Abstract
The internal ribosome entry site (IRES) element located at the 5´untranslated genomic region of various RNA viruses mediates cap-independent initiation of translation. Picornavirus IRES activity is highly dependent on both its structural organization and its interaction with host factors. Small molecules able to interfere with RNA function are valuable candidates for antiviral agents. Here we show that a small molecule based on benzimidazole (IRAB) inhibited foot-and-mouth disease virus (FMDV) IRES-dependent protein synthesis in cells transfected with infectious RNA leading to a decrease of the virus titer, which was higher than that induced by a structurally related benzimidazole derivative. Interestingly, IRAB preferentially inhibited IRES-dependent translation in cell free systems in a dose-dependent manner. RNA structural analysis by SHAPE demonstrated an increased local flexibility of the IRES structure upon incubation with IRAB, which affected 3 stem-loops (SL) of domain 3. Fluorescence binding assays conducted with individual aminopurine-labeled oligoribonucleotides indicated that the SL3A binds IRAB (EC50 18 μM). Taken together, the results derived from SHAPE reactivity and fluorescence binding assays suggested that the target site of IRAB within the FMDV IRES might be a folded RNA structure that involves the entire apical region of domain 3. Our data suggest that the conformational changes induced by this compound on a specific region of the IRES structure which is essential for its activity is, at least in part, responsible for the reduced IRES efficiency observed in cell free lysates and, particularly, in RNA-transfected cells.
Keywords: antiviral molecules, fluorescence binding assay, IRES elements, picornavirus, RNA ligands, RNA structure, SHAPE probing, translation initiation, 2-aminobenzimidazole
Introduction
In recent years RNA has received great attention being recognized a major player in many cellular processes.1 The vast majority of RNA functions depend on RNA structure,2 which on the other hand impacts on RNA-protein interactions and contributes to gene expression control in all living organisms.3 Common to most RNA regulatory elements, RNA structure dictates the function of internal ribosome entry site (IRES) elements.4 These elements, which were discovered in picornavirus genomic RNAs,5,6 are in charge of driving translation initiation in various RNA viruses and a subset of cellular mRNAs.7,8 IRES-driven translation initiation represents an alternative mechanism to start protein synthesis under situations that compromise cap-dependent translation initiation, typically occurring in infected cells. Under normal situations, however, most eukaryotic mRNAs initiate translation by a mechanism that depends on the recognition of the m7GpppN residue (termed cap) located at the 5' end of most mRNAs.9
IRES elements exhibit a wide diversity of nucleotide sequences, RNA secondary structures and trans-acting factors requirements, as illustrated by those present in the genome of picornaviruses and the hepatitis C virus (HCV).10 Foot-and-mouth disease virus (FMDV) is a member of the Picornaviridae family that causes a contagious disease in cattle and cloven-hoofed ungulates. The viral genome consists of a positive polarity single-stranded RNA of about 8500 bases, encoding a single open reading frame flanked by a highly structured untranslated region (UTR) at the 5´end of about 1200 nucleotides.7 Upon entry into the cell, the viral genome is immediately translated into a polyprotein that is processed by virus-encoded proteases. Translation and replication of the viral genome occurs in the cytoplasm of infected cells. As in all picornaviruses, the viral genome does not contain the cap structure (m7GpppN) typically present in cellular mRNAs at its 5’ end. Instead, picornavirus 5’UTR are longer and contain the IRES element, a heavily structured region controlling internal initiation of translation.5-6
Functional and structural analysis have shown that the FMDV IRES is organized in 5 structural domains of which domain 2 includes a polypyrimidine tract that provides the polypyrimidine tract-binding protein (PTB) binding site.11 Domain 3 consists of 210 nucleotides, the most apical region of which is arranged as a cruciform structure12 that harbors conserved motifs. Disruption of these motifs abrogates IRES activity, in addition to altering distant RNA-RNA interactions.13 The proximal part of domain 3 consists of a long stem interrupted with bulges that include several non-canonical base pairs and a helical structure essential for IRES activity. Domain 4 is organized into 2 hairpin-loops conserved between FMDV and encephalomyocarditis virus (EMCV), providing the binding site for the translation initiation factor eIF4G.14-16 Finally, domain 5 is composed of a hairpin-loop and a polypyrimidine-rich tract that provides the binding site for various RNA-binding proteins.17,18
Selective 2´-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) data revealed a modular organization of the FMDV IRES region.19 Furthermore, the RNA organization of this IRES region is affected by the concentration of divalent ions in the folding buffer20 with the peculiarity that the apical loop of domain 3 is more flexible at near physiological concentration of Mg2+ (0.5 mM) than at high concentration (6 mM). Given that RNA structure plays a critical role in the function of regulatory elements controlling expression of viral proteins, we took advantage of the knowledge of IRES structural features to design compounds potentially inhibiting protein synthesis.
Treatment of picornavirus infected cells with small molecules such as Ribavirin (a guanosine analog) or Quinacrine (9-aminoacridine derivative) effectively reduce virus titer.21,22 On the other hand, the nucleic acid-binding ligand acriflavine inhibits protein synthesis in cell free lysates with specificity for internal initiation of translation.23 However, the efficacy of most these molecules in animal models are not well documented and their mode of action are still poorly understood.
Small molecules capable of interacting with nucleic acids are candidates to interfere with gene expression.24,25 The benzimidazole scaffold is a privileged pharmacophore of many therapeutic agents.26 Benzimidazole derivatives have also been shown to elicit a variety of pharmacological effects by interacting with RNA structural elements. In a search for mimics of aminoglycoside antibiotics, an NMR-based screening of a compound library identified a series of benzimidazole hits that bind the bacterial A-site 16S rRNA.27 Recently, a 2-aminobenzimidazole library has been screened for binding to RNA internal loops,28 and a bisbenzimidazole derivative has been reported to bind the expanded r(CUG) repeats that cause myotonic dystrophy type 1 (DM1), and to some extent overcome DM1-associated defects.29
In the IRES context, 2-aminobenzimidazole derivatives have also been shown to interact with the SLIIa structural element of the HCV-like IRES,30,31 and to interfere with viral multiplication at low micromolar concentrations.32,33 However, to the best of our knowledge, their potential to bind other types of IRES elements as well as cap-dependent translation has not been examined.
Here we show that a small molecule based on benzimidazole (IRAB) induced a local modification of FMDV IRES RNA structure and inhibited IRES-dependent translation in RNA-transfected cells as well as in cell free systems. Our data suggest that the conformational changes induced by this compound on a region of the IRES structure which is essential for internal translation initiation is responsible for the reduced IRES activity observed in cell free lysates and in living cells.
Results
The 2-aminobenzimidazole derivative IRAB preferentially inhibits IRES-dependent translation in cell free systems
The IRES elements present in various RNA virus consist of specialized RNA structural elements that govern cap-independent initiation of translation.7 Because of their lack of conservation with cellular mRNAs these structural elements provide an attractive target for antiviral agents. A main goal of this study was to develop small molecules able to interfere with picornavirus IRES activity, with special emphasis on FMDV. However, since IRES elements differ considerably in primary sequence and RNA structure organization34 the interaction of a particular type of RNA ligand(s) with different types of IRES elements needs to be experimentally tested.
IRAB (IRES Aminobenzimidazole Binder) (Fig. 1A) was devised to be an achiral analog of the 2-aminobenzimidazole derivatives known as Isis-11 (Fig. 1A), an inhibitor of HCV replication that hinders HCV IRES-mediated translation.32 We reasoned that chirality does not appear to be a major determinant to account for the binding of Isis-11 to HCV IRES, since the racemic mixture and the single enantiomers do not significantly differ in their affinity for the target site.35 When compared with Isis-11, IRAB features an additional unsaturated bond at the oxygen-containing ring that eliminates the single stereocenter of Isis-11 and renders an aromatic furan ring. It is worth mentioning that an additional aim in IRAB design was to develop a shorter and more efficient synthetic route than that described for Isis-11 (Fig. S1).
Figure 1.
