heterogametic (ZW), while males are homogametic (ZZ).1,2 Classical genetic studies have revealed that B. mori sex is dominantly determined by the presence of the W chromosome.1,2 One copy of the W chromosome is sufficient for determining femaleness, regardless of the copy number of the Z chromosome.1,2 This observation originally led to the simple assumption that the W chromosome encodes a dominant feminizing gene. However, neither the female-determining gene nor its candidate was isolated until 2014. There are 2 major reasons for this insuperability. First, a conventional positional cloning strategy is not applicable to this species because of the lack of crossing-over events in females. Second, the B. mori W chromosome is almost fully occupied by nested transposable and repeat elements,3,4 which preventing construction of long accurate sequence scaffolds of this chromosome.
Controlling transposable elements in germ line cells is essential for the host genome to ensure a precise transmission of parental genomic information to the next generation.5,6 Animals have evolved an excellent defense system to combat invading non-self elements lying concealed in their own genomes. At the core of this immune system are PIWI proteins and associated PIWI-interacting RNAs (piRNAs). piRNAs are small RNAs that range 23–30 nucleotides (nts) in length.5,6 piRNAs potentially act as sequence-specific guides for PIWI proteins that cleave target RNAs.5,6 The biogenesis of piRNAs is Dicer-independent and initiates with fragmentation of long single-stranded piRNA precursors by the endonuclease MitoPLD/Zucchini.7-9 Fragmented RNAs that are longer than mature piRNAs are incorporated into a subset of PIWI proteins, Siwi in B. mori (Fig. 1A),10,11 with a specific nucleotide preference for uracil (1U) at the 5′ end of loaded RNAs.5,6,12-16 The 3′ ends of Piwi-associated RNAs are further trimmed to the mature length by an unidentified 3′ to 5′ exonuclease that we named Trimmer,16 followed by 2′-O-methylation catalyzed by the methyltransferase Hen1.17-19 This 1U piRNA is called primary piRNA. The Piwi-primary piRNA complexes cleave their targets from positions 10 and 11 of the guide piRNAs. The 3′ fragments of the cleavage products are incorporated into another subset of PIWI proteins, BmAgo3 in B. mori (Fig. 1A),10,11 which do not show a first nucleotide bias. This Ago3-associated RNA is processed into mature secondary piRNAs with adenine at position 10 (10A). The secondary piRNAs precisely overlap with primary 1U piRNAs by 10 nts.5,6,12-15 The secondary piRNA-Ago3 complexes, in turn, digest their complementary targets to generate secondary 1U piRNAs. This piRNA biogenesis pathway is called a ping-pong amplification cycle, and the 10-nt overlap is called a ping-pong signature5,6 (see Fig. 1A). In animals, Piwi-bound 1U piRNAs are usually antisense to repetitive sequences, whereas secondary 10A piRNAs associated with Ago3 are identical to the sense strands of repetitive sequences. In other words, piRNA production occurs concomitantly with the repression of transposable elements in this ping-pong amplification loop.
Figure 1.
