Abstract
Gene expression regulation relies on a variety of molecular mechanisms affecting different steps of a messenger RNA (mRNA) life: transcription, processing, splicing, alternative splicing, transport, translation, storage and decay. Light induces massive reprogramming of gene expression in plants. Differences in alternative splicing patterns in response to environmental stimuli suggest that alternative splicing plays an important role in plant adaptation to changing life conditions. In a recent publication, our laboratories showed that light regulates alternative splicing of a subset of Arabidopsis genes encoding proteins involved in RNA processing by chloroplast retrograde signals. The light effect on alternative splicing is also observed in roots when the communication with the photosynthetic tissues is not interrupted, suggesting that a signaling molecule travels through the plant. These results point at alternative splicing regulation by retrograde signals as an important mechanism for plant adaptation to their environment.
Keywords: alternative splicing, chloroplast, light, photoreceptors, retrograde signaling, RNA
Abbreviations
- mRNA
messenger RNA
- Pol II
RNA polymerase II
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-benzoquinone
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- PS
photosystem
- PQ
plastoquinone
- ROS
reactive oxygen species
The Life of a mRNA in Plants
The birth of mRNA molecules in plants does not differ much from that of other eukaryotes. Transcription from a DNA template is carried out by RNA polymerase II (Pol II) to form the precursor mRNA (pre-mRNA) molecule. The pre-mRNA undergoes 5’ end capping, splicing and 3’ end cleavage and polyadenylation, processes that have been shown to be coupled to transcription.1
Splicing is performed by the spliceosome, a ribonucleoprotein machinery that recognizes splice sites consisting of consensus sequences: strong splice sites are more adjusted to the consensus, and therefore more efficiently recognized compared to weak splice sites. Competition between strong and weak splice sites along the nascent pre-mRNA leads to alternative splicing.2 Transcriptome diversity is increased by alternative splicing since it allows a single gene to produce 2 or more mature mRNA variants that are similar but not identical, expanding the coding capacity of eukaryotic genomes.2,3 Alternative splicing can also regulate mRNA levels through degradation of specific alternative splicing isoforms by nonsense-mediated mRNA decay, introducing a quality control mechanism.4 Once in the cytoplasm, translatable mature mRNA is also subjected to different pathways of degradation. Each of these processes from transcription to degradation can be regulated, thus allowing multiple layers of gene expression regulation.
Transcription and Alternative Splicing
Alternative splicing regulation is mostly performed by splicing factors that recognize cis-acting elements present in the RNA molecule, known as splicing enhancers or silencers. However, many different approaches, both in vivo and in vitro, have revealed another layer of regulation that involves a mechanistic coupling between transcription and splicing machineries.5 Accumulated evidence indicates that pre-mRNA splicing occurs,6 or is committed to occur, co-transcriptionally. This means that the actual splicing reactions take place, or that the factors needed for splicing are recruited to the pre-mRNA target sequences, before RNA Pol II has reached the end of the gene and while the transcript is still associated to chromatin.7,8 Chromatin structure, nucleosome positioning, post-translational histone modifications and the existence of adaptor complexes between the chromatin and splicing machineries can regulate alternative splicing through its coupling with transcription.7 Changes in RNA Pol II elongation rate can also modulate alternative splicing: in most cases slow RNA Pol II elongation leads to higher inclusion of alternative exons;9 however, it has also been reported that slow elongation can cause exon skipping.10
Alternative Splicing in Plants
Approximately 60% of Arabidopsis genes produce different transcript isoforms due to alternative splicing.11 This percentage is lower than that of mammalian cells (95% in multiexonic genes),12 but still high enough to consider alternative splicing as a main contributor to expanding the repertoire of transcripts and proteins in plants.13
The frequency of different alternative splicing events in plants has been deeply studied. Intron retention is the most prominent event in plants, whereas cassette exons are the most common alternative splicing event in animals. However, the alternative splicing landscape is more complex since multiple splicing events may occur in the same transcript, raising the question of a possible coordination between them.11
Different cell types, tissues, developmental stages and environmental conditions show differences in the presence and abundance of splicing factors that contribute to modulate alternative splicing patterns.11 Widespread changes in the patterns of splicing mRNA isoforms in response to stress and developmental cues suggest an important role for alternative splicing in plant development and environmental responses.3 Alternative splicing is important for photosynthesis, flowering, defense responses and the circadian clock.13
Several abiotic stresses can regulate alternative splicing, such as high or low temperature, drought, salt stress or light.14 Global changes in alternative splicing were found under salt stress, which involve about 49% of all intron-containing genes. Among them, most genes that showed significant changes were associated with specific functional pathways, such as stress response and RNA splicing.15 Light/dark conditions affect alternative splicing of a subset of Arabidopsis genes preferentially encoding proteins involved in RNA processing. The alternative splicing of At-RS31, which encodes a serine/arginine-rich splicing factor, changes in different light conditions through a mechanism that involves the chloroplast (see below).16
Altogether, conspicuous and widespread differences in alternative splicing patterns in response to environmental stimuli suggest that alternative splicing plays important roles in plant adaptation to changing life conditions.
