Abstract
Compensatory islet response is a distinct feature of the pre-diabetic insulin resistant state in humans and rodents. To identify alterations in the islet proteome that characterize the adaptive response, we analyzed islets from five-month-old male control, high-fat diet fed (HFD) or obese ob/ob mice by LC-MS(/MS) and quantified ~1,100 islet proteins (at least two peptides) with a false discovery rate <1%. Significant alterations in abundance were observed for ~350 proteins between groups. A majority of alterations were common to both models, and the changes of a subset of ~40 proteins and 12 proteins were verified by targeted quantification using selected reaction monitoring and Western blots, respectively. The insulin resistant islets in both groups exhibited reduced expression of proteins controlling energy metabolism, oxidative phosphorylation, hormone processing, and secretory pathways. Conversely, an increased expression of molecules involved in protein synthesis and folding suggested effects in endoplasmic reticulum stress response, cell survival, and proliferation in both insulin resistant models. In summary, we report a unique comparison of the islet proteome that is focused on the compensatory response in two insulin resistant rodent models that are not overtly diabetic. These data provide a valuable resource of candidate proteins to the scientific community to undertake further studies aimed at enhancing β-cell mass in patients with diabetes. The data are available via the MassIVE repository, with accession MSV000079093.
Keywords: insulin resistance, pancreatic islets, proteome, proliferation, metabolism, function
INTRODUCTION
Type 2 diabetes has reached epidemic proportions worldwide and impacts multiple organ systems. Following the development of insulin resistance the onset of the disease is triggered when the residual functional β-cells fail to compensate for the increased metabolic needs of the individual1. Despite available insulin-based and oral hypoglycemic medications, the disease continues to spread worldwide and is predicted to affect over 360 million individuals globally by 20302. Genome wide association studies revealed that type 2 diabetes-linked genes are involved in regulating β-cell mass as well as function3 suggesting the relevance of targeting β-cells as a therapeutic strategy for type 2 diabetes. Although islet transplantation has achieved success in reversing the disease and limiting its complications4, the shortage of islets from donors has prompted a reconsideration of designing alternative therapies.
While it is still debatable whether therapies should target enhancing insulin secretion from residual β-cells or increasing the number of functional insulin-producing cells5, insights to design efficient therapeutics might emerge from an understanding of the processes by which β-cells compensate to chronic increased demands for insulin. Indeed, obese non-diabetic individuals develop compensatory islet β-cell response to adjust the levels of insulin to counteract insulin resistance and therefore maintaining normoglycemia. Generally speaking, humans with insulin resistance (e.g. impaired fasting glucose or pregnancy) exhibit increased insulin secretion as compared to controls6. However, whether this compensatory response is attributed to structural or functional adaptation of islet β-cells is incompletely understood. Although increased β-cell proliferation in metabolically challenged rodents is known as a major structural adaptive response within islets7, the proportional contribution of functional changes in islet cells is unclear. Most studies which investigated islet-cell function in the context of insulin resistance were performed in vivo1a where the islet-cell mass is a considerable confounder – and fewer in vitro metabolic studies have been undertaken in rat islets8. Several proteomics studies were performed on islets derived from insulin resistant diabetic mice9. However, these studies did not address adaptive functional molecular changes in islet-cells in response to insulin resistance but rather dysfunction of islet β-cells in diabetes. In one study, the diabetic MKR (a transgenic mouse with a dominant-negative IGF-1R in skeletal muscle) mouse was used to investigate deleterious effects of insulin resistance on β-cell function9c. The same group reported a combined proteomic and microarray screen to assess defects occurring in insulin resistance-induced β-cell failure9b. Interestingly, a proteomics screen was used to address the transition from obesity to diabetes in the Zucker Fatty (ZF) and Zucker Diabetic Fatty (ZDF) rat models9a. Finally, a two-dimensional gel electrophoresis approach was applied to identify proteomic changes in the entire pancreas derived from db/db or C57BL/6J mice challenged with high fat diet (HFD); however, a major limitation in these studies was a lack of distinction between acinar and islet cells9d, 10.
Herein we used a comparative proteomics approach to characterize changes in the islet proteome in two commonly used insulin resistant pre-diabetic models, the ob/ob (small or large islets) and HFD mice. Ingenuity pathway analysis of the significantly altered proteins revealed an intriguing down-regulation of major proteins involved in pathways critical for hormone secretion including glucose and amino acid metabolism, Krebs cycle, mitochondrial oxidative phosphorylation, hormone biosynthesis and the final steps of exocytosis, suggesting functional maladaptation of islet-cells in insulin resistance states. Moreover, an increased protein synthesis and vesicular transport was observed indicating endoplasmic reticulum (ER) stress in insulin resistant islets. Interestingly, several proteins known to control cell proliferation and survival were upregulated in both HFD and ob/ob islets as compared to controls. Finally, it is notable that most proteomic changes were observed in both models of insulin resistance, and in both small and large islets. These data provide a comprehensive view of proteomic changes occurring during obesity induced islet hyperplasia and provide potential opportunities for therapeutic strategies to address β-cell decline in diabetic states.
EXPERIMENTAL PROCEDURES
Islet isolation
Islets from 5-month old C57/Bl6 male high-fat diet (HFD) fed mouse and obese ob/ob mice (n=6) manifesting insulin-resistance and age-matched control C57/Bl6 males were isolated by the intraductal enzyme injection technique using liberase11. Briefly, the pancreas was inflated with collagenase and islets were isolated as reported previously12. All islets were cultured overnight at physiological glucose levels (7 mM glucose, 10% FBS) to allow the islets to recover from the effects of liberase digestion. Islets were then transferred to nuclease- and pyrogen-free tubes and washed with phosphate buffer. Following removal of the buffer, pellets were frozen at – 80°C prior to proteomic analyses.
Protein digestion
Islet samples were homogenized and digested using a 2,2,2-trifluoroethanol (TFE)-based protocol13. Briefly, islets were dissolved in 30 ul of 50% TFE / 50% 25 mM NH4HCO3 by 3 min sonication in 5510 Branson ultrasonic water bath (Branson Ultrasonics, Danbury, CT) with ice cold water bath. Protein concentration was determined by BCA assay. About 40 μg islet proteins from each mouse were denatured in 50% TFE for 105 min at 60 °C, reduced by 2 mM DTT for 60 min at 37 °C, diluted by 5 fold with 50mM NH4HCO3, and digested by 0.8 μg trypsin (1:50 w/w trypsin-to-protein ratio) for 3 hours at 37°C. The digestion was stopped by 0.1% TFA. All peptide samples were dried down in Speed Vac remove TFE, and resuspended in 25 mM NH4HCO3 for LC-MS/MS analysis.
LC-MS/MS analysis
LC-MS/MS analyses were performed on a custom-built automated LC system coupled on-line to an LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) via a nanoelectrospray ionization interface as previously described14. Briefly, 0.75 μg of peptides were loaded onto a home-made 65-cm-long reversed-phase capillary column with 75-μm-inner diameter packed using 3 μm Jupiter C18 particles (Phenomenex, Torrance, CA). The mobile phase was held at 100% A (0.1% formic acid) for 20 min, followed by a linear gradient from 0 to 60% buffer B (0.1% formic acid in 90% acetonitrile) over 85 min. The instrument was operated in data-dependent mode with an m/z range of 400–2000, in which a full MS scan with a resolution of 100K was followed by 6 MS/MS scans. The 6 most intensive precursor ions were dynamically selected in the order of highest intensity to lowest intensity and subjected to collision-induced dissociation using a normalized collision energy setting of 35% and a dynamic exclusion duration of 1 min. The heated capillary was maintained at 200 °C, while the ESI voltage was kept at 2.2 kV.
MS/MS data analysis
LC-MS/MS raw data were converted into .dta files using Extract_MSn (version 3.0) in Bioworks Cluster 3.2 (Thermo Fisher Scientific, Cambridge, MA), and the SEQUEST algorithm (version 27, revision 12) was used to search all MS/MS spectra against a mouse protein FASTA file that contains 16, 244 entries (Uniprot, released on April 20, 2010). The search parameters used were: dynamic oxidation of methionine, 3 Da tolerances for precursor ion masses, and 1 Da for fragment ion masses. The search parameter file did not include any enzyme cleavage restraints on the termini of the identified peptides, which means that both tryptic peptides and non-tryptic peptides were identified during database searching and tryptic rules were only applied during data filtering steps. Moreover, the search parameter file allowed a maximum of three trypsin miscleavage sites for any given peptide identification. MS Generating-Function (MSGF) scores were generated for each identified spectrum as described previously by computing rigorous p-values (spectral probabilities)15. Fully tryptic peptides with MSGF score <5E-10 and mass measurement errors <3 ppm were accepted as identifications. All peptides that passed the filtering criteria were input into the ProteinProphet program16 to generate a final non-redundant list of proteins. The decoy-database searching methodology13,17 was used to control the FDR at the unique peptide level to <0.5%. The LC-MS/MS raw data along with Sequest output files have been deposited into the MassIVE repository, with accession MSV000079093. It was also shared with ProteomeXchange, and assigned dataset identifier PXD002009.