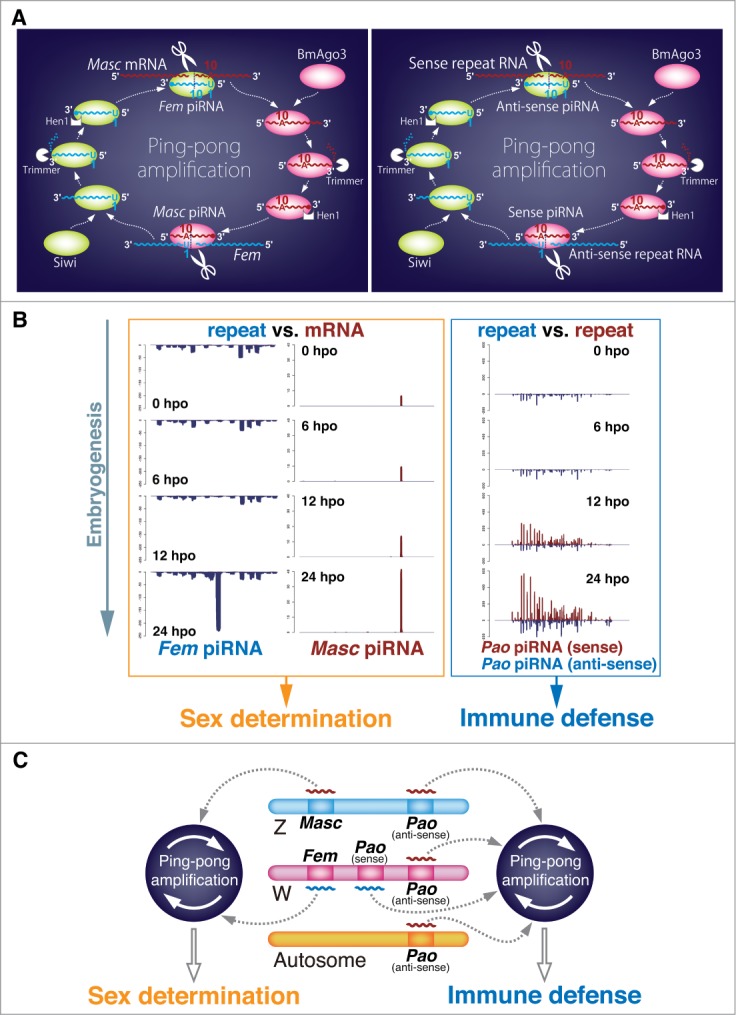
The B. mori sex is determined by the piRNA pathway. (A) Models of ping-pong amplification cycles of piRNAs in B. mori embryos. The left panel shows a ping-pong loop involving Fem and Masc piRNAs, which controls the sex determination pathway. The right panel shows a canonical ping-pong cycle, resulting in transposon silencing. (B) Zygotic amplification of repeat RNA- and protein-coding mRNA-derived piRNAs by maternally transmitted antisense piRNAs. Density plots of Fem-, Masc-, and Pao-derived piRNAs are shown at 0, 6, 12, and 24 hours post-oviposition (hpo), respectively. Pao was used as a representative transposable element. (C) Two different roles of the piRNA pathway in B. mori embryogenesis. In B. mori embryos, the piRNA system plays critical roles not only in immune defense against active transposable elements but also in the sex determination pathway. Full-length sense transposons, which are piRNA precursors, are transcribed frequently from the W chromosome.20 They are silenced by Siwi, which is complexed with anti-sense piRNAs derived from truncated transposable elements located on each chromosome. In the sex determination pathway, Fem RNAs are transcribed from the sex-determination regions and are cleaved by the maternally transmitted Masc piRNA-BmAgo3 complex. The Fem piRNA-Siwi complex cleaves Masc mRNA, resulting in the accumulation of Masc piRNA and feminization.
The B. mori W chromosome but not the Z chromosome is almost completely occupied by selfish repetitive elements, most of which are considered piRNA precursors.20 To determine if the difference in B. mori sex chromosome constitution affects piRNA profiles, we analyzed piRNA libraries prepared from pupal ovary and testis of wild-type and 3 B. mori strains with a unique truncated W chromosome.20 We found that the sex-determining region of the W chromosome produces female-enriched piRNAs that originate from transposons or repetitive sequences;20 however their biological functions remain to be elucidated. In 2014, we performed transcriptome analysis by deep sequencing (RNA-seq) using male and female RNAs from B. mori early embryos and discovered a single female-specific transcript. This transcript was a non-transposable repetitive sequence expressed from the sex-determining region of the W chromosome and was a precursor of a single female-specific piRNA.21 Injecting an RNA inhibitor for this piRNA into female embryos resulted in the production of the male-type splicing isoform of B. mori doublesex (Bmdsx), which acts at the downstream end of the sex differentiation cascade in B. mori.22,23 These results demonstrated that this piRNA is required for the female-type splicing of Bmdsx, and we named the precursor of this piRNA as Feminizer (Fem).