Nuclear Gene Expression Regulation by Light: the Role of Retrograde Signals
One key strategy for plants to adapt to challenges imposed by stress conditions and to cope with the changing environment is the fine tuning of gene expression in response to those changes.17,18 As briefly described before, gene expression regulation can rely on a wide variety of molecular mechanisms affecting different steps of the mRNA life like transcription, processing, splicing, alternative splicing, transport, translation, storage and decay.19 Light is one of the most important environmental cues for almost all living organisms and it is also the source of energy for plants.20 It is therefore not surprising that plants have adopted the ability to sense multiple parameters of light signals, including light quantity (fluence), quality (wavelength), direction and duration.21 Light signals are perceived through different families of photoreceptor proteins. Red and far-red lights are sensed by phytochromes. Blue and ultraviolet (UV)-A wavelengths are sensed by cryptochromes, phototropins, and members of the Zeitlupe family in Arabidopsis, whereas UV-B is perceived by the UVR8 photoreceptor. Light perception by these photoreceptors triggers many biological processes, including gene expression regulation by signal transduction or by nuclear relocalization of light-activated photoreceptors.22-24
Besides photoreceptor proteins, once a green seedling is established, chloroplasts play a key role in sensing light fluctuations and in the communication of these changes to the nucleus by retrograde signaling pathways.25,26 Two different modes can be established for retrograde signaling. First, there is a chloroplast biogenic control of nuclear genes during early plastidial development, triggered by signals related to the photosystems and pigment biogenesis, and, secondly, there is a chloroplast operational control associated with signals induced by the function of mature chloroplast to regulate plant responses to varying light conditions.27-30 Most of the operational retrograde signals are dependent on the quantity and/or quality of light. Changes in light quality might lead to preferential excitation of one of the photosystems (PS), leading to redox changes in intersystem electron carriers, particularly the plastoquinone (PQ) pool and the cytochrome b6f complex, whereas high light reduces the whole electron transfer chain and causes accumulation of reducing equivalents in the stroma as well as the production of reactive oxygen species (ROS).31,32 Redox signals from photosynthetic electron transport components have been shown to control the expression of genes in the chloroplast genome at both the transcriptional and translational levels, as well as the expression of nuclear genes mainly at the transcriptional level.33 The redox state of the PQ pool in particular has been strongly suggested as a prominent candidate for the origin of chloroplast redox signals. This has been demonstrated by using the photosynthetic electron transfer inhibitors 2,5-dibromo-3-methyl-6-isopropyl-benzoquinone (DBMIB) and 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and by modulating the redox state of the PQ pool by light that predominantly excites either PSII or PSI reducing or oxidizing the PQ pool, respectively.32,34 Interestingly, changes in temperature, light conditions, CO2 availability and nutrients impact on the photosynthesis efficiency, changing the photosynthetic electron flux and the redox state of the components involved in it.35 Therefore, signals arising from the photosynthetic electron transport chain components (i.e.: PQ pool) will be integrating diverse environmental cues to regulate gene expression in order to fine tune plant responses.36 While the transmission pathway followed by the redox signals remains still elusive, evidence for the effects of these signals on nuclear gene expression including the regulation of transcription, stability and translational efficiency is continuously growing.34,37,38 Other signals, not directly derived from the redox state of the components of the photosynthetic electron transport chain have been reported. Among these, ROS -continuously produced during photosynthesis by partial reduction of oxygen molecules or energy transfer to them- were shown to work as signaling molecules affecting nuclear gene expression.39-43
Finally, to add more complexity to this scenario, it is interesting to point out that different light signals are connected with signals from other pathways and both, light receptor proteins and chloroplast derived signals, interact in the same network and contribute to the control of plant responses and development.44-46
Regulation of Alternative Splicing by Light: a Novel Role for Retrograde Signals