Label-free quantification
Label-free MS intensity-based quantification was performed using the accurate mass and time (AMT) tag approach as previously described18. Briefly, the islet AMT tag database were populated based on all the confident peptide identifications from the MS/MS data and the theoretical masses and observed normalized elution time (NET) values for each identified peptide were included in the database. The AMT tag database essentially serves as a “look-up” table for LC-MS feature identifications. LC-MS datasets were automatically analyzed using an in-house-developed software package that included Decon2LS and VIPER informatics software tools19. Initial analysis of the raw LC-MS data involved the use of Decon2LS to perform a de-isotoping step, which generated a text file report for the detected masses and their corresponding intensities. Each dataset was then processed by using the feature-matching tool VIPER to identify and quantify peptides. LC-MS feature identification was achieved by matching the accurately measured masses and NET values of each detected feature to the islet AMT tag database. Only when the measured mass and NET for each given feature matched the calculated mass and NET of a peptide in the AMT tag database within a 2 ppm mass error and 2% NET error, the features were considered confidently identified as peptides.
The obtained abundance data (MS intensities) for all identified peptides from different dataset were further processed by statistical data analysis software tool DAnTE20. The peptide abundance data were initially log2 transformed, normalized using the central tendency approach. Protein abundance profiles across different conditions were generated by taking a rescaling procedure for peptide profiles for each protein against a reference peptide18a. Statistical analysis using nested ANOVA was applied to identify proteins with significant abundance changes between different biological conditions by considering both biological replicates (n=5) and technical replicates (n=2). Proteins with significant abundance changes across the biological groups were identified by requiring a p-value of <0.01 and log2 ratio (over control) >0.58 (corresponding to 50% change) in at least one of the conditions.
Preparation of 18O-labeled peptide reference sample
The 18O-labeled peptide reference sample was generated by trypsin-catalyzed 18O labeling at the peptide level was performed using a recently improved protocol21. Briefly, the reference sample pooled from all biological replicates was lyophilized to dryness and reconstituted in 100 μl of 50 mM NH4HCO3 in H 182O (97%; ISOTEC, Miamisburg, OH), pH 7.8. One μl of 1 M CaCl2 and solution phase trypsin dissolved in H 182O at a 1:50 trypsin/peptide ratio (w/w) were added to the samples. The tubes were wrapped in parafilm and mixed continuously for 5 h at 37 °C. The reaction was stopped by boiling the sample in a water bath for 10 min. After snap-freezing the sample in liquid nitrogen, the samples were acidified by adding 5 μl of formic acid, and final peptide concentrations were measured using a BCA assay.
Targeted quantification using selected reaction monitoring (SRM)
SRM-based targeted quantification using 18O-based reference22 was performed for 39 selected proteins. The peptides and SRM transitions was selected and screened as previously described22, and were listed in Supplemental Table 7. At least 6 transitions of each peptide were monitored in initial screening to ensure the confident identification and detection of the targeted peptides. The best two transitions (without interference) for each peptide were selected for final quantification. The predicted collision energies from Skyline were used for all peptides. Prior to LC-SRM analyses, the 18O-labeled reference sample was spiked into each peptide sample in 1:1 mixing ratio. All peptide samples were analyzed on a Waters nanoACQUITY UPLC system (Waters Corporation, Milford MA) directly coupled to coupled on-line to a triple quadrupole mass spectrometer (TSQ Vantage; Thermo Fisher Scientific) using a 25-cm-long, 75-μm-inner diameter fused silica capillary column. 1 μl aliquots of each sample containing ~ 0.5 μg/μl peptides were injected onto the analytical column with a 40-min linear gradient of 10–50% acetonitrile and 0.1% formic acid. A fixed dwell time of 10 ms and a scan window of 0.002 m/z were employed. All datasets were analyzed by Skyline software. The peak area ratios were used for the evaluation of protein abundance changes.
Western blot and antibodies
For western blotting, more than 150 isolated islets from six-month old male C57/Bl6 and age-matched male ob/ob mice were lysed in ice-cold M-PER buffer (Thermo Fisher Scientific) with protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma). After centrifugation, the extracts were subjected to western blotting with antibodies to CDK5Rap3 (Santa Cruz, sc-134627), Sel1I (Abcam, ab78298), Nucb2 (Abcam, ab30945), PCSK1 (Thermo Fisher, PA1-057), PCSK2 (Thermo Fisher, PA1-058), SYTL4 (Santa Cruz, sc-34446), UCN3 (Bioss, bs-2786R), VAMP2 (Thermo Fisher, PA1-766), COX7A2 (Life technologies, A21367), COX4I1 (Cell Signaling, #4850), MAOB (Abcam, ab125010), SDHB (Life technologies, A21345), or β-actin (Cell Signaling, #4697). Densitometry was performed using Image J software.
RESULTS
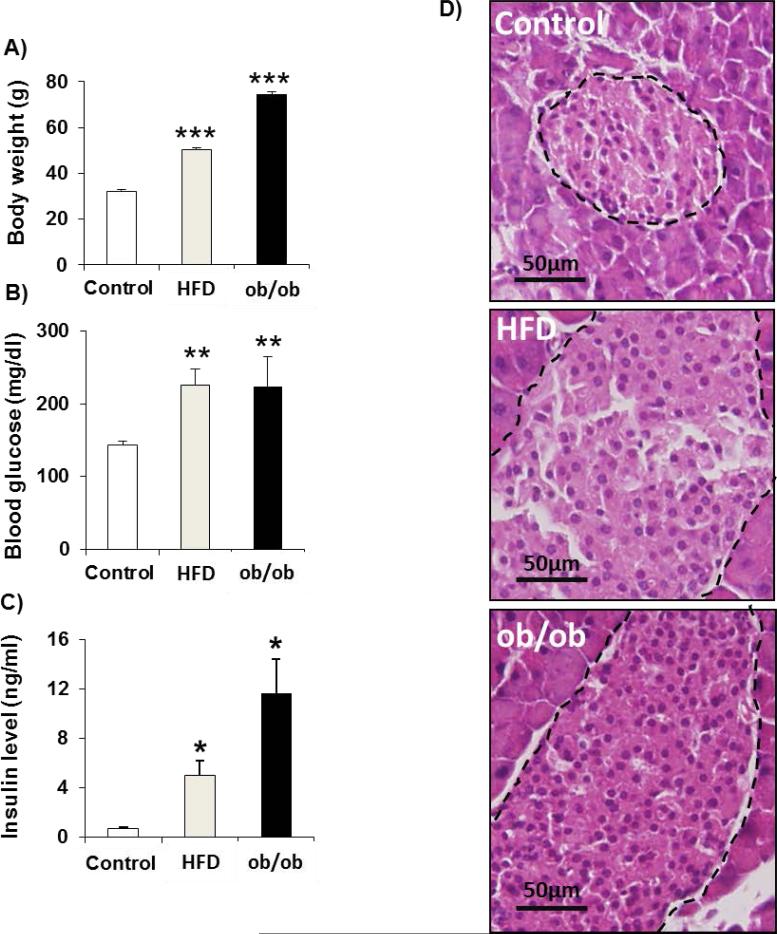
To examine islet proteome changes occurring during islet-cell adaptation in the course of insulin resistance, we performed comprehensive LC-MS-based quantitative proteomic profiling using freshly isolated islets from 5 month-old wild type, age matched leptin-deficient obese (ob/ob), or mice challenged for 12 weeks with 60% kcal High Fat Diet (HFD) beginning at age 8-weeks. Moreover, to elucidate whether insulin resistance-induced islet compensatory response is distinct in populations of islets with variable size, we also compared the proteome of small (S, ~50 microns) and large (L, 200 microns) islets from ob/ob mice where the variability in islet size was greater compared to the HFD model. Mice in both models exhibited increased body weight, and mild hyperglycemia (200 mg/dl), hyperinsulinemia in the fed state, and manifested islet hyperplasia as compared to controls (Fig. 1A-D).
Fig.1. Characteristics of HFD and ob/ob mice.
A. body weights. B. random fed blood glucose. C. random fed insulin levels. D. Hematoxylin and Eosin staining of pancreatic sections. Data represent mean ± SEM, * p < 0.05 based on Student's t-test, (n= 5-6 per group). HFD: high fat diet mice, ob/ob: leptin-deficient mice. The black dashed lines in Figure 1D indicate the islet contour, which shows the larger sizes of islets in HFD and ob/ob compared to the Control.