To understand how Fem piRNA transmits the feminizing signal to downstream cascades in female embryos, we searched for the targets of Fem piRNA by bioinformatic analyses. We identified only one genomic locus where the Fem piRNA sequence was extensively complementary (26/29, 5′ 14 nts are perfectly matched). This locus was present in the coding region of a CCCH-tandem zinc finger gene located on the Z chromosome. We named this gene Masculinizer (Masc).21 Injection of small interfering RNA (siRNA) of Masc into male embryos led to the production of the female-type Bmdsx transcripts, indicating that Masc is essential for silkworm masculinization. We also observed that the embryonic knockdown of Masc mRNA results in male-specific lethality. RNA-seq of Masc siRNA-injected embryos revealed that the Masc protein globally represses gene expression from the male Z chromosome at the embryonic stage, suggesting that a failure of dosage compensation on the Z chromosome possibly results in male-specific lethality in Masc mRNA-depleted male embryos. Taken together, the Masc protein controls both dosage compensation and masculinization.21
piRNA biogenesis occurs via a ping-pong mechanism, which is achieved by 2 different PIWI proteins, Siwi and BmAgo3, in B. mori (Fig. 1A).10,11 Bioinformatic analysis revealed that the Masc mRNA-derived piRNA (Masc piRNA) is a ping-pong partner of Fem piRNA (Fig. 1A). Deep sequencing of the immunoprecipitated piRNA libraries showed that Fem piRNA preferentially binds to Siwi, whereas Masc piRNA preferentially binds to BmAgo3 (Fig. 1A).21 Furthermore, a modified RNA ligase-mediated 5′ rapid amplification of cDNA ends (RACE) experiment indicated Fem piRNA-mediated cleavage of Masc mRNA.21 These findings indicate a ping-pong amplification model for Fem and Masc piRNAs (Fig. 1A). Fem piRNA is de novo produced during B. mori embryogenesis, whereas Masc piRNA is maternally transmitted at moderate levels (Fig. 1B). Western blot analysis of egg lysates revealed that BmAgo3 is also maternally transmitted,24 suggesting that the Masc piRNA-BmAgo3 complex is maternally inherited and exists in newly laid eggs, which cleaves zygotically synthesized Fem transcripts to generate Fem piRNA in female embryos. The resulting Fem piRNA then cleaves Masc mRNA by associating with Siwi (Fig. 1A), inhibiting masculinization of female embryos.21
The piRNA pathway is an evolutionarily conserved system that is found in a diverse range of organisms. Although this pathway was originally considered a limited system for protecting the host genome against non-self elements, it was recently revealed to be utilized in other biological roles. Rajasethupathy et al. reported that piRNAs are abundantly expressed in Aplysia brain, and some are markedly induced by serotonin, an important neuromodulator for learning and memory.25 Further experiments revealed that the Piwi-piRNA complex mediates serotonin-dependent CpG methylation and transcriptional silencing of CREB2, a key protein that functions as a transcriptional repressor of memory. These results demonstrate that neuronal piRNAs are involved in long-term memory. Watanabe et al. also reported that a non-coding RNA transcribed from the differentially methylated region (DMR) of the imprinted mouse Rasgrf1 locus is cleaved by repeat-derived piRNAs.26 Although the relationship between cleavage by the PIWI-piRNA complex and DMR methylation is unclear, this result suggests a role of the piRNA pathway in genomic imprinting by de novo DNA methylation in mice. In addition, recent studies have suggested that somatic piRNAs are involved in tumorigenesis,27 liver regeneration,28 and genomic heterogeneity in the brain.29 Together with our recent discovery that the piRNA pathway is required for determining femaleness in B. mori, piRNA plays crucial roles in diverse biological processes of various animals.