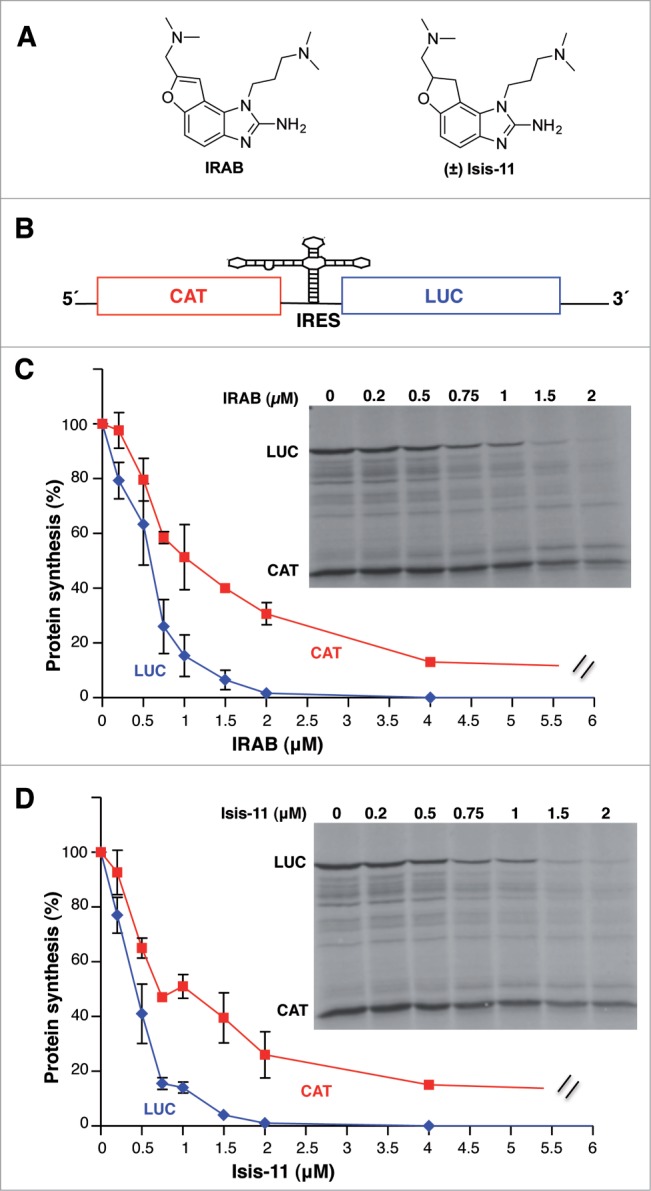
Inhibitory effect of IRAB on the in vitro translation efficiency driven by the FMDV IRES. (A). Chemical structure of IRAB and the structurally related compound Isis-11. (±) denotes racemic mixture (B) Schematic diagram of the bicistronic mRNA used in in vitro translation assays. Initiation of translation of luciferase (Luc) is IRES-dependent while translation of chloramphenicol acetyl transferase (CAT) is cap-dependent. (C) Inhibitory effect of IRAB on the in vitro translation efficiency. Autoradiography of translation products synthesized in reticulocyte lysates using the FMDV bicistronic RNA incubated with the indicated amounts of IRAB. The intensity of Luc and CAT polypeptides in each lane were determined by densitometry and shown relative to the total intensity of the lane with DMSO alone, which was set at 100%. Data corresponds to the mean (±SD) of 3 independent assays. (D) Inhibitory effect of Isis-11 on the in vitro translation efficiency. Autoradiography of translation products obtained from in vitro synthesized FMDV bicistronic RNA reporter in the presence of the indicated amounts of Isis-11. The intensity of the lanes were determined by densitometry and shown relative to the total intensity of the lane with DMSO alone, which was set at 100%. Data corresponds to the mean (±SD) of 3 independent assays.
To directly assess the effect of IRAB on IRES activity we set up in vitro translation assays using a bicistronic RNA harboring the picornavirus FMDV IRES between chloramphenicol acetyl transferase (CAT) and firefly luciferase (Luc) reporters that monitored on translation efficiency initiated via 5´-dependent or IRES-dependent, respectively (Fig. 1B). Increasing amounts of IRAB were added to the reticulocyte lysates (RRL) in the presence of a constant amount of the bicistronic RNA. Incorporation of 35S-methionine in the newly synthesized proteins allowed us to monitor the efficiency of protein synthesis encoded by the first cistron (CAT) or the second cistron (luciferase). A preferential decrease of luciferase protein synthesis was observed in the presence of IRAB, reaching about 50% inhibition at 0.5 μM and near complete inhibition at 2 μM (Fig. 1C). In contrast, only 20% inhibition of the first cistron (CAT) was observed at 0.5 μM concentration of the drug. According to two-tail Student´s t-test, the differences observed at 0.75, 1 and 2 μM of IRAB were statistically significant (P < 0.05). Higher concentration (up to 4 μM) of this compound still showed a preferential decrease of luciferase relative to CAT synthesis.
Further studies conducted to compare the effect of IRAB with Isis-11 in vitro indicated a similar inhibitory capacity of these compounds when tested in RRL (Fig. 1D). In support of the specificity of the inhibitory effect of these small molecules, unrelated potential RNA ligands, such as peptoids or peptide nucleic acids (PNAs), did not affect translation of any of these reporter genes in vitro (Fig. S2).
Additional in vitro translation studies using a bicistronic RNA that carries the HCV IRES indicated a similar inhibitory effect (Fig. S3), suggesting certain similarities between these genetically distant IRES elements. Furthermore, in both cases (IRAB at 0.5, 1, 2 μM, and Isis-11 at 1, 2 μM) there was a preferential statistically significant (P < 0.05) effect over IRES-dependent translation (monitored by luciferase) compared to 5´end-dependent translation (monitored by CAT), in agreement with the data obtained using the FMDV IRES (Fig. 1C and D). Moreover, the effect on IRES activity was also confirmed using a monocistronic HCV RNA (Fig. S4).
Taken together the results obtained with the bicistronic RNAs, we conclude that there is a preferential effect of IRAB on IRES-dependent translation initiation over cap-dependent translation, irrespectively of the IRES (FMDV or HCV) present in the RNA analyzed.
IRAB inhibits multiplication of FMDV
Next, in order to assess whether IRAB could effectively interfere with picornavirus IRES-dependent translation in living cells, and therefore affect virus multiplication, we transfected BHK-21 cells with infectious FMDV RNA synthesized in vitro (Fig. 2A). Virus yield was determined in the supernatant of transfected monolayers by plaque assays on fresh susceptible cell monolayers. In addition, cell lysates were recovered at different times to determine viral protein synthesis. Incubation of RNA transfected cells with IRAB induced a dose-dependent reduction of the virus titer 24 h post-transfection (Fig. 2B). Furthermore, the virus titer decrease induced by IRAB (2 μM) was higher than that observed in the presence of Isis-11 (P < 0.05). These results were fully supported by the extent of VP1 protein synthesis determined by western blot (Fig. 2C). While the cells transfected with FMDV RNA yielded high amounts of VP1, addition of IRAB (2 μM) to the transfected cells led to severe VP1 reduction 24 hpt. In contrast, transfected cells treated with Isis-11 showed a lower decrease of VP1 at the same post-transfection time. Equal amount of total protein loading control was verified using anti-tubulin antibody.
Figure 2.
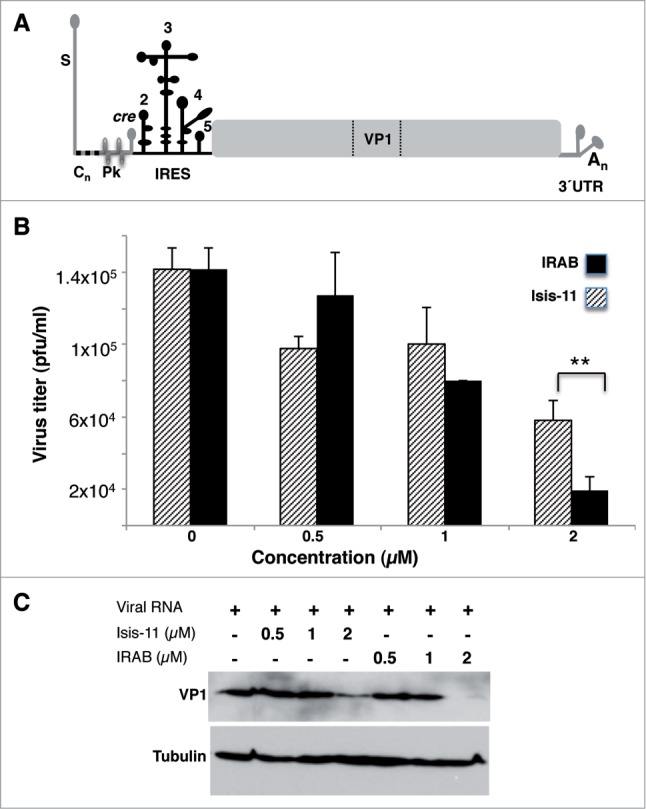
(A). Schematic representation of the FMDV viral genome. A gray line at the 5´end depicts structural elements, including the S hairpin, the polyC tract (Cn), 2–4 pseudoknots (Pk), and the cis replicative element (cre), followed by the IRES element depicted by a solid black line. A solid gray rectangle represents the viral polyprotein with indication of the region encoding the capsid protein VP1. The 3´UTR consists of 2 stem-loops followed by a poly(A) tail (An). (B) Analysis of virus multiplication in the presence of IRAB. Viral yield reduction induced by increasing amounts of IRAB (black bars) or Isis-11 (striped bars). In vitro synthesized FMDV RNA (50 pg) was transfected in IRAB treated BHK-21 cell monolayers, in duplicate wells. Virus yield was determined using fresh cells monolayers as the number of plaque forming units (PFU)/ml in the supernatant 24 hpt, shown relative to the control RNA which was set at 100%. Values represent the mean and standard deviation of triplicate assays. P values were calculated using two-tail Student´s t-test; differences were considered significant when P < 0.05. (C) Analysis of the effect of IRAB on viral proteins. Western blot of viral protein VP1 accumulated in transfected BHK-21 cells 24 hpt incubated with the indicated concentration of IRAB and Isis-11; anti-tubulin was used as loading control.