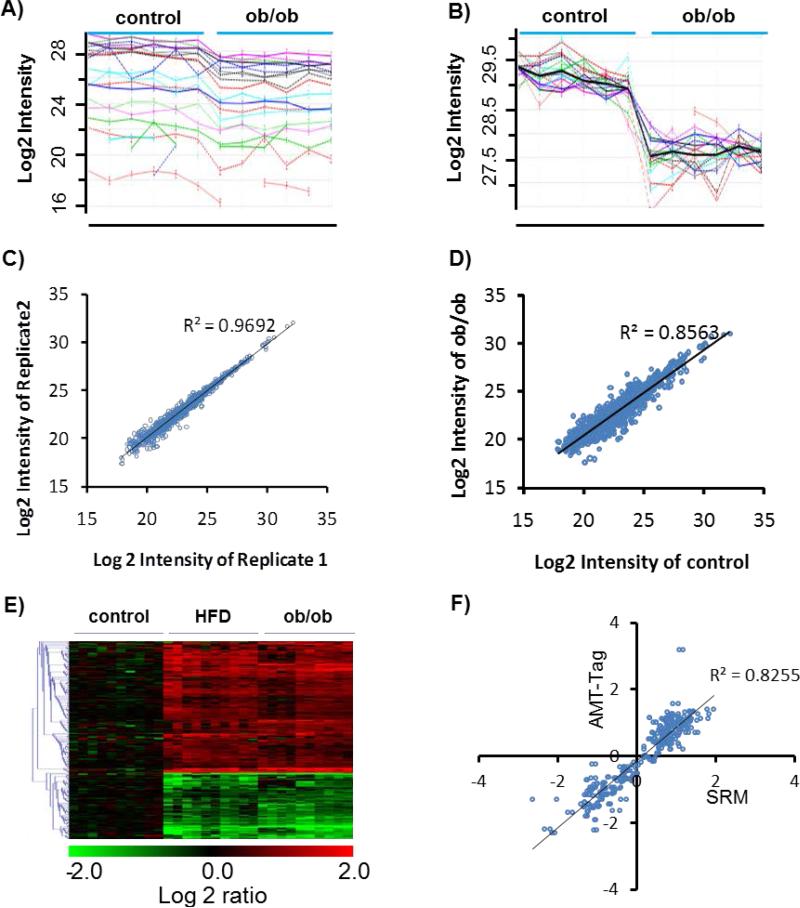
LC-MS based label-free quantification of samples isolated from control, ob/ob small, ob/ob large, or HFD islets (n=5 for each group) resulted in confident identification and quantification of ~6,900 unique peptides and ~1,100 proteins with at least two peptides per protein applying the accurate mass and time (AMT) tag approach18b (Supplementary Table 1 and Table 6). Fig. 2A and 2B illustrates the data analysis process where the raw peptide abundance profiles for a given protein obtained by the AMT tag approach was displayed in 2A using the data analysis tool DAnTE20. In Fig. 2B, a protein abundance profile was obtained for the protein (blank curve) after rescaling and rolling up to protein level. The high reproducibility of the quantitative approach was illustrated in the comparison of two technical replicates (Fig. 2C), while the comparison between control and ob/ob (small islets) conditions shows more biological variation (Fig. 2D). After subjecting the data to statistical analysis, approximately ~350 proteins were revealed to be significantly altered in either ob/ob or HFD mouse islets (Fig. 2E, and Supplemental Table 2). Among the ~350 proteins, the majority displayed a relative small changes (log2 ratio <1) for any given biological conditions (HFD, ob/ob small, or ob/ob large) and only ~100 proteins exhibited more than 2-fold changes (Supplemental Figure 1). Since we did not observe significant difference between small and large islets from ob/ob mice, only the data from the small islets from ob/ob mice are presented here.
Fig.2. Quantitative analysis strategy for the islet proteome.
A, Raw peptide intensity profile data (log2 transformed) of all peptides identified from glucagon. Each line profile represents a peptide from glucagon. 3 Control and 3 ob/ob (small islets) samples are presented with each sample analyzed in duplicated. B, Rescaled peptide intensity profile data. Dark line represents the protein abundance profile by averaging the intensity of all peptides after the rescaling process. C, Reproducibility of protein abundance quantification between technical replicates. D, Comparison of protein abundances between control and ob/ob (small islets). E, Heatmap of all proteins with significant changes. Each condition has 5 biological replicates, each replicate has duplicated runs. Values are normalized to the average of control. Only data from small islets from ob/ob mice were presented here. F, Validation of label-free quantitative data for selected proteins by selected reaction monitoring (SRM). Values are the log2 ratios to control. Each data represents one protein in one condition summarized from 5 biological replicates. Data points from both HFD and ob/ob mice were included here.
To further validate the global quantitative data, targeted quantification using selected reaction monitoring (SRM), a multiplexed quantitative technology providing similar quality as western blot or immunoassays23, was applied to validate a select list of 39 proteins from different functional categories (Supplemental Table 3). Selected examples of extracted ion chromatograms (XICs) for targeted peptides from SRM measurements are shown in Supplemental Figure S2. Fig.2F shows that SRM measured abundance ratios (HF or ob/ob divided by control) for the 39 proteins correlate well with the abundance ratio data obtained from AMT tag-based global profiling, supporting the overall high quality of the global quantitative profiling.
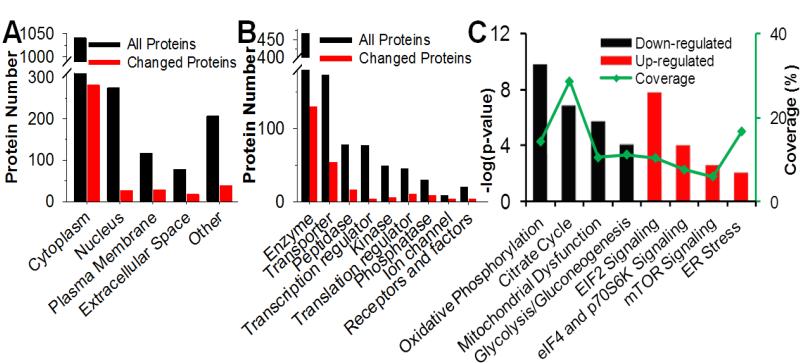
Among the altered proteins, most of the changes were common between HFD and ob/ob models (Fig. 2E). The subcellular components, molecular functions, and canonical pathways of the altered proteins were analyzed by Ingenuity Pathway Analysis (IPA) as shown in Fig.3. The distribution of altered proteins in subcellular components indicated that the majority of protein alterations occurred in the cytoplasm, which consisted of nearly a third of total quantified protein in the category (Fig. 3A). The observation that enzymes are the most altered category in molecular function (Fig. 3B) corroborates well with cytoplasm as the main component of protein alterations. The canonical pathway analysis (Fig. 3C) clearly indicated down-regulation in mitochondrial function and metabolism, and up-regulation in translational regulation and stress-related signaling. Selected regulated proteins implicated in different functional categories (Table 1) were further examined in detail.
Fig.3. Functional analysis of quantified proteins.
All quantified proteins were submitted to Ingenuity Pathway Analysis (IPA) to evaluate the biological function. Proteins were grouped based on subcellular location (A) or molecular functions (B). The major significantly down-regulated (black) and up-regulated (red) canonical pathways are presented in C. Green line in C is the proteome coverage of each canonical pathway.
Table 1.
List of selected altered proteins involved in different functional categories.