Light induces massive reprogramming of the plant transcriptome. Up to one third of the transcriptome can be regulated by light in Arabidopsis and similar results were obtained in other plant species.47,48 Noticeably, many of the genes affected by light conditions encode transcription factors or proteins with DNA binding domains.49 Besides the transcriptional effects, chromatin modifications are tightly regulated by light.50 As an example, acetylation of lysine 9 in histone 3 (H3K9ac), a mark that positively correlates with active transcription, is enriched in light regulated genes, in a light intensity-dependent manner.51 Alternative splicing is also under light control. Pioneer work of Mikio Nishimura, performed in pumpkin in the 90's, showed that light regulates alternative splicing patterns of the transcripts coding for a hydroxypyruvate reductase and an ascorbate peroxidase, determining different subcellular localizations of the protein products.52,53 More recently it was shown that alternative splicing regulation of circadian clock-related genes in Arabidopsis is important for the proper functioning of the biological clock,54,55 and that light is affecting the alternative splicing of splicing factor coding genes.56,57 It was also shown that an Arabidopsis mutant for a gene that codes for an ortholog of the human potential splicing factor SR140 fails in phytochrome B responses.58 Furthermore, a recent publication reveals that light-regulated alternative splicing is important in shaping transcriptome responses to light in the moss Physcomitrella patens.59 The authors proposed that alternative splicing is rapidly fine-tuned by light in this system and these responses are misregulated in P. patens mutants defective in red light sensing phytochromes. In summary, plant responses to light also include alternative splicing regulation, and the photoreceptor proteins might be key players in this scene59
In a recent publication, our laboratories showed that light regulates alternative splicing of a subset of Arabidopsis genes encoding proteins involved in RNA processing by chloroplast retrograde signals. Earlier evidence had shown that the mRNA for a negative regulator of tetrapyrrole biosynthesis of Chlamydomonas reinhardtii, the product of a FLU-like gene, undergoes alternative splicing whose pattern is controlled by light and plastid signals.60 Using the alternative splicing of At-RS31, a gene encoding a serine arginine-rich splicing factor61 as a model, we showed that light modulates the relative amounts of its mRNA isoforms in a way that the protein coding isoform (mRNA1) is more abundant in light. The participation of phytochrome and cryptochrome photosensory pathways was ruled out for the regulation of this event by light/dark transitions. Changes in At-RS31 alternative splicing occur in seedlings both under prolonged light/dark regimes and under natural short day (8:16 hr light:dark) photoperiod conditions but not in long day photoperiods. Photosynthetically active different light qualities produce similar effects than white light, in a light intensity-dependent manner. The light signaling pathway does not involve the circadian clock since the light/dark effect on splicing is still observed in several clock mutants. Since photoreceptor proteins are not involved in this regulatory mechanism we tested whether the chloroplast, which is able to sense and to communicate light signals to the nucleus, was triggering the alternative splicing changes in response to light. Consequently, drugs that disrupt chloroplast function like DCMU and DBMIB abolish the splicing responses to light showing the involvement of the organelle in the pathway. Both DCMU and DBMIB inhibit the overall electron transport chain, but whereas DCMU increases the oxidized PQ pool by blocking the electron transfer from the PSII to PQ, DBMIB keeps the PQ pool reduced by preventing the electron transfer to cytochrome b6f. DCMU duplicates the effect of darkness but DBMIB mimics the light effect on At-RS31, revealing that the reduced PQ pool upregulated by light initiates the retrograde signaling pathway. The nature of the retrograde signal is4 still unknown, however. Interestingly, we showed that alternative splicing regulation of At-RS31 by light would be important for plant survival when facing suboptimal light conditions. Furthermore, the light effect on At-RS31 alternative splicing is also observed in roots when the communication with the photosynthetic tissues is not interrupted, suggesting that a signaling molecule travels through the plant.16 These results pointed at alternative splicing regulation by retrograde signals as an important mechanism for plants to adapt to the environment. Effort must be made to identify the signal that travels from the chloroplast to the nucleus as well as the nature of the molecule that communicates the light effect to the roots.