Cells treated with either Isis-11 or IRAB (1 or 2 μM) continued proliferating during more than 48 hours, although with a slower doubling time than the untreated cells (Fig. S5A). This inhibitory effect on cell growth was not accompanied by cell monolayer detachment at the concentration inducing decrease in viral protein synthesis and virus yield (Fig. S5B).
These results enable us to conclude that IRAB exerts an inhibitory effect on FMDV multiplication, which is consistent with the capacity of this compound to inhibit IRES-driven protein synthesis.
IRAB induces a local structural reorganization of the IRES element
RNA structure plays a key role in IRES-dependent translation.10 Since incubation of IRAB with RNAs effectively inhibited internal initiation driven by the FMDV IRES in in vitro translation assays, we used SHAPE chemistry to analyze whether this compound had any influence on the conformation of the IRES structural organization. SHAPE probing is a quantitative methodology that reports on RNA flexibility at nucleotide-resolution, allowing the study of long RNA molecules in solution.36,37 RNA transcripts synthesized in vitro carrying the IRES element upstream of the luciferase ORF were incubated with increasing concentrations of IRAB (2, 10 and 20 μM) prior to NMIA treatment, which selectively reacts with ribose 2’-hydroxyl groups of conformational flexible nucleotides forming adducts.38 Reactive positions were subsequently detected as RT stops in primer extension reactions conducted with radiolabeled primers. Four primers spaced all along the IRES sequence generated overlapping readings allowing the study of the entire region.20 In all cases the background values observed in the NMIA (-) RNA were subtracted from the NMIA-treated RNA prior to SHAPE data normalization.39
The mean SHAPE reactivity values obtained for free RNA and IRAB-treated RNA (20 μM) is shown in Figure 3A. Nucleotides with normalized reactivity >0.5 (red shaded) are considered to be highly flexible, likely unpaired. Nucleotides that showed very low reactivity values (<0.2) are putatively involved in base pairing or in tertiary interactions. Nucleotides with SHAPE reactivity between 0.2 and 0.5 (yellow shaded) are considered to be moderately reactive.
Figure 3.
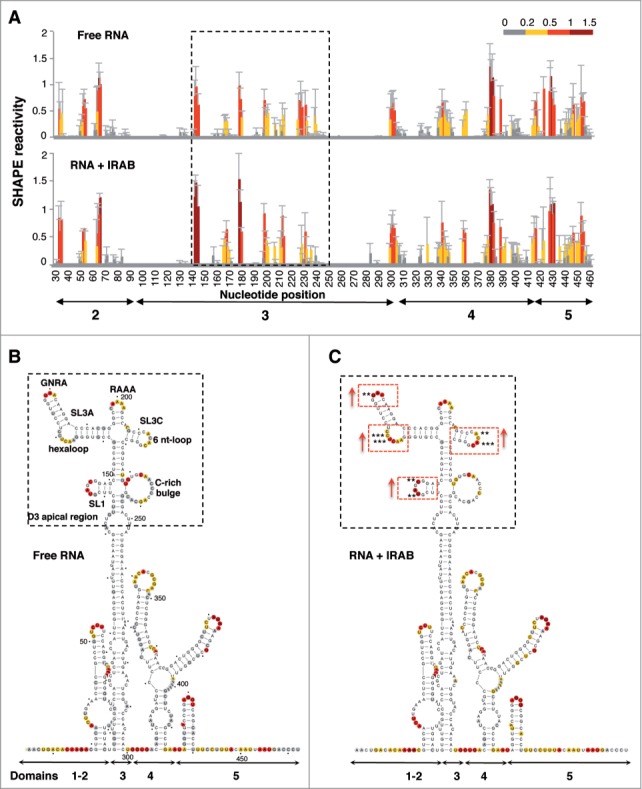
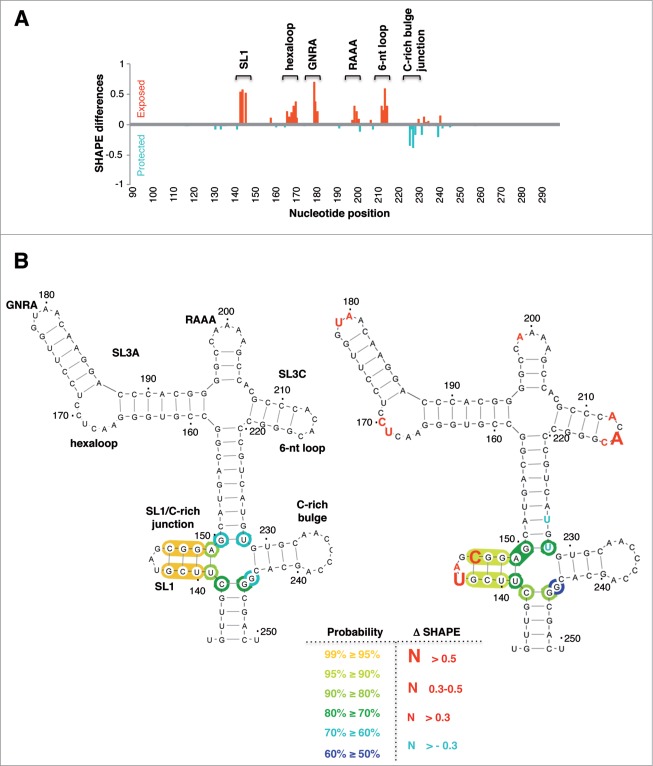
Modification of SHAPE reactivity profile upon incubation of the IRES RNA with IRAB. (A) SHAPE reactivity profiles of the IRES folded at 0.5 mM Mg2+ in the absence (free RNA) or in the presence of IRAB (20 μM). Values correspond to the mean SHAPE reactivity (±SD) of at least 3 independent assays. SHAPE reactivity is represented in a colored scale in which 0 indicates unreactive nucleotides and the average intensity at highly reactive nucleotides is set to 1.0. Nucleotide positions are indicated on the x-axis. Domains 1, 2, 3, 4 and 5 of the IRES are indicated below the corresponding nt position. A broken rectangle depicts the region harboring main reactivity changes. (B) Secondary structure of free RNA according to SHAPE reactivity. Nucleotides are colored according to their reactivity, a dot is positioned every 10 nt, numbers are indicated every 50 nt. Stem-loops (SL1, SL3A and SL3C) and motifs (hexaloop, GNRA within SL3A, RAAA, 6 nt-loop within SL3C, and the C-rich bulge), located in domain 3 (D3) apical region referred to in the text are indicated. (C) SHAPE reactivity for the RNA treated with IRAB (20 μM). Red rectangles depict location of nts with statistically significant reactivity modification relative to free RNA. p values were calculated using the two-tail Student´s t-test; differences were considered significant when P < 0.05 or P < 0.01, denoted by 2 or 3 asterisks, respectively.
The SHAPE reactivity profile obtained for the free RNA plotted on the secondary RNA structure (Fig. 3B) is in full agreement with earlier data20 as well as with chemical and enzymatic RNA probing.40 Predicted base-paired regions showed no or low reactivity while most of the predicted loops and internal bulges showed moderate to high reactivity. In particular, the apical region of domain 3 (D3) showed SHAPE reactive nucleotides within specific motifs located in the loop of SL1, the hexaloop and the GNRA motif of SL3A, the RAAA motif, the 6 nt-loop of SL3C and the C-rich bulge (Fig. 3B).
Upon treatment of the IRES with IRAB, the apical region of domain 3 showed significant reactivity changes relative to free RNA (Fig. 3C, depicted by red dashed rectangles, nts marked with 2 or 3 asterisks denoting P values <0.05 or <0.01, respectively). The statistical significance of the reactivity data obtained in the presence and absence of IRAB was determined by the two-tail Student´s t-test (Fig. 3B, C). Specifically, significant increased reactivity (P < 0.05 or <0.01) was observed in positions U143, C146, U169C170, U179, and A212A214. Statistically significant changes were not observed in the remaining IRES regions. Incubation of the RNA with lower IRAB concentration IRAB solutions (2 and 10 μM) showed a dose–dependent reactivity in the same IRES region (Fig. S6).
To quantify the effect of IRAB using an unbiased approach for visualizing the compound-induced modification of the IRES local flexibility, we obtained the difference profile39,41 by subtracting the reactivity of the free RNA from that of the IRAB treated RNA (Fig. 4A). The difference profile showed increased reactivity or protected residues that were reported as positive and negative values, respectively. Nucleotides showing absolute difference reactivity values ≥0.3 and P values <0.05 were located in SL1, the hexaloop and the GNRA motif of SL3A, and the 6-nt loop of SL3C (Fig. 4A). Furthermore, although no statistical significant reactivity difference was observed in the residues at the junction of the C-rich bulge, a partial protection relative to free RNA was detected in U226U228. We hypothesized that binding of IRAB to domain 3 could give rise to a less constrained apical region structure induced by a disrupted interaction among the motifs mentioned above.
Figure 4.