Maternally inherited antisense piRNAs trigger acute amplification of secondary sense piRNA production in B. mori embryos, which is coupled with zygotic transcription of sense transposable RNAs (Fig. 1B).24 This suggests that the Siwi-antisense piRNA complexes cleave sense transposable RNAs to reduce transposon activity, and sense piRNAs are generated as a consequence (Figs. 1B, C). Combined with the finding of the Fem piRNA-Masc pathway, the B. mori piRNA system plays 2 critical roles simultaneously during embryogenesis: immune defense and sex determination (Figs. 1B, C). How does B. mori sense Masc mRNA as a target of the Siwi-Fem piRNA complex? Previous studies in B. mori, Drosophila, and mice revealed that transcription from a piRNA cluster in the host genome possibly initiates de novo piRNA production against new insertion of “non-self” elements.30-32 However, Masc is not likely to be localized within a distinct piRNA cluster of the B. mori genome. Furthermore, Masc is a single-copy protein-coding gene and is not a repeat-like element. Although Drosophila piRNAs are supposed to be involved in the silencing of the 2 protein-coding genes, FasIII33 and nos,34 the genomic regions targeted by these piRNAs are intronic and 3′-untranslated regions, respectively. The predicted cleavage site of Masc mRNA by the Fem piRNA-Siwi complex is located within the translated region (9th exon) of Masc, and this cleavage has been experimentally verified by a modified RACE procedure,21 indicating that the B. mori embryonic piRNA pathway recognizes a protein-coding exonic region as a non-self element. We are now addressing the question of why the B. mori genome considers “self” Masc mRNA as “non-self” RNA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry and Grants-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for non-coding RNAs”) to SK.
References
- 1.Hasimoto H. The role of the W-choromosome in the sex deternination of Bombyx mori. Jpn J Genet 1933; 8:245-7. [in Japanese]; http://dx.doi.org/10.1266/jjg.8.245 [Google Scholar]
- 2.Fujii T, Shimada T. Sex determination in the silkworm, Bombyx mori: a female determinant on the W chromosome and the sex-determining gene cascade. Semin Cell Dev Biol 2007; 18:379-88; PMID:17446095; http://dx.doi.org/10.1016/j.semcdb.2007.02.008 [DOI] [PubMed] [Google Scholar]
- 3.Abe H, Mita K, Yasukochi Y, Oshiki T, Shimada T. Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet Genome Res 2005; 110:114-51. [DOI] [PubMed] [Google Scholar]
- 4.Abe H, Fujii T, Tanaka N, Yokoyama T, Kakehashi H, Ajimura M, Mita K, Banno Y, Yasukochi Y, Oshiki T. et al. Identification of the female-determining region of the W chromosome in Bombyx mori. Genetica 2008; 133:269-82; PMID:17901928; http://dx.doi.org/10.1007/s10709-007-9210-1 [DOI] [PubMed] [Google Scholar]
- 5.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009; 10:94-108; PMID:19148191; http://dx.doi.org/10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136:656-68; PMID:19239887; http://dx.doi.org/10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320-4; PMID:16809489; http://dx.doi.org/10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- 8.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J. et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012; 491:284-7; PMID:23064230; http://dx.doi.org/10.1038/nature11509 [DOI] [PubMed] [Google Scholar]
- 9.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012; 491:279-83; PMID:23064227; http://dx.doi.org/10.1038/nature11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun 2008; 367:755-60; PMID:18191035; http://dx.doi.org/10.1016/j.bbrc.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 11.Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 2009; 15:1258-64; PMID:19460866; http://dx.doi.org/10.1261/rna.1452209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089-103; PMID:17346786; http://dx.doi.org/10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- 13.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5´ end formation in Drosophila. Science 2007; 315:1587-90; PMID:17322028; http://dx.doi.org/10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- 14.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 2008; 18:136-48; PMID:18282709; http://dx.doi.org/10.1016/j.tcb.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development 2008; 135:3-9; PMID:18032451; http://dx.doi.org/10.1242/dev.006486 [DOI] [PubMed] [Google Scholar]