| Gene symbol | Protein name | Log2 ratio ± SEM | ||
|---|---|---|---|---|
| HFD vs. Control | ob/ob (S) vs. Control | ob/ob (L) vs. Control | ||
| Glycolysis/Gluconeogenesis | ||||
| AKR1A1 | Alcohol dehydrogenase [NADP+] | 0.17±0.08 | 0.47±0.11 | 0.64±0.04 |
| LDHA | L-lactate dehydrogenase A chain | 0.12±0.26 | 1.70±0.15 | 2.10±0.13 |
| PGM2 | Phosphoglucomutase-2 | 0.90±0.06 | 0.77±0.21 | 1.32±0.05 |
| ALDH3A2 | Fatty aldehyde dehydrogenase | −0.03±0.11 | −0.28±0.12 | −0.6±0.06 |
| ALDOA | Fructose-bisphosphate aldolase A | −0.41±0.07 | −0.65±0.08 | −0.45±0.03 |
| ENO2 | Gamma-enolase | −0.45±0.10 | −1.01±0.10 | −0.82±0.16 |
| PFKL | 6-phosphofructokinase, liver type | −0.65±0.09 | −0.72±0.06 | −0.73±0.06 |
| Citrate cycle | ||||
| DLD | Dihydrolipoyl dehydrogenase | −0.39±0.11 | −0.78±0.08 | −0.70±0.08 |
| IDH1 | Isocitrate dehydrogenase | −0.66±0.10 | −0.67±0.13 | −0.23±0.13 |
| IDH2 | Isocitrate dehydrogenase | −0.81±0.15 | −1.25±0.09 | −1.20±0.09 |
| PC | Pyruvate carboxylase, mitochondrial | −0.8±0.12 | −1.10±0.19 | −1.42±0.15 |
| PCK2 | Phosphoenolpyruvate carboxykinase | −0.65±0.11 | −0.52±0.11 | −0.87±0.10 |
| SDHB | Succinate dehydrogenase | −0.34±0.08 | −0.45±0.05 | −0.66±0.08 |
| Oxidative phosphorylation and mitochondrial dysfunction | ||||
| ATP5I | ATP synthase subunit e, mitochondrial | −0.35±0.12 | −0.61±0.13 | −0.34±0.02 |
| ATP5J2 | ATP synthase subunit f, mitochondrial | −0.02±0.07 | −0.13±0.13 | −0.60±0.09 |
| ATP6V1A | V-type proton ATPase catalytic | −0.32±0.09 | −0.54±0.05 | −0.62±0.03 |
| COX5A | Cytochrome c oxidase subunit 5A | −0.34±0.18 | −0.59±0.07 | −0.58±0.10 |
| COX7A2 | Cytochrome c oxidase subunit 7A2 | −0.45±0.16 | −0.87±0.14 | −0.56±0.07 |
| UQCRFS1 | Cytochrome b-c1 subunit Rieske | −0.26±0.23 | −0.99±0.16 | −0.60±0.08 |
| UQCRQ | Cytochrome b-c1 complex subunit 8 | −0.78±0.17 | −1.12±0.12 | −1.65±0.28 |
| GPD2 | Glycerol-3-phosphate dehydrogenase | −1.06±0.17 | −1.36±0.14 | −1.76±0.08 |
| MAOB | Amine oxidase [flavin-containing] B | −1.38±0.16 | −2.10±0.22 | −2.55±0.14 |
| NDUFB3 | NADH dehydrogenase 1 beta subunit 3 | −0.22±0.06 | −0.28±0.09 | −0.59±0.05 |
| PRDX3 | Peroxide reductase | −0.06±0.09 | −0.74±0.10 | −0.52±0.13 |
| SOD2 | Superoxide dismutase [Mn] | −0.59±0.05 | −0.80±0.06 | −0.65±0.05 |
| Protein synthesis | ||||
| EIF3C | EIF 3 subunit C | 0.63±0.04 | 0.46±0.03 | 0.44±0.05 |
| EIF3E | EIF 3 subunit E | 0.60±0.06 | 0.56±0.05 | 0.60±0.07 |
| KHSRP | Far upstream element-binding protein 2 | 0.16±0.12 | 0.53±0.05 | 0.59±0.07 |
| RPL5 | 60S ribosomal protein L5 | 0.60±0.11 | 0.29±0.06 | 0.43±0.06 |
| RRBP1 | Ribosome-binding protein 1 | 1.06±0.12 | 1.03±0.11 | 0.78±0.09 |
| RPS19 | 40S ribosomal protein S19 | 0.60±0.05 | 0.27±0.05 | 0.39±0.08 |
| DARS | Aspartyl-tRNA synthetase, cytoplasmic | 0.62±0.02 | 0.45±0.10 | 0.55±0.09 |
| Protein folding and transport | ||||
| ERP29 | ER resident protein 29 | 0.82±0.04 | 0.32±0.05 | 0.20±0.06 |
| LMAN1 | Protein ERGIC-53 | 0.75±0.07 | 0.62±0.04 | 0.65±0.03 |
| MOGS | Mannosyl-oligosaccharide glucosidase | 1.15±0.05 | 1.16±0.10 | 0.80±0.14 |
| PDIA6 | Protein disulfide-isomerase A6 | 1.04±0.09 | 0.61±0.09 | 0.48±0.11 |
| MIA3 | Melanoma inhibitory activity protein 3 | 1.13±0.08 | 0.97±0.08 | 0.90±0.12 |
| NME2 | Nucleoside diphosphate kinase B | 0.64±0.05 | 0.19±0.16 | 0.50±0.06 |
| ARCN1 | Coatomer subunit delta | 0.81±0.006 | 0.65±0.04 | 0.56±0.05 |
| COPA | Coatomer subunit alpha | 0.76±0.03 | 0.70±0.04 | 0.56±0.05 |
| SEC23A | Protein transport protein Sec23A | 0.57±0.08 | 0.66±0.05 | 0.64±0.02 |
| Processing and hormone secretion | ||||
| GCG | Glucagon | −1.14±0.29 | −1.50±0.06 | −1.23±0.07 |
| PCSK1 | Neuroendocrine convertase 1 | −1.03±0.20 | −0.74±0.24 | −0.35±0.14 |
| PCSK2 | Neuroendocrine convertase 2 | −1.11±0.15 | −0.21±0.28 | −0.16±0.17 |
| PYY | Peptide YY | −1.35±0.37 | −1.30±0.07 | −0.93±0.1 |
| RTN4 | Reticulon-4 | −1.05±0.12 | −1.27±0.13 | −1.23±0.09 |
| SST | Somatostatin | −1.58±0.30 | −2.32±0.07 | −2.42±0.09 |
| STXBP1 | Syntaxin-binding protein 1 | −0.47±0.06 | −0.83±0.12 | −0.91±0.08 |
| UCN3 | Urocortin-3 | −2.40±0.14 | −1.47±0.32 | −1.19±0.27 |
| VAMP2 | Vesicle-associated membrane protein 2 | −0.91±0.14 | −0.28±0.14 | −0.05±0.1 |
| Anti-apoptosis | ||||
| TXNDC5 | Thioredoxin domain-containing protein 5 | 1.23±0.08 | 0.95±0.10 | 0.75±0.11 |
| TPT1 | Translationally-controlled tumor protein | 0.85±0.06 | 0.82±0.07 | 0.60±0.12 |
| HSPA5 | 78 kDa glucose-regulated protein | 0.82±0.06 | 0.57±0.09 | 0.55±0.08 |
| HSP90B1 | Endoplasmin | 0.67±0.08 | 0.18±0.08 | 0.06±0.11 |
| TMX1 | Thioredoxin-related membrane protein 1 | 0.85±0.08 | 0.52±0.15 | 0.25±0.16 |
| ANXA4 | Annexin A4 | 0.15±0.03 | 0.53±0.06 | 0.70±0.05 |
| Pro-apoptosis | ||||
| HSPD1 | 60 kDa heat shock protein, mitochondrial | −0.46±0.06 | −0.56±0.07 | −0.69±0.03 |
| HSPA9 | Stress-70 protein, mitochondrial | −0.63±0.10 | −0.66±0.06 | −0.84±0.03 |
| RTN4 | Reticulon-4 | −1.05±0.12 | −1.27±0.13 | −1.23±0.09 |
| Proliferation | ||||
| CDK5rap3 | CDK5 regulatory subunit-associated protein | 0.83±0.14 | 0.65±0.09 | 0.60±0.09 |
| PRDX6 | Peroxiredoxin-6 | 0.43±0.04 | 0.65±0.05 | 0.74±0.02 |
| Sel1I | Protein sel-1 homolog 1 | 1.35±0.10 | 1.55±0.18 | 1.61±0.16 |
| Nucb2 | Nucleobindin-2 | 1.13±0.11 | 1.14±0.08 | 1.18±0.08 |
| SEPT5 | Septin-5 | 1.34±0.12 | 1.75±0.07 | 1.54±0.08 |
| SEPT7 | Septin-7 | 0.34±0.06 | 0.44±0.06 | 0.71±0.05 |
| NPM | Nucleophosmin | 0.40±0.07 | 0.68±0.10 | 0.45±0.07 |
All changes are presented as log2 ratio between HFD or ob/ob versus control. Standard errors of the mean (SEM) for log2ratio were also included. Proteins are grouped by functional analysis results using Ingenuity Pathway Analysis (IPA).