Impact of IRAB on IRES local flexibility. (A) Domain 3 SHAPE difference plot of IRAB treated RNA relative to the untreated RNA. Red bars depict positions with positive value of SHAPE differences while blue bars depict positions with negative values. (B) RNA structure model predicted by RNAstructure software imposing the SHAPE reactivity values (nts 133 to 250) for the free RNA (left) and the IRAB-treated RNA (right). The position of motifs refer to in the text are indicated on the free RNA structure. Red letters depict nts with absolute SHAPE differences higher than 0.3; font size reflects the magnitude of SHAPE difference. Blue letters indicate nts with absolute difference > −0.3. Unmarked nts have a probability >99% of adopting the predicted conformation, yellow circles indicate probabilities >95%, light to dark green circles indicate probabilities >90, >80 or >70% while light to dark blue circles reflect probabilities > 60 or >50%, respectively.
RNA structure models predicted for the free RNA and the IRAB-treated RNA using RNAstructure software42 incorporating the SHAPE reactivity indicated local modification of the secondary structure of the apical region of domain 3 (Fig. 4B) resulting in free energy differences (ΔG -149.3 kcal/mol for the free RNA versus -154.4 kcal/mol for the IRAB treated RNA). This difference may arise from the penalty for base pairing of highly reactive nucleotides imposed by the pseudo-free energy change term integrated in RNAstructure.43
Together, these results enable us to conclude that incubation of IRAB with RNAs bearing the FMDV IRES induces a conformational change of domain 3, likely responsible for the inhibitory effect on IRES activity.
Hydroxyl radical cleavage reveals a more solvent accessible domain 3 structure upon IRAB treatment
The changes in the pattern of SHAPE reactivity observed in RNA treated with IRAB prompted us to determine the solvent accessibility of the IRES by hydroxyl radical cleavage footprinting, a methodology that reveals nucleotides buried within the folded RNA structure.44 Solvent accessibility of the IRES was probed using Fe(II)-EDTA and H2O2 in a solution of RNA renatured in the presence or absence of IRAB (20 μM) followed by primer extension analysis with fluorescently labeled primers, resolved by capillary electrophoresis. The results yielded cleavage information at single nucleotide resolution, reporting on solvent exposure of the IRES nucleotides (Fig. 5A). Yellow and not-shaded residues indicate buried residues due to structural compaction, while pale and dark gray indicate solvent accessible positions.
Figure 5.
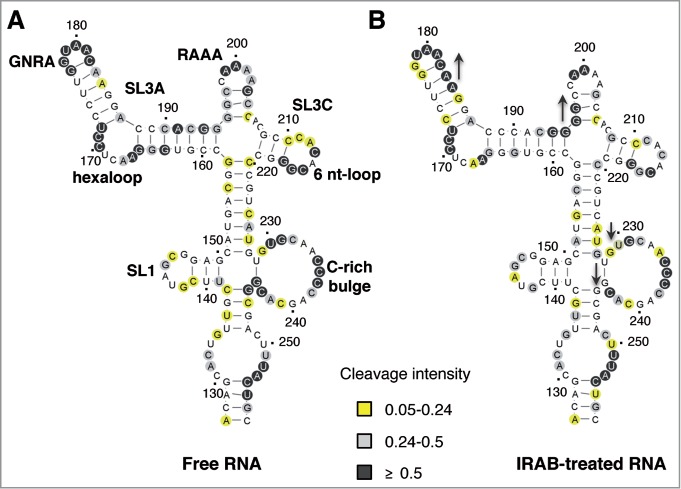
Hydroxyl radical probing of the IRES upon addition of IRAB. Secondary RNA structure of the apical region of domain 3 in the absence (A) or the presence (B) of IRAB. Hydroxyl radical cleavage intensities were normalized to a scale from 0 to 1.5, where 1.0 is defined as the average intensity of highly reactive residues. Nucleotides with reactivity values lower than half the mean reactivity are defined as solvent-inaccessible. Solvent-inaccessible positions are shown in white (cleavage intensity < 0.05) or yellow (cleavage intensity < 0.24), while accessible nucleotides are shown in pale gray (cleavage intensity > 0.24 to 0.5) or dark gray (cleavage intensity > 0.5). The position of motifs refer to in the text are indicated on the free RNA structure. Upside or upside-down arrows on the IRAB-treated RNA depict nucleotides with increased or decreased solvent accessibility, respectively.
Upon folding of the RNA in the presence of IRAB, residues A181CAA184 and G194GG196 within SL3A of domain 3 became more accessible to solvent (Fig. 5B, dark gray nucleotides) indicating a lower structural compaction. In contrast, residues U230, G244GC246 at the C-rich junction became less solvent accessible (Fig. 5B, light gray, yellow or unmarked nts). These results together with those shown on Figures 3 and 4, suggest that IRAB treatment of the IRES induces a less compact RNA structure of domain 3, supporting the hypothesis that this RNA ligand perturbs the tertiary structure of this region.
Binding of IRAB to the FMDV IRES requires at least the apical region of domain 3
Although HCV and FMDV IRES elements are genetically distant, short structural motifs appear to be conserved between these RNA regulatory elements. For instance, the HCV IIa and the FMDV SL3A subdomains share partial RNA structure similarity, such as the AACUA pentaloop (A53-A57) and the AACUCC hexaloop (A166-C171), respectively, within a stable stem-loop (Fig. 6A). This striking conservation, together with the results of SHAPE probing, prompted us to determine whether subdomain SL3A could be the target of IRAB. To this end, an oligoribonucleotide (SL3A/AP167, Fig. 6A) bearing the FMDV SL3A sequence and labeled with 2-aminopurine at the A167 position was used in fluorescence assays (Fig. 6B). When the FMDV SL3A/AP167 oligoribonucleotide was titrated with IRAB, a dose-dependent increase in fluorescence was observed, consistent with a conformational change that displaces and unstacks the fluorescent base analog from the interior of the RNA bulge. Positive binding of both IRAB and Isis-11 to SL3A/AP167 was determined (EC50 of 18.62 and 15.49 μM, respectively, Fig. 6D). Next, we attempted to determine whether other structural motifs (Fig. 6A) of the apical region of the FMDV IRES could behave as the target of these compounds. As shown in Figure 6D, IRAB did not bind to SL3A/AP180 (labeled in the GNRA motif). Since SL3A/AP167 and SL3A/AP180 differ exclusively in the AP-labeled position, the lack of binding to SL3A/AP180 (EC50 >100 ) indicates that IRAB is not an unselective RNA ligand. Rather, it shows a preferential binding to the AACUCC hexaloop of the SL3A subdomain over the GNRA loop.
Figure 6.
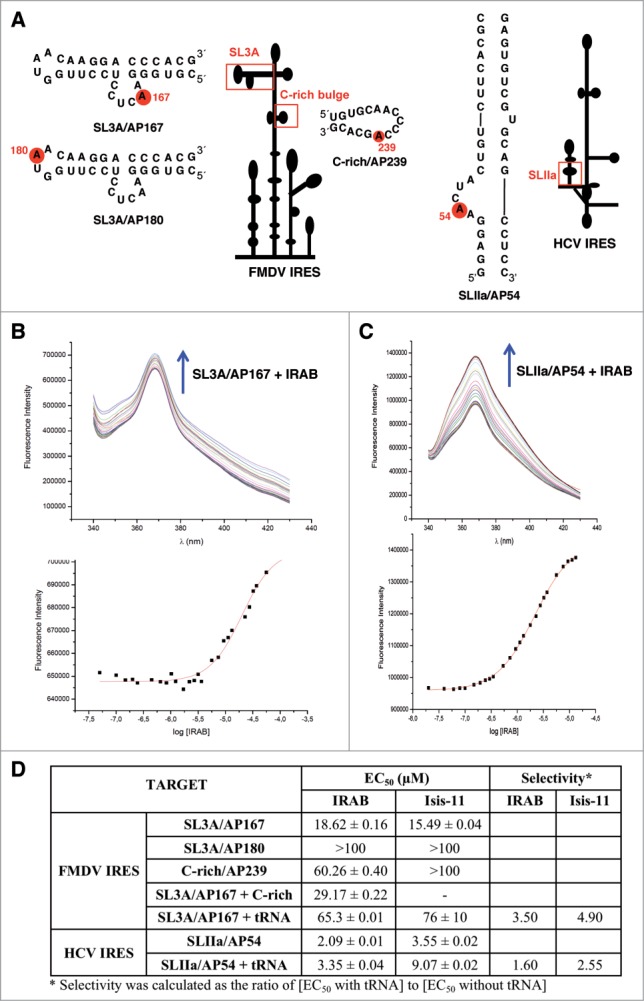
Fluorescence binding assays. (A) 2-Aminopurine labeled oligonucleotides within the FMDV and HCV IRES used in this study. A red circle on each oligonucleotide depicts the labeled residue, and a red rectangle depicts the position of each oligonucleotide within the corresponding IRES element. (B) Fluorescence profile for the titration of 0.5 μM SL3A/AP167 with IRAB (from 50 nM to 130 μM). The arrow indicates the changes in SL3A/AP167 fluorescence produced by increasing amounts of IRAB (top panel). Fitting of the corresponding dose-response curve for the calculation of the EC50 value (bottom panel). (C) Fluorescence profile for the titration of 0.5 μM SLIIa/AP54 with IRAB (from 20 nM to 13 μM). The arrow indicates the changes in SL3A/AP167 fluorescence produced by increasing amounts of IRAB (top panel). Fitting of the corresponding dose-response curve for the calculation of the EC50 value (bottom panel). (D) Binding affinities (EC50 ± standard deviation) and selectivities of IRAB and Isis-11 for the 2-aminopurine labeled oligonucleotides shown in Figure 6A. The EC50 of SL3A/AP167 + C-rich was determined using equimolar amounts of labeled SL3A/AP167 and unlabeled C-rich oligonucleotide. The EC50 of SL3A/AP167 + tRNA and SLIIa/AP54 + tRNA were determined by performing fluorescence titrations in the presence of a 10-fold molar excess of competitor tRNAmix. The later data were used to calculate the selectivities of the ligands for the given oligonucleotide, expressed as the ratio of [EC50 with tRNA] to [EC50 without tRNA].