Perspectives: Searching for the Signal(s)
In the next years effort has to be done to clarify how much of the genome is regulated at the level of alternative splicing in response to light by the different pathways, the role of photoreceptor proteins, plastid signals, and the possible interactions between them.
The chloroplast and the sensitivity of the photosynthetic mechanisms make this organelle ideal for the integration of different signals,33 not only from the environment, but also derived from the different tissues, developmental and nutritional states of plants. Besides the chloroplast retrograde signals derived from the redox state of the photosynthetic electron transport chain components, and those related to ROS, other retrograde signals related to chloroplast biogenesis have been studied:25,28 tetrapyrroles,30 those linked to thioredoxins,62,63 and other mechanisms.64,65 Sugars produced by photosynthesis, besides being the main source of biochemical energy, are also sensed as signals in the plant cell.66,67 The interaction of chloroplast signals with retrograde signals generated in mitochondria deserves further investigation.68 We are just starting to understand the way in which the different pieces work together: which genes are regulated, which processes are affected (i.e., chromatin, transcription, splicing, translation), and the possible connections and cross regulations of the different layers (Fig. 1).
Figure 1.
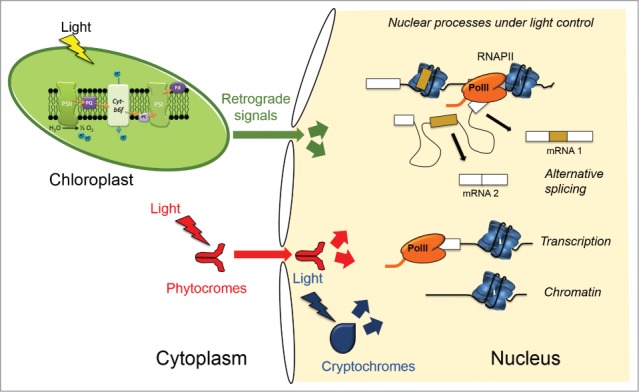
Scheme of light triggered signals affecting different aspects of nuclear gene expression. Photoreceptor proteins like phytochromes and cryptochromes absorb specific light wavelengths and transduce these signals into chromatin, transcriptional and post-transcriptional changes (see text for further details). The chloroplast is also able to act as a light sensor. Chloroplast derived retrograde signals, longer known for transcriptional regulation, are also able to regulate alternative splicing decisions in the nucleus.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Barta, Srebrow and Kornblihtt laboratories for constant support and input. M.G.H. is a fellow and A.R.K. is a career investigator from the Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina. E.P. is a Marie Curie postdoctoral fellow. A.R.K. is a senior international research scholar of the Howard Hughes Medical Institute.
Funding
This work was supported by the Agencia Nacional de Promoción de Ciencia y Tecnología of Argentina, the University of Buenos Aires, the King Abdulaziz University, and the Austrian Science Fund FWF (P26333 to M.K.; DK W1207, SFBF43-P10, ERA-NET I254 to A.B.).