- 16.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 2011; 43:1015-22; PMID:21925389; http://dx.doi.org/10.1016/j.molcel.2011.07.029 [DOI] [PubMed] [Google Scholar]
- 17.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 2007; 17:1265-72; PMID:17604629; http://dx.doi.org/10.1016/j.cub.2007.06.030 [DOI] [PubMed] [Google Scholar]
- 18.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 2007; 14:347-8; PMID:17384647; http://dx.doi.org/10.1038/nsmb1218 [DOI] [PubMed] [Google Scholar]
- 19.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 2007; 14:349-50; PMID:17384646; http://dx.doi.org/10.1038/nsmb1220 [DOI] [PubMed] [Google Scholar]
- 20.Kawaoka S, Kadota K, Arai Y, Suzuki Y, Fujii T, Abe H, Yasukochi Y, Mita K, Sugano S, Shimizu K. et al. The silkworm W chromosome is a source of female-enriched piRNAs. RNA 2011; 17:2144-51; PMID:22020973; http://dx.doi.org/10.1261/rna.027565.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014; 509:633-6; PMID:24828047; http://dx.doi.org/10.1038/nature13315 [DOI] [PubMed] [Google Scholar]
- 22.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol 2003; 213:345-54; PMID:12733073; http://dx.doi.org/10.1007/s00427-003-0334-8 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev 2005; 7:58-68; PMID:15642090; http://dx.doi.org/10.1111/j.1525-142X.2005.05007.x [DOI] [PubMed] [Google Scholar]
- 24.Kawaoka S, Arai Y, Kadota K, Suzuki Y, Hara K, Sugano S, Shimizu K, Tomari Y, Shimada T, Katsuma S. Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA 2011; 17:1401-07; PMID:21628432; http://dx.doi.org/10.1261/rna.2709411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012; 149:693-707; PMID:22541438; http://dx.doi.org/10.1016/j.cell.2012.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A. et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011; 332:848-52; PMID:21566194; http://dx.doi.org/10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K. et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015; 29:196-206; PMID:24732595; http://dx.doi.org/10.1038/leu.2014.135. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Rizzo F, Hashim A, Marchese G, Ravo M, Tarallo R, Nassa G, Giurato G, Rinaldi A, Cordella A, Persico M. et al. Timed regulation of P-Element-Induced wimpy testis-interacting RNA expression during rat liver regeneration. Hepatology 2014; 60:798-806; PMID:24930433; http://dx.doi.org/10.1002/hep.27267 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013; 340:91-5; PMID:23559253; http://dx.doi.org/10.1126/science.1231965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaoka S, Mitsutake H, Kiuchi T, Kobayashi M, Yoshikawa M, Suzuki Y, Sugano S, Shimada T, Kobayashi J, Tomari Y. et al. A role for transcription from a piRNA cluster in de novo piRNA production. RNA 2012; 18:265-73; PMID:22194309; http://dx.doi.org/10.1261/rna.029777.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. Production of artificial piRNAs in flies and mice. RNA 2012; 18:42-52; PMID:22096018; http://dx.doi.org/10.1261/rna.029769.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto Y, Watanabe T, Hoki Y, Shirane K, Li Y, Ichiiyanagi K, Kuramochi-Miyagawa S, Toyoda A, Fujiyama A, Oginuma M. et al. Targeted gene silencing in mouse germ cells by insertion of a homologous DNA into a piRNA generating locus. Genome Res 2013; 23:292-9; PMID:23132912; http://dx.doi.org/10.1101/gr.137224.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009; 461:1296-9; PMID:19812547; http://dx.doi.org/10.1038/nature08501 [DOI] [PubMed] [Google Scholar]
- 34.Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010; 467:1128-32; PMID:20953170; http://dx.doi.org/10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]