Additional fluorescence experiments were carried out using the oligoribonuclotide C-rich/AP239, which contains a large loop rich in AC sequences. The results indicated that C-rich/AP239 was a poor ligand of both IRAB (EC50 60.26) and Isis-11 (>100). Further studies aimed to determine whether the binding affinities could be improved in the presence of a different structural motif as a consequence of a tertiary interaction were done using equimolar mixtures of two FMDV oligoribonucleotides. The results of these binding assays showed that the unlabeled C-rich oligoribonucleotide (same sequence as C-rich/AP239 but is not labeled) behaved as a competitor for the IRAB binding to SL3A/AP167, giving rise to higher EC50 values (29.17, Fig. 6D).
In parallel, an HCV subdomain IIa oligoribonucleotide construct (SLIIa/AP54, Fig. 6A), used earlier for binding assays of Isis-11,30 was also employed to assess IRAB affinity. Not surprisingly, IRAB was found to bind SLIIa/AP54 with an affinity similar to that of Isis-11, even a bit higher (Fig. 6C and D). This result suggests that the 2 inhibitor compounds likely induce similar conformational changes in the HCV IRES subdomain IIa.30,35
To assess the target selectivity of IRAB and Isis-11 for both HCV and FMDV IRES subdomains, fluorescence titrations were also performed in the presence of a 10-fold molar excess of competitor tRNAmix, which corresponds to about 27-fold nucleotide excess in the case of SL3A/AP167 and 23-fold in the case of SLIIa/AP54. The EC50 determined for the mixtures of tRNA and either SL3A/AP167 or SLIIa/AP54 are higher than those found for the oligonucleotides alone, as often in this kind of competition experiments.25 From these data, the degree of selectivity of the ligands for their respective targets can be calculated as the ratio of [EC50 with tRNA] to [EC50 without tRNA] (Fig. 6D). The selectivities found rule out the possibility of IRAB and Isis-11 being promiscuous ligands. Interestingly, and contrary to what could be speculated for the more extended aromatic system of IRAB as compared to Isis-11, IRAB shows better selectivity than Isis-11 on both IRES subdomains. This result highlights that the so far unexplored extended aromaticity of IRAB-like compounds should not be a handicap to allow for the development of more selective IRES ligands.
These results, in conjunction with the SHAPE reactivity data, suggest that the target site of IRAB within the FMDV IRES might be a folded RNA structure that involves the entire apical region of domain 3. It is worth noting that the conformational changes induced by the IRAB compound on FMDV IRES are dissimilar to those described for the interaction of HCV IRES with Isis-11 (and most probably IRAB). Isis-11 showed binding to a chimeric HCV subdomain IIa bulge, leading to a conformational change that widens the interhelical angle.30,35,45 It remains to be established whether there is a different flexibility of the bulges of HCV IIa and FMDV 3a subdomains that may account for the observed differences in binding affinities.
Discussion
In this work we have used a molecular design approach aimed at interfering with internal initiation translation driven by the FMDV IRES. We have prepared a novel benzimidazole compound (IRAB) and found that i), IRES activity is preferentially inhibited upon addition of IRAB to cell-free lysates; ii), incubation of IRAB with BHK-21 cells transfected with in vitro synthesized viral RNA leads to a decline in virus yield and viral protein synthesis; iii), IRAB induces a local perturbation of the IRES structure that correlates with the inhibitory effect of this compound; iv), the target site of IRAB on the HCV and the FMDV IRES differs in sequence composition and RNA structure.
Our strategy for the design of an Isis-11 analog as IRES-dependent translation inhibitor has proven to be relevant, since IRAB is much easier to obtain than Isis-11 (and other Isis-related molecules), and its lack of chirality does not hamper its binding to RNA secondary structural motifs but rather increases its selectivity for the target sites, at least in the cases here reported.
The observation that IRES activity is inhibited in in vitro translation assays at lower concentration of IRAB than cap-dependent translation initiation emphasizes the usefulness of this RNA ligand to specifically interfere viral RNA multiplication. Indeed, different synthetic molecules targeting specific RNA motifs essential for viral RNA multiplication have been exploited as therapeutic agents.46-51 Furthermore, the IRES-inhibitory property of IRAB was observed both in in vitro translation assays using cell free lysates and in cells transfected with infectious RNA revealing that the primary targeted event is viral RNA translation, which is IRES-dependent.7 In agreement with this, a specific decrease of viral protein synthesis occurred in transfected cells concomitantly to a reduction in virus yield in the presence of IRAB (2 μM). Noteworthy, cells continued dividing in the presence of either Isis-11 or IRAB during 72 hours, although with a slower doubling time than the untreated cells (Fig. S5A). This inhibitory effect on cell proliferation, however, was not accompanied by cell monolayer detachment at the concentration inducing decrease in viral protein synthesis and virus yield reduction (Fig. S5B).
The inhibition of gene expression observed in susceptible cells transfected with infectious RNA may depend on the interaction of the IRES with trans-acting factors present in different concentrations within the cell cytoplasm, interactions with other cis-acting RNA elements such as the viral 3´UTR,52 and modification of host factors in infected cells.53-55 Thus, we could not discard that this compound could be acting at various levels affecting proteins needed for IRES-dependent translation initiation. However, the inhibition observed in transfected cells are unlikely due to type I interferon (IFN) production since hamster BHK-21 and swine IBRS-2 cell lines have an inactive type I IFN system in response to FMDV infection.56
Our study provides evidences for changes in local flexibility of the IRES structure upon incubation of the RNA with IRAB, leading to a local perturbation of the RNA structure of the apical region of domain 3. This IRES region, which plays a critical role in IRES function, harbors conserved motifs that have been proposed to be involved in distant interactions likely generating a compact RNA structure.13,57,58 Specifically, SHAPE probing of the IRES upon incubation with IRAB showed increased local flexibility involving residues of the hexaloop and the GNRA motif of SL3A, in addition to SL3C and SL1, while U226U228 (located at the junction of the C-rich loop) were partially protected. Hence, disruption of distant interactions among the loops mentioned above would be accompanied by a less accessible C-rich bulge toward NMIA (internally base-paired, as predicted by RNAstructure software). The increase in SHAPE reactivity of residues within D3 apical loops after IRAB-treatment suggests a perturbation of RNA structure according to RNAstructure software. Differences in SHAPE reactivity induced by IRAB in SL1 (U143 and C146) and the junction with the C-rich bulge (U226U228) would result in structural rearrangements as a consequence of the penalty imposed by the RNAstructure software for highly or moderately reactive nucleotides to form base pairs. In support of this hypothesis, a mutational analysis carried out on SL1 residues showed that disruption of the stem reduced IRES activity whereas compensatory mutations restoring the secondary structure recovered translation.59
Although the nucleotides involved in tertiary interactions remain elusive, our results support the existence of a complex network of long-distant interactions within this IRES region. This hypothesis is in agreement with earlier proposals in which nucleotides belonging to the GNRA motif, the RAAA stem-loop and G240 within the C-rich bulge are linked in the three-dimensional RNA structure.12,57 As demonstrated by mutational and RNA probing studies, domain 3 plays a fundamental role in dictating the formation of a constrained structure likely providing the correct orientation to recruit the ribosome subunits to the initiation site.10 Therefore, the conformational changes induced by IRAB would result in a misfolded IRES structure leading to diminished internal translation initiation, as observed in the translation assays reported here.
Attempts to identify specific target sites of IRAB within the IRES using short stem-loops showed that the FMDV SL3A alone is enough to support binding to this compound. However, the EC50 value (18.62 μM) of the SL3A/AP167-IRAB interaction measured by fluorescence assays, together with the results of SHAPE reactivity of the IRAB-treated IRES RNA, strongly support the hypothesis that the entire apical region of domain 3 may be important for IRAB binding.