Metabolic and mitochondrial dysfunction
IPA analysis of the altered proteins revealed significant changes in glycolysis, gluconeogenesis and Krebs cycle pathways (Table 1). Although Aldo-Keto Reductase family 1, member A1 (AKR1A1), lactate dehydrogenase A (LDHA) and phosphoglucomutase 2 (PGM2) were found upregulated in insulin resistance-derived islets, proteins regulating glycolysis/gluconeogenesis were observed to be down-regulated. The down-regulated proteins including aldehyde dehydrogenase 3 family, member A2 (ALDH3A2), aldolase A (ALDOA), dihydrolipoamide S-acetyltransferase (DLAT), dihydrolipoamide dehydrogenase (DLD), enolase 2 (ENO2) and phosphofructokinase, liver (PFKL). Moreover, several enzymes regulating citrate cycle were affected. Proteins belonging to isocitrate dehydrogenase family (IDH1, IDH2 and IDH3A), pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase 2 (PCK2), and succinate dehydrogenase complex, subunit B (SDHB) were down-regulated in islets derived from both HFD and ob/ob models. Down-regulation in mitochondrial function was detected in islets derived from HFD and ob/ob animals since the expression levels of several components of ATP synthase machinery, including ATP5I, ATP6V1D, ATP5J2, ATP6V1A, ATP6V1B2, ATP6V1F, and ATP6V1H were lower compared to control animals (Table 1 and Supplemental Table 2). Furthermore, members of cyclooxygenase family were also affected. Thus, expression of COX4I1, COX5A, COX6A1 and COX7A2, cytochrome C1 (CYC1) and components of ubiquinol-cytochrome c reductase complex (UQCRC1, UQCRC2, UQCRFS1 and UQCRQ) were decreased in hyperplastic islets (Table 1 and Supplemental Table 2). Notably, we found SOD2, a major enzyme of defense against oxidative damage, to be down-regulated in all insulin resistant islets
Protein synthesis and transport and endoplasmic reticulum stress
A remarkable feature observed in this study is the up-regulation of key components of the translational machinery (Table 1 and Supplemental Table 2). Eukaryotic translation initiation factors (eIFs) such as eIF3C, eIF3E, eIF3F, eIF3G, eIF3H, eIF2S1 and subsets of ribosomal proteins including RPL5, RPL7 and RPS19 were up-regulated and several proteins implicated in biogenesis of ribosomal and/or transfer RNAs such as keratin 7 (KRT7), nucleophosmin (NMP1), ribosomal binding protein 1 (RRBP1), Aspartyl-tRNA synthetase (DARS) and phenylalanyl-tRNA synthetase beta subunit (FARSB) were found to be over-expressed in islets from HFD and ob/ob as compared to control islets. ER stress proteins, including endoplasmic proteins 29 and 44 (ERP29 and ER44), lectin mannose-binding 1 (LMAN1), mannosyl-oligosaccharide glucosidase (MOGS), protein disulfide isomerase family A, member 3 and 6 (PDIA3 and PDIA6) and selenoprotein 15 (SEP15) were up-regulated in insulin resistant islets. Significant upregulation of key proteins implicated in facilitating intracellular protein transport were also observed, including endoplasmic reticulum protein 29 (ERP29), eukaryotic initiation factor 5A (EIF5A), melanoma inhibitory activity 3 (MIA3), new molecular entity 2 (NME2), nucleophosmin (NPM1), protein disulfide isomerase 3 (PDIA3) and SEC23-interacting protein (SEC23IP). Additionally, several members of clathrin-ordered proteins family (COP) involved in protein transport and cell membrane organization manifested substantial increases in their protein levels in insulin resistant islets.
Insulin processing and exocytosis
In contrast to the marked increase in the machinery of protein biosynthesis, folding and transport, a substantial down-regulation of proteins involved in hormone processing PCSK1 and PCSK224 was observed in both models of insulin resistance models although more pronounced in HFD islets. Consistent with the latter observation, glucagon and somatostatin were down-regulated in islets derived from HFD and ob/ob mice. Finally, several proteins implicated in vesicular transport and exocytosis of hormone granules such as VAMP225, RAB5C , RAB7A, STYL4 and UCN326 were also down-regulated (Table 1 and Supplemental Table 2).
Apoptosis and proliferation
Increased β-cell mass in rodents is a major structural compensation to insulin resistance that involves both an enhancement of cell proliferation and inhibition of cell death7a, 27. However, the downstream intracellular targets mediating these effects have not been fully identified. Using IPA analysis, we focused on identifying factors that are relevant to cell survival and proliferation. We observed that several anti-apoptotic factors, including TXNDC5, TPT1, HSPA5, HSP90B1, TXM1 and ANXA4 were found to be commonly up-regulated while pro-apoptotic factors such as HSPDA1, HSPA9 and RTN4 are down-regulated in islets derived from either HFD or ob/ob models. On the other hand, and in agreement with the compensatory role of cell proliferation in insulin resistant islets, we found that several proliferation-linked proteins were up-regulated in insulin resistant islets including, isoform 1 of protein SEL-1 homolog1 (Sel1I), previously reported for its mitogenic action on β-cells28. We also found Nucleobindin-2 (Nucb2)29, CDK5 regulatory subunit associated protein 3 (CDK5rap3)30 and Peroxiredoxin 6 (PRDX6)31 up-regulated in both HFD and ob/ob suggesting their potential in promoting proliferation of insulin resistant islet cells. Moreover, a notable increase was observed in SEPT5 and SEPT7, members of septin family known to control cell division32 and Nucleophosmin-1 (NPM), a protein described to promote c-Myc-mediated proliferation33 (Table 1).
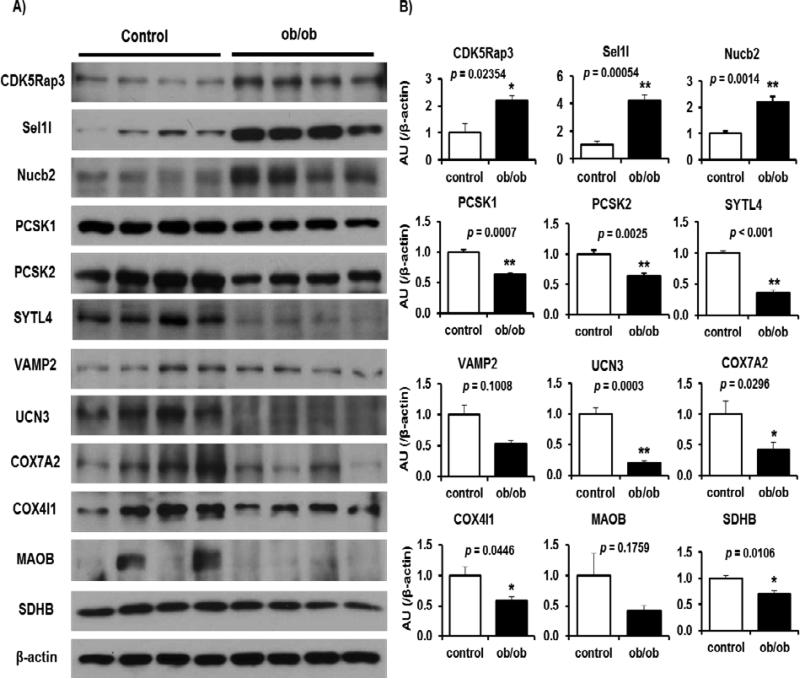
Western blot validation of selected protein targets
Considering that most islet proteome changes occur in both HFD and ob/ob models and in both small and large islets, we focused on the ob/ob mouse model to validate key regulated proteins in various biological processes in insulin resistant states. To this end, we isolated islets from five month-old wild type or age matched ob/ob mice and subjected the extracted proteins to Western blotting and quantification (Fig 4A and 4B). Consistent with proteomics data, we observed that expression of CDK5rap3, Sel1I and Nucb2, proteins reported to be linked to proliferation, are increased in ob/ob islets as compared to the control group. Moreover, expression of PCSK1 and PCSK2, involved in hormone biosynthesis were down-regulated, in insulin resistant ob/ob islets, in Western blot experiments, similar to the proteomics data. Additionally, a decrease in expression of SYTL4, UCN3 and VAMP2 exocytosis-regulating proteins was validated in ob/ob islets. A subset of proteins involved in mitochondrial function and oxidative phosphorylation, including COX4I1 and COX7A2 were also decreased consistent with the proteomics data. Finally, we confirmed by Western blot approach that two metabolic enzymes, MAOB and SDHB, are down-regulated in ob/ob islets as compared to controls (Fig. 4).
Fig.4. Validation of key regulated proteins in ob/ob mice.
A, The total cell extracts from the islets were subjected to immunoblotting as indicated. B, Intensity of the signals quantified by densitometry (image J) (n = 4-6). Data are normalized to actin. Individual p values from Student's t-test for each quantification are indicated in Figure 4B.
Discussion
This study was designed to interrogate changes occurring in the islet proteome of insulin resistant models prior to the development of overt diabetes. We used a genetic leptin deficient (ob/ob) and dietary-induced insulin resistance (HFD) mouse models to elucidate whether compensatory islet-cell response to insulin resistance is mediated by morphological or functional adaptation. Furthermore, we used small and large islets to uncover potentially distinct signatures in the adaptation of islet subpopulations to insulin resistance.
Surprisingly, most of the changes noted in our proteomic study were common between HFD and ob/ob and only a subset of proteins appeared to be differentially regulated. One possibility for the observed similarities in the proteome phenotypes is that HFD mice develop leptin resistance in insulin resistant settings34, and become blind to the leptin as naturally occurring in ob/ob mice. The alterations in proteins specific to HFD model were mainly involved in protein processing, translation, regulation of secretion and exocytosis; while those specific to the ob/ob were associated to processes such as sugar metabolism, oxidation and reduction processes, chromatin and nucleosome assembly (Supplemental Table 4 and 5). These observations are intriguing and require further investigation.