References
- 1. Perales R, Bentley D. “Cotranscriptionality:" the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 2009; 36:178-91; PMID:19854129; http://dx.doi.org/ 10.1016/j.molcel.2009.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013; 14(3):153-65; PMID:23385723; http://dx.doi.org/ 10.1038/nrm3525 [DOI] [PubMed] [Google Scholar]
- 3. Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013; 25(10):3657-83 ; PMID:24179125; http://dx.doi.org/ 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012; 40(6):2454-69; PMID:22127866; http://dx.doi.org/ 10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kornblihtt AR, Schor IE, Allo M, Blencowe BJ. When chromatin meets splicing. Nat. Struct. Mol. Biol 2009; 16(9):902-3; PMID:19739285; http://dx.doi.org/ 10.1038/nsmb0909-902 [DOI] [PubMed] [Google Scholar]
- 6. Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012; 22:1616-25; PMID:22955974; http://dx.doi.org/ 10.1101/gr.134445.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell 2011; 144(1):16-26; PMID:21215366; http://dx.doi.org/ 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gómez Acuña LI, Kornblihtt AR. Long range chromatin organization: a new layer in splicing regulation? Transcription 2014; 5:PMID:24802896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 2003; 12(2):525-32; PMID:14536091 [DOI] [PubMed] [Google Scholar]
- 10. Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 2014; 54(4):683-90; PMID:24793692; http://dx.doi.org/ 10.1016/j.molcel.2014.03.044 [DOI] [PubMed] [Google Scholar]
- 11. Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 2012; 22(6):1184-95; PMID:22391557; http://dx.doi.org/ 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413-5; PMID:18978789; http://dx.doi.org/ 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 13. Kornblihtt AR. A long noncoding way to alternative splicing in plant development. Dev Cell 2014; 30:117-9; http://dx.doi.org/ 10.1016/j.devcel.2014.07.010in press [DOI] [PubMed] [Google Scholar]
- 14. Staiger D, Brown JW. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013; 25(10):3640-56; PMID:24179132; http://dx.doi.org/ 10.1105/tpc.113.113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding F, Cui P, Wang Z, Zhang S, Ali S, Xiong L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 2014; 15:431; PMID:24897929; http://dx.doi.org/ 10.1186/1471-2164-15-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JW, Barta A, Kalyna M, et al. A chloroplast retrograde signal regulates nuclear alternative splicing. Science 2014; 344:(6182):427-30; PMID:24763593; http://dx.doi.org/ 10.1126/science.1250322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baena-González E. Energy signaling in the regulation of gene expression during stress. Mol Plant 2010; 3:300-13; PMID:20080814 [DOI] [PubMed] [Google Scholar]
- 18. Lauria M. and Rossi V. Epigenetic control of gene regulation in plants. Biochim Biophys Acta 2011; 1809:369-78; PMID:21414429; http://dx.doi.org/ 10.1016/j.bbagrm.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 19. Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B. Post-transcriptional regulation of gene expression in plants during abiotic stress. Int J Mol Sci 2009; 10:3168-85; PMID:19742130; http://dx.doi.org/ 10.3390/ijms10073168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmoll M, Tian C, Sun J, Tisch D, Glass N. Unravelling the molecular basis for light modulated cellulase gene expression - the role of photoreceptors in Neurospora crassa. BMC Genomics 2012; 13:127; PMID:22462823; http://dx.doi.org/ 10.1186/1471-2164-13-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet 2007; 8:217-30; PMID:17304247; http://dx.doi.org/ 10.1038/nrg2049 [DOI] [PubMed] [Google Scholar]
- 22. Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol 2010; 91:29-66; PMID:20705178; http://dx.doi.org/ 10.1016/S0070-2153(10)91002-8 [DOI] [PubMed] [Google Scholar]
- 23. Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 2013; 64:403-27; PMID:23373700; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120221 [DOI] [PubMed] [Google Scholar]