An additional question is whether the affinity measured with a single stem-loop of the IRES can explain the translation inhibition activities of this compound. As previously observed with other RNA ligands, a relative weak EC50 (around 10 μM) does not imply poor inhibitor activity, as shown for TAR-Argininamide or neomycin-16S rRNA complexes.60,61 Although the inhibition induced by the IRAB benzimidazole compound is detected at a concentration of 1 to 2 μM in in vitro translation assays, the conformational changes induced on the RNA structure of the full-length IRES in the context of a functional mRNA were detectable at 10-fold higher concentration (20 μM) by SHAPE methodology. Taken together, these results indicate that subtle structural changes of the IRES conformation could lead to drastic effects over its activity. Moreover, it also underscores the difficulty in correlating biological activities with data obtained using 2 techniques with different sensitivities, SHAPE probing and fluorescence assays, that provide information on structural features of RNAs on distinct context (the entire IRES element or short oligoribonucleotides).
Taken together, these data suggest that the changes in local RNA flexibility induced by IRAB on a specific region of the IRES structure which is essential for its activity is, at least in part, responsible for the reduced translation efficiency observed in cell free lysates and, particularly, in viral RNA-transfected cells. Further studies will be focused at understanding the mechanistic insights of this small molecule as IRES inhibitor.
Materials and Methods
IRAB synthesis
IRAB (Fig. 1A) was synthesized in 8 steps from commercially available 2-chloro-6-fluorobenzaldehyde (Sigma-Aldrich) as outlined in Figure S1. Isis-11 was prepared as a mixture of enantiomers according to a previously published procedure with minor modifications.33 The identity and purity of all the compounds were confirmed by mass spectrometry as well as 1H- and 13C-NMR spectroscopy. Full experimental details can be provided upon request.
RNA synthesis
The constructs expressing the FMDV and HCV IRES RNAs were previously described.62 For RNA structural analysis RNA was synthesized in vitro using SphI linearized plasmid; to prepare RNA for translation assays, the plasmid was linearized with Not1. Transcription was performed using T7 RNA polymerase. The plasmid pMT28, encoding a cDNA copy of FMDV C-S8c1 genome, was described previously.63 Following NdeI linearization, RNA was synthesized in vitro using SP6 RNA polymerase. DNA template was removed by RQ1 DNase treatment, followed by phenol extraction and ethanol precipitation. Synthesis of full-length products was verified by gel electrophoresis.
In vitro translation assays
In vitro translation was conducted in a reaction mix containing 6.5 μl of nuclease-treated rabbit reticulocyte lysates (RRL) (Promega), 0.5 μl (0.5 mM) amino acid mix less methionine, 0.5 μl (6 μCi) 35S-methionine and RNA (250 ng) in a final volume of 10 μl, incubated at 30°C for 60 min. The inhibitory capability of IRAB was determined using a concentration range (0.1 to 20 μM) dissolved in DMSO. The reaction was then treated with RNase A and fractionated in sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels. The intensity of polypeptides synthesized was quantified by densitometry. Efficiency of protein synthesis measured in 3 independent assays was shown relative to the intensity of luciferase (LUC) and chloramphenicol acetyl transferase (CAT) polypeptides translated in the control RNA reaction, without IRAB.
RNA transfection
Viral RNA (50 pg) synthesized in vitro was transfected into BHK-21 cell monolayers (5 × 105 cells) as described.47 Immediately prior to transfect cells with RNA, a solution of IRAB in DMSO [ranging from 0.1 to 4 μM in Dulbecco´s modified Eagle´s medium (DMEM)] was added to the cell monolayer medium. A mock-transfected cell monolayer was treated in parallel with the same concentration of the compound. Triplicates of BHK-21 cells grown in 35 mm well dishes, were transfected with a mixture of RNA-lipofectine (Invitrogen). Three hours post-transfection (hpt), cell monolayers were washed with DMEM 3 times prior to addition of fresh DMEM supplemented with 5% fetal calf serum (FCS) and the same concentration of the chemical compounds. Twenty-four and 48 hpt, 200 μl of the cell culture supernatant was collected to determine virus yield. Transfection experiments were repeated at least 3 times.
Cytoplasmic cell extracts were prepared for the determination of VP1 viral protein 24 hpt, prior to cell detachment, using lysis buffer (100 μl of 50 mM Tris-HCl, pH 7.8, 120 mM NaCl, 0.5% NP40). Cell lysates were centrifuged at 14,000 RPM for 5 min to remove cellular debris.
Cell viability was monitored by measuring cell number during 72 hours of treatment with Isis-11 or IRAB relative to untreated cells. For this, cells were seeded at low density in DMEM supplemented with FCS in the absence or presence of IRAB and Isis-11 (1 or 2 μM). At the indicated times, cell monolayers were washed twice with tripsine, and resuspended in DMEM. The number of cells/ml was determined using a Neubauer chamber. Assays were done in triplicate.
PFU inhibition assay
Virus yield from 3 independent RNA infectivity assays were titrated in fresh susceptible IBRS-2 cell monolayers to determine the capacity of IRAB to inhibit plaque-forming units (PFU). Monolayers of IBRS-2 cells were infected with serial dilutions of the supernatant from transfected BHK-21 cells. One hour after adsorption, the viral inoculum was removed and the cells were washed 3 times, overlaid with DMEM medium with 0.5% agar supplemented with 2% FCS. Virus titer (PFU/ml) was scored 24 h post-infection (hpi) by fixing the cells with 2% formaldehyde solution and stained with 0.3% crystal violet in 2% formaldehyde solution. The viral titer from the 24 and 48 hpt supernatant was determined by counting the viral plaques. The virus yield was calculated as the mean of PFU/ml of 3 independent assays of FMDV RNA transfected BHK-21 cells incubated with increasing amounts of IRAB relative to the PFU/ml of FMDV RNA transfected with no drug, which was set at 100%. Cell monolayers were used to determine cytotoxicity of the chemical compounds by cell staining at the end of the treatment.
Western blot analysis
Equal amounts of total protein prepared from BHK-21 transfected cells were resolved in 10% SDS-PAGE, transferred to a PVDF membrane (Bio-Rad). VP1 protein was detected using a mouse monoclonal antibody (1:1000)64 followed by goat-anti-mouse secondary antibody coupled to horseradish peroxidase (1:2000) (Thermo Scientific), and enhanced chemiluminescence (Amersham). After stripping using restore western blot stripping buffer (Thermo Scientific) the same membrane was used to detect α-tubulin as a loading control using anti-tubulin antibody (1:5000) (Sigma).
SHAPE reactions
RNA (2 pmol) was treated with N-methylisatoic anhydride (NMIA) (Invitrogen) as described.57 Prior to NMIA treatment, in vitro synthesized RNA was renatured by heating at 95°C for 2 min, snap cooling on ice for 2 min, and subsequently incubated in a final volume of 18 μl of folding mix (100 mM HEPES pH 8.0, 0.5 mM MgCl2, 100 mM NaCl) for 20 min at 37°C. Folded RNAs were incubated with 2, 10 or 20 μM of IRAB dissolved in DMSO for 5 min at room temperature. Then, RNAs were incubated with DMSO (untreated RNA) or 6.5 mM NMIA for 45 min at 37°C, precipitated and finally resuspended in 50 μl of 0.5X TE.
For primer extension reactions, 10 μl of treated and untreated RNAs were incubated with the appropriate antisense 5’-32P-labeled primer (5´CTACGAAGCAACAGTG, 5´CCCGGGTGTGGGTACC, 5´GGAATGGGATCCTCGAGCTCAGGGTC, 5´GGCCTTTCTTTATGTTTTTGGCG). Primer extension reactions were conducted in a final volume of 16 μl containing reverse transcriptase (RT) buffer (50 mM Tris–HCl, pH 8.3, 3 mM MgCl2, 75 mM KCl, 8 mM DTT) and 1 μM each dNTP. The mix was heated at 52°C for 1 min, prior to addition of 100 U of Superscript III RT (Invitrogen) and incubated at 52°C for 30 min. cDNA products were fractionated in 6% acrylamide, 7 M urea gels, in parallel to a sequence obtained with the corresponding primer.
SHAPE analysis
For SHAPE data processing, the intensities of RT-stops were quantified by densitometry using Quantity One software (Bio-Rad). The reactivity values observed in the untreated RNA (NMIA-) were subtracted from all the NMIA-treated samples (free RNA and IRAB-treated RNAs). Quantitative SHAPE reactivity for individual data sets were normalized to a scale spanning 0 to 1.5, in which 0 indicates an unreactive nucleotide and the average intensity at highly reactive nucleotides is set to 1.0. The normalization factor for each dataset was determined by excluding the most-reactive 2% of peak intensities, followed by calculating the average for the next 8% of peak intensities. All reactivity values were normalized to this average value to generate the corresponding SHAPE reactivity profiles.39
Data from 3 independent assays were used to calculate the mean (±SD) SHAPE reactivity. Differences in SHAPE reactivity induced by incubation of the IRES with IRAB were analyzed by unpaired two-tail Student´s t-test; differences were considered significant when P < 0.05.
Absolute differences in SHAPE reactivity ≥ 0.3 arbitrary units and a P-value < 0.05 were taken to be statistically significant.41
RNA structure modeling
RNA structure models incorporating the SHAPE reactivity values were predicted using RNAstructure and SHAPEKnots software.42,65 SHAPE reactivity data was imposed as a pseudo-free energy change constraint together with nearest neighbor thermodynamic parameters.