A unique feature of our approach is the comparison between small and large islets in the ob/ob model. Although several previous studies have suggested functional differences in islet subpopulations in different species35 that may occur in normal states, we observed that in the case of insulin resistance nearly all the changes observed in proteins involved in hormone processing and secretory pathways, energy metabolism, mitochondrial function, protein synthesis and ER stress were affected to a similar extent in both small and large islets in the ob/ob model. One interpretation of these data is that impairment of β-cell function likely precedes the proliferation, and that alterations in proliferation are unlikely to promote β-cell dysfunction. The cause of β-cell secretory dysfunction is likely due to a combination of the deleterious effects of hyperglycemia and hyperlipidemia and/or pro-inflammatory cytokines36. Consistently, several studies have reported alterations in function following chronic in vitro treatment of β-cells with glucose, FFA or cytokines37.
In the global context, our data suggest that insulin resistant conditions limit islet-cell energy metabolism and shut down the ability of cells to produce sufficient amounts of key metabolic intermediates. This is illustrated by a decline in the abundances of proteins controlling various metabolic pathways, including glycolysis, Krebs cycle, amino acid metabolism, and mitochondrial oxidative phosphorylation. Moreover, several mitochondrial proteins were down-regulated suggesting mitochondrial dysfunction in islet-cells derived from obese insulin resistant animals. The reduction of anti-oxidant proteins such as PRDX3 and SOD2 is suggestive of oxidative stress consistent with the protective role of these proteins is islet-cells, particularly in β-cells where the levels of these molecules are low, to mitigate oxidative stress38. However, islet cells showed increased PRDX6, another antioxidant member of the Peroxiredoxin family known (as for PRDX3) to be down-regulated by inflammation38b,39. It is possible that PRDX6 is upregulated by other stimulatory molecules to restore a mitochondrial redox state and protect the mice from developing overt diabetes40.
Several components of protein synthetic machinery, including proteins facilitating rRNA/tRNA biogenesis, initiation of translation factors, and regulators of protein transport and cell membrane organization were found to be activated and potentially participating in enhancing the biosynthetic capacity of insulin and other hormones. Consistent with the latter possibility, ER overloading by newly synthesized proteins increased protein expression of ER stress-induced chaperones in an attempt to limit ER stress.
It is of interest that several proteins involved in oxidative metabolism that were decreased in islets from HFD or ob/ob were also reported to be down-regulated in islets derived from 10-week old diabetic MKR mice9b. Commonly decreased proteins in HFD, ob/ob and MKR models include PFKL, DLD, IDH1, COX5A, GPD2 and MAOB. The alterations in expression of proteins in islets from models of “pre-diabetes” (e.g. HFD and ob/ob) that is also detectable in islets from diabetic MKR mice suggests their causal role in defective β-cell metabolism. We also observed that UCN3, a marker of β-cell maturation26, 41, was decreased in HFD, ob/ob and MKR models and is consistent with its previously reported role in impaired GSIS42.
A previous study used differential islet proteome analyses of Zucker Fatty (ZF) and Zucker Diabetic Fatty (ZDF) rats to reveal changes in the expression of proteins involved in insulin secretion, mitochondrial dysfunction, extracellular matrix proteins, or microvascular ischemia9a. Similar to the HFD or ob/ob models, islets derived from obese ZF rats, also exhibited increased protein levels of ATP5l, COPB, NME2, or PGM2, and decreased levels of GCG, GOT2, IDH2, or PCSK19a. This cross-species observation provides a valuable set of proteins associated with the transition from insulin resistance to type 2 diabetes in the context of islets. It is important, however, to note that some changes in the expression of secretory proteins such as SCG2, RAB5C and RAB7A, were commonly found in ZF or ZDF rats but not in HFD or ob/ob9a, suggesting that the regulation of expression of proteins involved in hormone secretion is not conserved in the mouse and rat.
As expected several proteins involved in cell proliferation were increased in insulin resistant islets43. We observed that the ER membrane protein Suppressor of lin-12-like protein 1 (Sel1l) is upregulated in insulin resistant islets. Sel1l is the ortholog of C. elegans gene sel-1, which is a negative regulator of LIN-12/NOTCH receptor proteins, previously implicated in β-cell growth and function. Heterozygote Sel1l (+/-) mice exhibit decreased β-cell mass due to reduced β-cell proliferation, and are predisposed to hyperglycemia upon a high-fat diet28, 44. Our observation of a substantial increase in the amounts of CDK5rap3 in the hyperplastic islets is interesting in the context of recent findings that over-expression of CDK5rap3 is positively correlated with cell proliferation of hepatocytes30 and lung cells45 both of which along with pancreatic cells share a common endodermal origin. The increased expression of Nucb2, a protein reported to be expressed in human and rodent islet β-cells and shown to be decreased in islets derived from type 2 diabetic patients29b is relevant because Nucb2 was reported to enhance cell proliferation via EGF-stimulated MAPK kinase/Erk signaling46. The decreased islet levels of Nucb2 are decreased in patients with type 2 diabetes29b warrants studies to explore a role for this protein to enhance pancreatic β-cell proliferation. The increased amounts of EIF5A in insulin resistant compared to control islets in our studies suggests that this protein which was initially described as an “initiator” of translation may be also relevant in proliferation. For example, EGF stimulates proliferation of corneal epithelial cells through PI3K-Akt-EIF5A signaling pathway47 and knockdown of EIF5A by small interfering RNAs abolishes the stimulatory action of EGF on cell proliferation47.
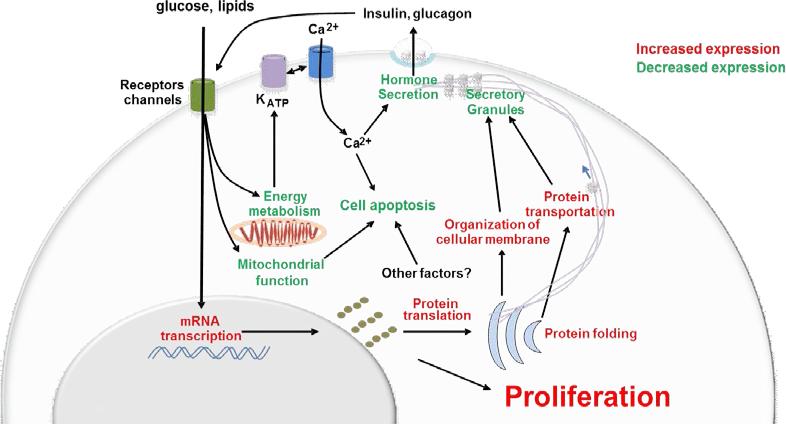
In summary, during the progression of insulin resistance, the secretory capacity of islet-cells tend to decline upon down-regulation of key proteins controlling multiple steps of insulin synthesis and release, including energy metabolism, mitochondrial function and hormone biosynthesis/exocytosis. Increased cell survival and proliferation of the endocrine pancreas appear to be central features that enable islet cells to meet chronic elevated demands of insulin and potentially other hormones (Fig. 5). The candidates identified and validated in this report could be considered for strategies aimed at developing new anti-diabetic therapeutics to enhance β-cell mass in efforts to counter diabetes.
Fig.5. Islet compensatory response in insulin resistance.
Schematic illustrating major regulated pathways in insulin resistant islets. Down- and upregulated biological processes are shown in green and red, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 DK074795, R01 DK067536, R01 DK103215, K99 DK090210, Société Francophone du Diabète (to A.E.), Association Française des Diabétiques (to A.E.), American Diabetes Association (to A.E.) and JDRF advanced postdoctoral fellowship (to A.E.). Samples were analyzed using capabilities developed under the support of the NIH Biomedical Technology Research Resource for integrative biology (P41 GM103493). Significant portions of the work were performed in the Environmental Molecular Science Laboratory, a DOE/BER national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is operated for the DOE by Battelle under contract DE-AC05-76RLO-1830.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available
Supplemental tables and figures are available free of charge via the Internet at http://pubs.acs.org.
Table S1: List of all quantified proteins. All values in each sample are log2 of peptide intensities after normalization.
Table S2: List of proteins showing significant change. All ratios are in log2 scale.
Table S3: SRM validation of candidate proteins.
Table S4: Protein candidates only showing significant change in HFD model.
Table S5: Protein candidates only showing significant change in ob/ob model.
Table S6: List of all identified peptides.
Table S7: List of all SRM peptides and associated transitions
Figure S1: Histograms of the extent of changes for altered proteins.
Figure S2: Examples of extracted ion chromatograms of SRM measurements.