- 24. Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdán PD. Arabidopsis thaliana life without phytochromes. PNAS 2010; 107:4776-81; PMID:20176939; http://dx.doi.org/ 10.1073/pnas.0910446107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woodson J, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 2008; 9:383-95; PMID:18368053; http://dx.doi.org/ 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM. Plastids are major regulators of light signaling in Arabidopsis. Plant Phys 2012; 159:366-90; PMID:22383539; http://dx.doi.org/ 10.1104/pp.112.193599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan K, Crisp P, Estavillo G, Pogson B. Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav 2010; 5:1575-82; PMID:21512326; http://dx.doi.org/ 10.4161/psb.5.12.13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nature reviews Mol Cell Biol 2013; 14:787-802; PMID:24263360; http://dx.doi.org/ 10.1038/nrm3702 [DOI] [PubMed] [Google Scholar]
- 29. Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J. Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 2012; 63:1637-61; PMID:22371324; http://dx.doi.org/ 10.1093/jxb/ers013 [DOI] [PubMed] [Google Scholar]
- 30. Terry M, Smith A. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 2013; 4:14; PMID:23407626; http://dx.doi.org/ 10.3389/fpls.2013.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szechyńska-Hebda M, Karpiński S. Light intensity-dependent retrograde signalling in higher plants. J Plant Phys 2013; 170:1501-16; PMID:23850030; http://dx.doi.org/ 10.1016/j.jplph.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 32. Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta U-M, Battchikova N, Aro E-M. Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol Genomics 2006; 25:142-52; PMID:16403842 [DOI] [PubMed] [Google Scholar]
- 33. Pfannschmidt T. Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci 2003; 8:33-41; PMID:12523998; http://dx.doi.org/ 10.1016/S1360-1385(02)00005-5 [DOI] [PubMed] [Google Scholar]
- 34. Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 1997; 9:627-40; PMID:9144965; http://dx.doi.org/ 10.1105/tpc.9.4.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfalz J, Liebers M, Hirth M, Grübler B, Holtzegel U, Schröter Y, Dietzel L, Pfannschmidt T. Environmental control of plant nuclear gene expression by chloroplast redox signals. Front Plant Sci 2012; 3:257; PMID:23181068; http://dx.doi.org/ 10.3389/fpls.2012.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bräutigam K, Dietzel L, Kleine T, Ströher E, Wormuth D, Dietz K-JJ, Radke D, Wirtz M, Hell R, Dörmann P, et al. Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 2009; 21:2715-32; PMID:19737978; http://dx.doi.org/ 10.1105/tpc.108.062018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. JBC 2005; 280:5318-28; PMID:15561727; http://dx.doi.org/ 10.1074/jbc.M406358200 [DOI] [PubMed] [Google Scholar]
- 38. Petracek ME, Dickey LF, Huber SC, Thompson WF. Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 1997; 9:2291-300; PMID:9437868; http://dx.doi.org/ 10.1105/tpc.9.12.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Phys 2006; 141:436-445; PMID:16603662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol 2008; 66:361-78; PMID:18158584; http://dx.doi.org/ 10.1007/s11103-007-9274-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki N, Koussevitzky S, Mittle R, Miller G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 2012; 35(2):259-270; PMID:21486305; http://dx.doi.org/ 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- 42. Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999; 284(5414):654-657; PMID:10213690; http://dx.doi.org/ 10.1126/science.284.5414.654 [DOI] [PubMed] [Google Scholar]
- 43. Karpinski S, Gabrys H, Mateo A, Karpinska B, Mullineaux P. Light perception in plant disease defence signalling. Curr Opin Plant Biol 2003; 6:390-6; PMID:12873535; http://dx.doi.org/ 10.1016/S1369-5266(03)00061-X [DOI] [PubMed] [Google Scholar]
- 44. Ruckle ME, Larkin RM. (2009). Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol 182:367-79; PMID:19140931; http://dx.doi.org/ 10.1111/j.1469-8137.2008.02729.x [DOI] [PubMed] [Google Scholar]
- 45. Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 2007; 19:3944-60; PMID:18065688; http://dx.doi.org/ 10.1105/tpc.107.054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lepistö A, Rintamäki E. Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 2012; 5:799-816; PMID:22199239; http://dx.doi.org/ 10.1093/mp/ssr106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 2001; 13:2589-607; PMID:11752374; http://dx.doi.org/ 10.1105/tpc.13.12.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiao Y, Ma L, Strickland E, Deng XW. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 2005; 17:3239-56; PMID:16284311; http://dx.doi.org/ 10.1105/tpc.105.035840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Casal JJ, Yanovsky MJ. Regulation of gene expression by light. Int J Dev Biol 2005; 49:501-11; PMID:16096960; http://dx.doi.org/ 10.1387/ijdb.051973jc [DOI] [PubMed] [Google Scholar]