Hydroxyl radical footprinting
RNA (2 pmol) was denatured, chilled on ice and incubated in folding buffer (40 mM MOPS pH 8.0, 80 mM KOAc, 0.5 mM MgCl2). Fe(II)–EDTA [7.5 mM Fe(SO4)2(NH4)2·6H2O and 11.25 mM EDTA, pH 8.0], 0.3% hydrogen peroxide and 150 mM sodium ascorbate solutions were freshly prepared before each experiment.39 Hydroxyl radical cleavage was initiated by adding 1 μl of Fe(II)–EDTA complex, 1 μl of sodium ascorbate and 1 μl of hydrogen peroxide solution to the reaction. A control reaction lacking Fe(II)–EDTA was performed in parallel. After incubation at 37°C for 30 sec, reactions were quenched and precipitated by addition of 75% glycerol (1/3 volume), 1 μl of 20 mg/ml glycogen, 1 μl of 3 M NaCl, 2 μl 0.5 M EDTA and 2.5 volumes ice-cold ethanol. RNAs were re-suspended and reverse transcribed. Primer extension reactions were conducted using fluorescent-labeled primer 5’-TAGCCTTATGCAGTTGCTCTCC-3’. NED fluorophore (Applied Biosystems) was used for both treated and untreated samples while VIC fluorophore (Applied Biosystems) was used for the sequencing ladder, as described.20 cDNA products were resolved by capillary electrophoresis. Hydroxyl radical cleavage intensities were normalized to a scale from 0 to 1.5, where 1.0 is defined as the average intensity of highly reactive residues. Nucleotides with cleavage intensity values lower than half the mean intensity are defined as solvent-inaccessible.
Fluorescence binding assays
Unlabeled RNA oligonucleotides were custom synthesized and HPLC purified. 2-Aminopurine (AP)-labeled and HPLC-purified RNAs were purchased from Integrated DNA Technologies (IDT). Stock solutions were prepared by dissolving lyophilized oligonucleotides in 10 mM sodium cacodylate buffer, 150 mM NaCl, pH 7.5. RNAs (SLIIa/AP54, SL3A/AP167, SL3a/AP180 or C-rich/AP239) were heated at 90°C for 5 min followed by slow cooling to room temperature. Fluorescence measurements were performed on a thermostatted Quanta-Master (PTI) spectrofluorometer at 20°C. Titrations were carried out with 0.5 μM RNA and increasing ligand concentration, from 20 nM to 130 μM depending on the target RNA. All fluorescence measurements were dilution corrected. Excitation wavelenght was fixed at 327 nm, and emission spectra were recorded over a range of 340–430 nm with a maximum fluorescence signal at 368 nm. It is worth mentioning that both IRAB and Isis-11 are fluorescent compounds, especially the latter.35 If the excitation wavelenght were fixed at 310 nm, which is the one commonly employed for 2-aminopurine binding assays and used in previous assays of Isis-11,30,45 the dose-response curves could not be properly adjusted, and the errors became too high (Fig. S7). By fixing the excitation wavelenght at 327 nm, the interference of IRAB and Isis-11 in the binding assays was substantially reduced, even though the desired complete saturation could not be fully achieved (Fig. 6). EC50 data, which can be equivalent to dissociation constants only in 1:1 ligand-target interactions, are the average of at least 2 titrations.
Titrations were also performed in the presence of a 10-fold molar excess of tRNAmix (Escherichia coli type XXI, strain W) (Sigma-Aldrich) in order to determine the selectivity ratios shown in Figure 6D. Experiments were carried out on mixtures containing tRNAmix and the corresponding RNA (SLIIa/AP54 or SL3A/AP167) in 10 mM sodium cacodylate buffer, 150 mM NaCl at pH 7.5. In these conditions, the ratios of tRNAmix base to RNA base were estimated to be 27 in the case of SL3A/AP167 and 23 with respect to SLIIa/AP54.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We thank C. Gutierrez for valuable comments on the manuscript, B. Borrego (INIA-CISA, Valdeolmos) for help with viral RNA transfection assays, and Elisabet Fernandez for kindly providing PNAs for testing.
Funding
This work was supported by grants CSD2009–00080, BFU2011–25437 and CTQ2010–21567-C02–01 from Ministerio de Economia y Competitividad (MINECO), and by an Institutional grant from Fundación Ramón Areces.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Sharp PA. The centrality of RNA. Cell 2009; 136:577-80; PMID:19239877; http://dx.doi.org/ 10.1016/j.cell.2009.02.007 [DOI] [PubMed] [Google Scholar]
- 2. Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell 2009; 136:604-9; PMID:19239882; http://dx.doi.org/ 10.1016/j.cell.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 3. Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 2013; 14:275-87; PMID:23478349; http://dx.doi.org/ 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- 4. Thompson SR. Tricks an IRES uses to enslave ribosomes. Trends Microbiol 2012; 20:558-66; PMID:22944245; http://dx.doi.org/ 10.1016/j.tim.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988; 334:320-5; PMID:2839775; http://dx.doi.org/ 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- 6. Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 1988; 62:2636-43; PMID:2839690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol 2008; 89:611-26; PMID:18272751; http://dx.doi.org/ 10.1099/vir.0.83426-0 [DOI] [PubMed] [Google Scholar]
- 8. Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell 2010; 40:228-37; PMID:20965418; http://dx.doi.org/ 10.1016/j.molcel.2010.09.028 [DOI] [PubMed] [Google Scholar]
- 9. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol 2008; 16:230-7; PMID:18420413; http://dx.doi.org/ 10.1016/j.tim.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol 1991; 65:6486-94; PMID:1658355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 2003; 9:1333-44; PMID:14561883; http://dx.doi.org/ 10.1261/rna.5950603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 2006; 12:223-34; PMID:16373480; http://dx.doi.org/ 10.1261/rna.2153206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 2000; 6:1380-92; PMID:11073214; http://dx.doi.org/ 10.1017/S1355838200000753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 1998; 273:18599-604; PMID:9660832; http://dx.doi.org/ 10.1074/jbc.273.29.18599 [DOI] [PubMed] [Google Scholar]
- 16. Clark AT, Robertson ME, Conn GL, Belsham GJ. Conserved nucleotides within the J domain of the encephalomyocarditis virus internal ribosome entry site are required for activity and for interaction with eIF4G. J Virol 2003; 77:12441-9; PMID:14610168; http://dx.doi.org/ 10.1128/JVI.77.23.12441-12449.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pineiro D, Fernandez N, Ramajo J, Martinez-Salas E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res 2013; 41:1017-28; PMID:23221641; http://dx.doi.org/ 10.1093/nar/gks1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernandez-Chamorro J, Pineiro D, Gordon JM, Ramajo J, Francisco-Velilla R, Macias MJ, Martinez-Salas E. Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res 2014; 42:5742-54; PMID:24598255; http://dx.doi.org/ 10.1093/nar/gku177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fernandez N, Garcia-Sacristan A, Ramajo J, Briones C, Martinez-Salas E. Structural analysis provides insights into the modular organization of picornavirus IRES. Virology 2011; 409:251-61; PMID:21056890; http://dx.doi.org/ 10.1016/j.virol.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 20. Lozano G, Fernandez N, Martinez-Salas E. Magnesium-dependent folding of a picornavirus IRES element modulates RNA conformation and eIF4G interaction. FEBS J 2014; 281:3685-700; PMID:24961997; http://dx.doi.org/ 10.1111/febs.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de la Torre JC, Alarcon B, Martinez-Salas E, Carrasco L, Domingo E. Ribavirin cures cells of a persistent infection with foot-and-mouth disease virus in vitro. J Virol 1987; 61:233-5; PMID:3023704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gasparian AV, Neznanov N, Jha S, Galkin O, Moran JJ, Gudkov AV, Gurova KV, Komar AA. Inhibition of encephalomyocarditis virus and poliovirus replication by quinacrine: implications for the design and discovery of novel antiviral drugs. J Virol 2010; 84:9390-7; PMID:20631142; http://dx.doi.org/ 10.1128/JVI.02569-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malina A, Khan S, Carlson CB, Svitkin Y, Harvey I, Sonenberg N, Beal PA, Pelletier J. Inhibitory properties of nucleic acid-binding ligands on protein synthesis. FEBS letters 2005; 579:79-89; PMID:15620694; http://dx.doi.org/ 10.1016/j.febslet.2004.06.103 [DOI] [PubMed] [Google Scholar]
- 24. Aboul-ela F. Strategies for the design of RNA-binding small molecules. Future Med Chem 2010; 2:93-119; PMID:21426048; http://dx.doi.org/ 10.4155/fmc.09.149 [DOI] [PubMed] [Google Scholar]
- 25. Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev 2008; 108:1171-224; PMID:18361529; http://dx.doi.org/ 10.1021/cr0681546 [DOI] [PubMed] [Google Scholar]
- 26. Bansal Y, Silakari O. The therapeutic journey of benzimidazoles: a review. Bioorg Med Chem 2012; 20:6208-36; PMID:23031649; http://dx.doi.org/ 10.1016/j.bmc.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 27. Yu L, Oost TK, Schkeryantz JM, Yang J, Janowick D, Fesik SW. Discovery of aminoglycoside mimetics by NMR-based screening of Escherichia coli A-site RNA. J Am Chem Soc 2003; 125:4444-50; PMID:12683814; http://dx.doi.org/ 10.1021/ja021354o [DOI] [PubMed] [Google Scholar]