REFERENCES
- 1.a Assmann A, Hinault C, Kulkarni RN. Growth factor control of pancreatic islet regeneration and function. Pediatr Diabetes. 2009;10(1):14–32. doi: 10.1111/j.1399-5448.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Weir GC, Bonner-Weir S. Five stages of evolving beta-cell dysfunction during progression to diabetes. Diabetes. 2004;53(Suppl 3):S16–21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 2.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 3.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8(3):186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Shapiro AM, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, Robertson RP, Secchi A, Brendel MD, Berney T, Brennan DC, Cagliero E, Alejandro R, Ryan EA, DiMercurio B, Morel P, Polonsky KS, Reems JA, Bretzel RG, Bertuzzi F, Froud T, Kandaswamy R, Sutherland DE, Eisenbarth G, Segal M, Preiksaitis J, Korbutt GS, Barton FB, Viviano L, Seyfert-Margolis V, Bluestone J, Lakey JR. International trial of the Edmonton protocol for islet transplantation. The New England journal of medicine. 2006;355(13):1318–30. doi: 10.1056/NEJMoa061267. [DOI] [PubMed] [Google Scholar]; b Thompson DM, Meloche M, Ao Z, Paty B, Keown P, Shapiro RJ, Ho S, Worsley D, Fung M, Meneilly G, Begg I, Al Mehthel M, Kondi J, Harris C, Fensom B, Kozak SE, Tong SO, Trinh M, Warnock GL. Reduced progression of diabetic microvascular complications with islet cell transplantation compared with intensive medical therapy. Transplantation. 2011;91(3):373–8. doi: 10.1097/TP.0b013e31820437f3. [DOI] [PubMed] [Google Scholar]
- 5.a Buteau J, Shlien A, Foisy S, Accili D. Metabolic diapause in pancreatic beta-cells expressing a gain-of-function mutant of the forkhead protein Foxo1. J Biol Chem. 2007;282(1):287–93. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]; b de Koning EJ, Bonner-Weir S, Rabelink TJ. Preservation of beta-cell function by targeting beta-cell mass. Trends Pharmacol Sci. 2008;29(4):218–27. doi: 10.1016/j.tips.2008.02.001. [DOI] [PubMed] [Google Scholar]; c Hansen JB, Arkhammar PO, Bodvarsdottir TB, Wahl P. Inhibition of insulin secretion as a new drug target in the treatment of metabolic disorders. Curr Med Chem. 2004;11(12):1595–615. doi: 10.2174/0929867043365026. [DOI] [PubMed] [Google Scholar]; d Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150(6):1223–34. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Jones CN, Pei D, Staris P, Polonsky KS, Chen YD, Reaven GM. Alterations in the glucose-stimulated insulin secretory dose-response curve and in insulin clearance in nondiabetic insulin-resistant individuals. J Clin Endocrinol Metab. 1997;82(6):1834–8. doi: 10.1210/jcem.82.6.3979. [DOI] [PubMed] [Google Scholar]; b Parsons JA, Brelje TC, Sorenson RL. Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology. 1992;130(3):1459–66. doi: 10.1210/endo.130.3.1537300. [DOI] [PubMed] [Google Scholar]; c Polonsky KS, Given BD, Hirsch L, Shapiro ET, Tillil H, Beebe C, Galloway JA, Frank BH, Karrison T, Van Cauter E. Quantitative study of insulin secretion and clearance in normal and obese subjects. J Clin Invest. 1988;81(2):435–41. doi: 10.1172/JCI113338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88(4):561–72. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]; b Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6(1):87–97. [PubMed] [Google Scholar]
- 8.a Chen C, Hosokawa H, Bumbalo LM, Leahy JL. Mechanism of compensatory hyperinsulinemia in normoglycemic insulin-resistant spontaneously hypertensive rats. Augmented enzymatic activity of glucokinase in beta-cells. J Clin Invest. 1994;94(1):399–404. doi: 10.1172/JCI117335. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Liu YQ, Jetton TL, Leahy JL. beta-Cell adaptation to insulin resistance. Increased pyruvate carboxylase and malate-pyruvate shuttle activity in islets of nondiabetic Zucker fatty rats. J Biol Chem. 2002;277(42):39163–8. doi: 10.1074/jbc.M207157200. [DOI] [PubMed] [Google Scholar]
- 9.a Han D, Moon S, Kim H, Choi SE, Lee SJ, Park KS, Jun H, Kang Y, Kim Y. Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. J Proteome Res. 2011;10(2):564–77. doi: 10.1021/pr100759a. [DOI] [PubMed] [Google Scholar]; b Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB. Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes. Diabetes. 2010;59(2):448–59. doi: 10.2337/db09-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Mol Cell Proteomics. 2008;7(8):1434–51. doi: 10.1074/mcp.M700478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Qiu L, List EO, Kopchick JJ. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol Cell Proteomics. 2005;4(9):1311–8. doi: 10.1074/mcp.M500016-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Vazquez V, Guzman-Flores JM, Mares-Alvarez D, Hernandez-Ortiz M, Macias-Cervantes MH, Ramirez-Emiliano J, Encarnacion-Guevara S. Differential proteomic analysis of the pancreas of diabetic db/db mice reveals the proteins involved in the development of complications of diabetes mellitus. International journal of molecular sciences. 2014;15(6):9579–93. doi: 10.3390/ijms15069579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni RN, Bruning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96(3):329–39. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni RN, Winnay JN, Daniels M, Bruning JC, Flier SN, Hanahan D, Kahn CR. Altered function of insulin receptor substrate-1-deficient mouse islets and cultured beta-cell lines. J Clin Invest. 1999;104(12):R69–75. doi: 10.1172/JCI8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG, 2nd, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4(6):2397–403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou JY, Schepmoes AA, Zhang X, Moore RJ, Monroe ME, Lee JH, Camp DG, Smith RD, Qian WJ. Improved LC-MS/MS spectral counting statistics by recovering low-scoring spectra matched to confidently identified peptide sequences. J Proteome Res. 2010;9(11):5698–704. doi: 10.1021/pr100508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7(8):3354–63. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 17.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 18.a Qian WJ, Liu T, Petyuk VA, Gritsenko MA, Petritis BO, Polpitiya AD, Kaushal A, Xiao W, Finnerty CC, Jeschke MG, Jaitly N, Monroe ME, Moore RJ, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, Smith RD. Large-scale multiplexed quantitative discovery proteomics enabled by the use of an (18)O-labeled “universal” reference sample. J Proteome Res. 2009;8(1):290–9. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25(3):450–82. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics. 2007;23(15):2021–3. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 20.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24(13):1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petritis BO, Qian WJ, Camp DG, 2nd, Smith RD. A simple procedure for effective quenching of trypsin activity and prevention of 18O-labeling back-exchange. J Proteome Res. 2009;8(5):2157–63. doi: 10.1021/pr800971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JS, Fillmore TL, Liu T, Robinson E, Hossain M, Champion BL, Moore RJ, Camp DG, 2nd, Smith RD, Qian WJ. 18O-labeled proteome reference as global internal standards for targeted quantification by selected reaction monitoring-mass spectrometry. Mol Cell Proteomics. 2011;10(12):M110 007302. doi: 10.1074/mcp.M110.007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim JS, Jones N, Moore RJ, Pasa-Tolic L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG, 2nd, Smith RD, Qian WJ. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A. 2012;109(38):15395–400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shi T, Qian WJ. Antibody-free PRISM-SRM for multiplexed protein quantification: is this the new competition for immunoassays in bioanalysis? Bioanalysis. 2013;5(3):267–9. doi: 10.4155/bio.12.336. [DOI] [PubMed] [Google Scholar]
- 24.Neerman-Arbez M, Cirulli V, Halban PA. Levels of the conversion endoproteases PC1 (PC3) and PC2 distinguish between insulin-producing pancreatic islet beta cells and non-beta cells. The Biochemical journal. 1994;300(Pt 1):57–61. doi: 10.1042/bj3000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regazzi R, Sadoul K, Meda P, Kelly RB, Halban PA, Wollheim CB. Mutational analysis of VAMP domains implicated in Ca2+-induced insulin exocytosis. The EMBO journal. 1996;15(24):6951–9. [PMC free article] [PubMed] [Google Scholar]