- 50. Fisher AJ, Franklin KA. Chromatin remodelling in plant light signalling. Physiol Plant 2011; 142:305-13; PMID:21457270; http://dx.doi.org/ 10.1111/j.1399-3054.2011.01476.x [DOI] [PubMed] [Google Scholar]
- 51. Guo L, Zhou J, Elling AA, Charron JB, Deng XW. Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Phys 2008; 147:2070-83; PMID:18550682; http://dx.doi.org/ 10.1104/pp.108.122929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mano S, Yamaguchi K, Hayashi M, Nishimura M. Stromal and thylakoid-bound ascorbate peroxidases are produced by alternative splicing in pumpkin. FEBS Lett. 413:21-6; PMID:9287110; http://dx.doi.org/ 10.1016/S0014-5793(97)00862-4 [DOI] [PubMed] [Google Scholar]
- 53. Mano S, Hayashi M, Nishimura M. Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J 1999; 17:309-20; PMID:10097389; http://dx.doi.org/ 10.1046/j.1365-313X.1999.00378.x [DOI] [PubMed] [Google Scholar]
- 54. Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010; 468(7320):112-6; PMID:20962777; http://dx.doi.org/ 10.1038/nature09470 [DOI] [PubMed] [Google Scholar]
- 55. Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 2012; 24:3278-95; PMID:22942380; http://dx.doi.org/ 10.1105/tpc.112.100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A, Brown JW. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J 2008; 53:1035-48; PMID:18088312; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03392.x [DOI] [PubMed] [Google Scholar]
- 57. Tanabe N, Yoshimura K, Kimura A, Yabuta Y, Shigeoka S. Differential Expression of Alternatively Spliced mRNAs of Arabidopsis SR Protein Homologs, atSR30 and atSR45a, in Response to Environmental Stress. Plant Cell Phys 2007; 48:1036-1049; PMID:17556373; http://dx.doi.org/ 10.1093/pcp/pcm069 [DOI] [PubMed] [Google Scholar]
- 58. Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, Matsuoka K, Lin C, Matsushita T. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J 2012; 70:727-38; PMID:22324426; http://dx.doi.org/ 10.1111/j.1365-313X.2012.04937.x [DOI] [PubMed] [Google Scholar]
- 59. Wu H-P, Su Y-S, Chen H-C, Chen Y-R, Wu C-C, Lin W-D, Tu S-L. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol 2014; 15:R10; PMID:24398233; http://dx.doi.org/ 10.1186/gb-2014-15-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Falciatore A, Merendino L, Barneche F, Ceol M, Meskauskiene R, Apel K, Rochaix J-D. The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev 2005; 19:176-187; PMID:15630026; http://dx.doi.org/ 10.1101/gad.321305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lopato S, Waigmann E, Barta A. Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 1996; 8:2255-64; PMID:8989882; http://dx.doi.org/ 10.1105/tpc.8.12.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010; 22:1498-515; PMID:20511297; http://dx.doi.org/ 10.1105/tpc.109.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barajas-López J, Blanco N, Strand Å. Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim. Biophys. Acta 2013; 1833:425-37; PMID:22749883; http://dx.doi.org/ 10.1016/j.bbamcr.2012.06.020 [DOI] [PubMed] [Google Scholar]
- 64. Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 2011; 23:3992-4012; PMID:22128124; http://dx.doi.org/ 10.1105/tpc.111.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun 2011; 2:477; PMID:21934661; http://dx.doi.org/ 10.1038/ncomms1486 [DOI] [PubMed] [Google Scholar]
- 66. Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 2006; 57:675-709; PMID:16669778; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105441 [DOI] [PubMed] [Google Scholar]
- 67. Hausler R, Heinrichs L, Schmitz J, Flugge U. How Sugars Might Coordinate Chloroplast and Nuclear Gene Expression during Acclimation to High Light Intensities. Mol Plant 2014; 7:1121-1137; PMID:25006007; http://dx.doi.org/ 10.1093/mp/ssu064 [DOI] [PubMed] [Google Scholar]
- 68. Blanco NE, Guinea-Díaz M, Whelan J, Strand Å. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Phil Trans R Soc B 2014; 369:20130231; PMID:24591717; http://dx.doi.org/ 10.1098/rstb.2013.0231 [DOI] [PMC free article] [PubMed] [Google Scholar]