- 28. Velagapudi SP, Pushechnikov A, Labuda LP, French JM, Disney MD. Probing a 2-aminobenzimidazole library for binding to RNA internal loops via two-dimensional combinatorial screening. ACS Chem Biol 2012; 7:1902-9; PMID:22958065; http://dx.doi.org/ 10.1021/cb300213g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc 2012; 134:4731-42; PMID:22300544; http://dx.doi.org/ 10.1021/ja210088v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Nat Acad Sci USA 2012; 109:5223-8; PMID:22431596; http://dx.doi.org/ 10.1073/pnas.1118699109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc Nat Acad Sci USA 2014; 111:15952-7; PMID:25349403; http://dx.doi.org/ 10.1073/pnas.1414678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S, Nelson CA, Xiao L, Lu L, Seth PP, Davis DR, Hagedorn CH. Measuring antiviral activity of benzimidazole molecules that alter IRES RNA structure with an infectious hepatitis C virus chimera expressing Renilla luciferase. Antiv Res 2011; 89:54-63; PMID:21075143; http://dx.doi.org/ 10.1016/j.antiviral.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ, et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem 2005; 48:7099-102; PMID:16279767; http://dx.doi.org/ 10.1021/jm050815o [DOI] [PubMed] [Google Scholar]
- 34. Plank TD, Kieft JS. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA 2012; 3:195-212; PMID:22215521; http://dx.doi.org/ 10.1002/wrna.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paulsen RB, Seth PP, Swayze EE, Griffey RH, Skalicky JJ, Cheatham TE, 3rd, Davis DR. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc Nat Acad Sci USA 2010; 107:7263-8; PMID:20360559; http://dx.doi.org/ 10.1073/pnas.0911896107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr., Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009; 460:711-6; PMID:19661910; http://dx.doi.org/ 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu B, Grigull J, Ore MO, Morin S, White KA. Global organization of a positive-strand RNA virus genome. PLoS Pathog 2013; 9:e1003363; PMID:23717202; http://dx.doi.org/ 10.1371/journal.ppat.1003363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 2008; 6:e96; PMID:18447581; http://dx.doi.org/ 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGinnis JL, Duncan CD, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Meth Enzymol 2009; 468:67-89; PMID:20946765; http://dx.doi.org/ 10.1016/S0076-6879(09)68004-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fernandez-Miragall O, Lopez de Quinto S, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res 2009; 139:172-82; PMID:18692097; http://dx.doi.org/ 10.1016/j.virusres.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 41. Steen KA, Rice GM, Weeks KM. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J Am Chem Soc 2012; 134:13160-3; PMID:22852530; http://dx.doi.org/ 10.1021/ja304027m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 2010; 11:129; PMID:20230624; http://dx.doi.org/ 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Nat Acad Sci USA 2009; 106:97-102; PMID:19109441; http://dx.doi.org/ 10.1073/pnas.0806929106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Nat Acad Sci USA 2003; 100:1574-9; PMID:12574513; http://dx.doi.org/ 10.1073/pnas.0337743100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol 2009; 5:823-5; PMID:19767736; http://dx.doi.org/ 10.1038/nchembio.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laxton C, Brady K, Moschos S, Turnpenny P, Rawal J, Pryde DC, Sidders B, Corbau R, Pickford C, Murray EJ. Selection, optimization, and pharmacokinetic properties of a novel, potent antiviral locked nucleic acid-based antisense oligomer targeting hepatitis C virus internal ribosome entry site. Antimicrob Agents Chemother 2011; 55:3105-14; PMID:21502629; http://dx.doi.org/ 10.1128/AAC.00222-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fajardo T, Jr., Rosas MF, Sobrino F, Martinez-Salas E. Exploring IRES Region Accessibility by Interference of Foot-and-Mouth Disease Virus Infectivity. PloS One 2012; 7:e41382; PMID:22815996; http://dx.doi.org/ 10.1371/journal.pone.0041382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Romero-Lopez C, Berzal-Herranz B, Gomez J, Berzal-Herranz A. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antiv Res 2012; 94:131-8; PMID:22426470; http://dx.doi.org/ 10.1016/j.antiviral.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 49. Vagnozzi A, Stein DA, Iversen PL, Rieder E. Inhibition of foot-and-mouth disease virus infections in cell cultures with antisense morpholino oligomers. J Virol 2007; 81:11669-80; PMID:17728223; http://dx.doi.org/ 10.1128/JVI.00557-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu YP, von Eije KJ, Schopman NC, Westerink JT, Brake OT, Haasnoot J, Berkhout B. Combinatorial RNAi Against HIV-1 Using Extended Short Hairpin RNAs. Mol Ther 2009; 17:1712-23; PMID:19672247; http://dx.doi.org/ 10.1038/mt.2009.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dausse E, Belair C, Staedel C, Toulme JJ. Regulating mRNA translation with a kiss. Nucleic Acids Symp Ser 2008; 52:711-2; PMID:18776578; http://dx.doi.org/ 10.1093/nass/nrn359 [DOI] [PubMed] [Google Scholar]
- 52. Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3' end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5' end region. J Gen Virol 2006; 87:3013-22; PMID:16963760; http://dx.doi.org/ 10.1099/vir.0.82059-0 [DOI] [PubMed] [Google Scholar]
- 53. Pineiro D, Ramajo J, Bradrick SS, Martinez-Salas E. Gemin5 proteolysis reveals a novel motif to identify L protease targets. Nucleic Acids Res 2012; 40:4942-53; PMID:22362733; http://dx.doi.org/ 10.1093/nar/gks172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gradi A, Foeger N, Strong R, Svitkin YV, Sonenberg N, Skern T, Belsham GJ. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: identification of the L-protease cleavage site in vitro. J Virol 2004; 78:3271-8; PMID:15016848; http://dx.doi.org/ 10.1128/JVI.78.7.3271-3278.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lawrence P, Schafer EA, Rieder E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology 2012; 425:40-52; PMID:22280896; http://dx.doi.org/ 10.1016/j.virol.2011.12.019 [DOI] [PubMed] [Google Scholar]
- 56. Rodriguez-Pulido M, Borrego B, Sobrino F, Saiz M. RNA structural domains in noncoding regions of the foot-and-mouth disease virus genome trigger innate immunity in porcine cells and mice. J Virol 2011; 85:6492-501; PMID:21525336; http://dx.doi.org/ 10.1128/JVI.00599-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fernandez N, Fernandez-Miragall O, Ramajo J, Garcia-Sacristan A, Bellora N, Eyras E, Briones C, Martinez-Salas E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res 2011; 39:8572-85; PMID:21742761; http://dx.doi.org/ 10.1093/nar/gkr560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jung S, Schlick T. Candidate RNA structures for domain 3 of the foot-and-mouth-disease virus internal ribosome entry site. Nucleic Acids Res 2013; 41:1483-95; PMID:23275533; http://dx.doi.org/ 10.1093/nar/gks1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fernandez N, Buddrus L, Pineiro D, Martinez-Salas E. Evolutionary conserved motifs constrain the RNA structure organization of picornavirus IRES. FEBS letters 2013; 587:1353-8; PMID:23507141; http://dx.doi.org/ 10.1016/j.febslet.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 60. Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 1992; 257:76-80; PMID:1621097; http://dx.doi.org/ 10.1126/science.1621097 [DOI] [PubMed] [Google Scholar]
- 61. Fourmy D, Recht MI, Puglisi JD. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA. J Mol Biol 1998; 277:347-62; PMID:9514735; http://dx.doi.org/ 10.1006/jmbi.1997.1552 [DOI] [PubMed] [Google Scholar]
- 62. Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N, Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 2009; 37:582-90; PMID:19066202; http://dx.doi.org/ 10.1093/nar/gkn979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garcia-Arriaza J, Manrubia SC, Toja M, Domingo E, Escarmis C. Evolutionary transition toward defective RNAs that are infectious by complementation. J Virol 2004; 78:11678-85; PMID:15479809; http://dx.doi.org/ 10.1128/JVI.78.21.11678-11685.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rosas MF, Vieira YA, Postigo R, Martin-Acebes MA, Armas-Portela R, Martinez-Salas E, Sobrino F. Susceptibility to viral infection is enhanced by stable expression of 3A or 3AB proteins from foot-and-mouth disease virus. Virology 2008; 380:34-45; PMID:18694581; http://dx.doi.org/ 10.1016/j.virol.2008.06.040 [DOI] [PubMed] [Google Scholar]
- 65. Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Nat Acad Sci USA 2013; 110:5498-503; PMID:23503844; http://dx.doi.org/ 10.1073/pnas.1219988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.