- 26.Blum B, Hrvatin SS, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature biotechnology. 2012;30(3):261–4. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.a Chen Z, Morris DL, Jiang L, Liu Y, Rui L. SH2B1 in beta cells regulates glucose metabolism by promoting beta cell survival and islet expansion. Diabetes. 2013 doi: 10.2337/db13-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jetton TL, Lausier J, LaRock K, Trotman WE, Larmie B, Habibovic A, Peshavaria M, Leahy JL. Mechanisms of compensatory beta-cell growth in insulin-resistant rats: roles of Akt kinase. Diabetes. 2005;54(8):2294–304. doi: 10.2337/diabetes.54.8.2294. [DOI] [PubMed] [Google Scholar]; c Okada T, Liew CW, Hu J, Hinault C, Michael MD, Krtzfeldt J, Yin C, Holzenberger M, Stoffel M, Kulkarni RN. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proc Natl Acad Sci U S A. 2007;104(21):8977–82. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francisco AB, Singh R, Sha H, Yan X, Qi L, Lei X, Long Q. Haploid insufficiency of suppressor enhancer Lin12 1-like (SEL1L) protein predisposes mice to high fat diet-induced hyperglycemia. The Journal of biological chemistry. 2011;286(25):22275–82. doi: 10.1074/jbc.M111.239418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.a Oh IS, Shimizu H, Satoh T, Okada S, Adachi S, Inoue K, Eguchi H, Yamamoto M, Imaki T, Hashimoto K, Tsuchiya T, Monden T, Horiguchi K, Yamada M, Mori M. Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature. 2006;443(7112):709–12. doi: 10.1038/nature05162. [DOI] [PubMed] [Google Scholar]; b Riva M, Nitert MD, Voss U, Sathanoori R, Lindqvist A, Ling C, Wierup N. Nesfatin-1 stimulates glucagon and insulin secretion and beta cell NUCB2 is reduced in human type 2 diabetic subjects. Cell and tissue research. 2011;346(3):393–405. doi: 10.1007/s00441-011-1268-5. [DOI] [PubMed] [Google Scholar]
- 30.Mak GW, Chan MM, Leong VY, Lee JM, Yau TO, Ng IO, Ching YP. Overexpression of a novel activator of PAK4, the CDK5 kinase-associated protein CDK5RAP3, promotes hepatocellular carcinoma metastasis. Cancer research. 2011;71(8):2949–58. doi: 10.1158/0008-5472.CAN-10-4046. [DOI] [PubMed] [Google Scholar]
- 31.Chang XZ, Li DQ, Hou YF, Wu J, Lu JS, Di GH, Jin W, Ou ZL, Shen ZZ, Shao ZM. Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast cancer research : BCR. 2007;9(6):R76. doi: 10.1186/bcr1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nature reviews. Molecular cell biology. 2012;13(3):183–94. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene. 2012 doi: 10.1038/onc.2012.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enriori PJ, Evans AE, Sinnayah P, Jobst EE, Tonelli-Lemos L, Billes SK, Glavas MM, Grayson BE, Perello M, Nillni EA, Grove KL, Cowley MA. Diet-induced obesity causes severe but reversible leptin resistance in arcuate melanocortin neurons. Cell Metab. 2007;5(3):181–94. doi: 10.1016/j.cmet.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 35.a Bonner-Weir S, Like AA. A dual population of islets of Langerhans in bovine pancreas. Cell and tissue research. 1980;206(1):157–70. doi: 10.1007/BF00233616. [DOI] [PubMed] [Google Scholar]; b Farhat B, Almelkar A, Ramachandran K, Williams SJ, Huang HH, Zamierowksi D, Novikova L, Stehno-Bittel L. Small human islets comprised of more beta-cells with higher insulin content than large islets. Islets. 2013;5(2):87–94. doi: 10.4161/isl.24780. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Huang HH, Novikova L, Williams SJ, Smirnova IV, Stehno-Bittel L. Low insulin content of large islet population is present in situ and in isolated islets. Islets. 2011;3(1):6–13. doi: 10.4161/isl.3.1.14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson RP. Beta-cell deterioration during diabetes: what's in the gun? Trends in endocrinology and metabolism: TEM. 2009;20(8):388–93. doi: 10.1016/j.tem.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.a Chen X, Cui Z, Wei S, Hou J, Xie Z, Peng X, Li J, Cai T, Hang H, Yang F. Chronic high glucose induced INS-1beta cell mitochondrial dysfunction: a comparative mitochondrial proteome with SILAC. Proteomics. 2013;13(20):3030–9. doi: 10.1002/pmic.201200448. [DOI] [PubMed] [Google Scholar]; b D'Hertog W, Overbergh L, Lage K, Ferreira GB, Maris M, Gysemans C, Flamez D, Cardozo AK, Van den Bergh G, Schoofs L, Arckens L, Moreau Y, Hansen DA, Eizirik DL, Waelkens E, Mathieu C. Proteomics analysis of cytokine-induced dysfunction and death in insulin-producing INS-1E cells: new insights into the pathways involved. Mol Cell Proteomics. 2007;6(12):2180–99. doi: 10.1074/mcp.M700085-MCP200. [DOI] [PubMed] [Google Scholar]; c Maris M, Ferreira GB, D'Hertog W, Cnop M, Waelkens E, Overbergh L, Mathieu C. High glucose induces dysfunction in insulin secretory cells by different pathways: a proteomic approach. J Proteome Res. 2010;9(12):6274–87. doi: 10.1021/pr100557w. [DOI] [PubMed] [Google Scholar]; d Maris M, Robert S, Waelkens E, Derua R, Hernangomez MH, D'Hertog W, Cnop M, Mathieu C, Overbergh L. Role of the saturated nonesterified fatty acid palmitate in beta cell dysfunction. J Proteome Res. 2013;12(1):347–62. doi: 10.1021/pr300596g. [DOI] [PubMed] [Google Scholar]
- 38.a Kang L, Dai C, Lustig ME, Bonner JS, Mayes WH, Mokshagundam S, James FD, Thompson CS, Lin CT, Perry CG, Anderson EJ, Neufer PD, Wasserman DH, Powers AC. Heterozygous SOD2 deletion impairs glucose-stimulated insulin secretion, but not insulin action, in high-fat-fed mice. Diabetes. 2014;63(11):3699–710. doi: 10.2337/db13-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wolf G, Aumann N, Michalska M, Bast A, Sonnemann J, Beck JF, Lendeckel U, Newsholme P, Walther R. Peroxiredoxin III protects pancreatic ss cells from apoptosis. The Journal of endocrinology. 2010;207(2):163–75. doi: 10.1677/JOE-09-0455. [DOI] [PubMed] [Google Scholar]
- 39.Paula FM, Ferreira SM, Boschero AC, Souza KL. Modulation of the peroxiredoxin system by cytokines in insulin-producing RINm5F cells: down-regulation of PRDX6 increases susceptibility of beta cells to oxidative stress. Molecular and cellular endocrinology. 2013;374(1-2):56–64. doi: 10.1016/j.mce.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Pacifici F, Arriga R, Sorice GP, Capuani B, Scioli MG, Pastore D, Donadel G, Bellia A, Caratelli S, Coppola A, Ferrelli F, Federici M, Sconocchia G, Tesauro M, Sbraccia P, Della-Morte D, Giaccari A, Orlandi A, Lauro D. Peroxiredoxin 6, a novel player in the pathogenesis of diabetes. Diabetes. 2014;63(10):3210–20. doi: 10.2337/db14-0144. [DOI] [PubMed] [Google Scholar]
- 41.van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, Huising MO. Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PloS one. 2012;7(12):e52181. doi: 10.1371/journal.pone.0052181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Chen P, Vaughan J, Lee KF, Vale W. Urocortin 3 regulates glucose-stimulated insulin secretion and energy homeostasis. Proc Natl Acad Sci U S A. 2007;104(10):4206–11. doi: 10.1073/pnas.0611641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.a Bock T, Pakkenberg B, Buschard K. Increased islet volume but unchanged islet number in ob/ob mice. Diabetes. 2003;52(7):1716–22. doi: 10.2337/diabetes.52.7.1716. [DOI] [PubMed] [Google Scholar]; b Hull RL, Kodama K, Utzschneider KM, Carr DB, Prigeon RL, Kahn SE. Dietary-fat-induced obesity in mice results in beta cell hyperplasia but not increased insulin release: evidence for specificity of impaired beta cell adaptation. Diabetologia. 2005;48(7):1350–8. doi: 10.1007/s00125-005-1772-9. [DOI] [PubMed] [Google Scholar]; c Stamateris RE, Sharma RB, Hollern DA, Alonso LC. Adaptive beta-cell proliferation increases early in high-fat feeding in mice, concurrent with metabolic changes, with induction of islet cyclin D2 expression. Am J Physiol Endocrinol Metab. 2013;305(1):E149–59. doi: 10.1152/ajpendo.00040.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonca N, Liu X, Locasale JW, Simpson KW, Duhamel GE, Kersten S, Yates JR, 3rd, Long Q, Qi L. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc Natl Acad Sci U S A. 2014;111(5):E582–91. doi: 10.1073/pnas.1318114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. The International journal of biological markers. 2007;22(2):108–13. doi: 10.1177/172460080702200204. [DOI] [PubMed] [Google Scholar]
- 46.Tagaya Y, Miura A, Okada S, Ohshima K, Mori M. Nucleobindin-2 is a positive modulator of EGF-dependent signals leading to enhancement of cell growth and suppression of adipocyte differentiation. Endocrinology. 2012;153(7):3308–19. doi: 10.1210/en.2011-2154. [DOI] [PubMed] [Google Scholar]
- 47.Ding L, Gao LJ, Gu PQ, Guo SY, Cai YQ, Zhou XT. The role of eIF5A in epidermal growth factor-induced proliferation of corneal epithelial cell association with PI3-k/Akt activation. Molecular vision. 2011;17:16–22. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.