Abstract
Bile acids are synthesized from cholesterol in the liver. Some cytochrome P450 (CYP) enzymes play key roles in bile acid synthesis. Bile acids are physiological detergent molecules, so are highly cytotoxic. They undergo enterohepatic circulation and play important roles in generating bile flow and facilitating biliary secretion of endogenous metabolites and xenobiotics and intestinal absorption of dietary fats and lipid soluble vitamins. Bile acid synthesis, transport and pool size are therefore tightly regulated under physiological conditions. In cholestasis, impaired bile flow leads to accumulation of bile acids in the liver, causing hepatocyte and biliary injury and inflammation. Chronic cholestasis is associated with fibrosis, cirrhosis and eventually liver failure. Chronic cholestasis also increases the risk of developing hepatocellular or cholangiocellular carcinomas. Extensive research in the last two decades has shown that bile acids act as signaling molecules that regulate various cellular processes. The bile acid-activated nuclear receptors are ligand-activated transcriptional factors that play critical roles in the regulation of bile acid, drug and xenobiotic metabolism. In cholestasis, these bile acid-activated receptors regulate a network of genes involved in bile acid synthesis, conjugation, transport and metabolism to alleviate bile acid-induced inflammation and injury. Additionally, bile acids are known to regulate cell growth and proliferation, and altered bile acid levels in diseased conditions have been implicated in liver injury/regeneration and tumorigenesis. We will cover the mechanisms that regulate bile acid homeostasis and detoxification during cholestasis, and the roles of bile acids in the initiation and regulation of hepatic inflammation, regeneration and carcinogenesis.
Keywords: cytochrome P450, nuclear receptor, cholestasis, liver cancer, liver regeneration
Introduction
Bile acids are physiological detergent molecules synthesized from cholesterol exclusively in the liver (Li and Chiang, 2014; Russell and Setchell, 1992). Under physiological conditions, most bile acids exist as glycine or taurine conjugates and are therefore referred to as “bile salts”. Bile acid synthesis has several important functions in the liver. Conversion of cholesterol into bile acids in the liver accounts for a major fraction of daily cholesterol turnover in humans (Chiang, 2009). Biliary bile acid secretion generates bile flow, and facilitates hepatobiliary secretion of various endogenous metabolites and xenobiotics (Trauner and Boyer, 2003). In the gallbladder, bile acids form mixed micelles with phospholipids and cholesterol to increase cholesterol solubility and decrease bile acid toxicity. Once released into the small intestine, bile acids facilitate the intestinal digestion and absorption of dietary cholesterol, fat and other lipophilic nutrients. In addition, bile acids are signaling molecules that activate intracellular ligand-activated nuclear receptors and cell surface G protein coupled receptors to regulate various cellular processes from lipid and glucose metabolism, drug metabolism to immunity (Li and Chiang, 2014). Impaired bile flow, which can be caused by both genetic and environmental factors, can lead to cholestasis, in which accumulation of bile acids in the liver causes hepatic inflammation and injury (Zollner and Trauner, 2008). Patients with cholestasis over time develop fibrosis, cirrhosis and eventually liver failure and increased risk of hepatocellular or cholangiocellular carcinomas. In cholestasis, a number of detoxification mechanisms, mainly mediated by bile acid-activated nuclear receptors, are activated to alleviate bile acid-induced inflammation and injury. In addition, bile acids are known to regulate cell growth and proliferation, and dysregulation of bile acid homeostasis and signaling can have an impact on liver regeneration and tumorigenesis (Fan et al., 2015; Vacca et al., 2013). In this chapter, we will cover the roles of major CYP enzymes involved in hepatic bile acid synthesis, the mechanisms that regulate bile acid homeostasis, and the roles of bile acid metabolism and signaling in the regulation of hepatic inflammation, regeneration and carcinogenesis.
10.1. Bile acid synthesis and transport
10.1.1. Primary bile acid synthesis
Primary bile acids are synthesized from cholesterol in the liver (Li and Chiang, 2014). Some primary bile acids are converted to secondary bile acids by bacterial enzymes in small and large intestine. In humans, the bile acid pool consists of the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) and the secondary bile acid deoxycholic acid (DCA). In the liver, two pathways, namely the classic pathway and the alternative pathway, are responsible for the synthesis of primary bile acids (Figure 1). The conversion of cholesterol to bile acids involves hydroxylation, saturation of the double bond at C5-C6, epimerization of the 3-hydroxyl group, and oxidative cleavage of a 3-carbon unit, and these reactions are catalyzed by a number of CYP enzymes localized in the endoplasmic reticulum, mitochondria, cytosol and peroxisomes (Russell, 2003). All bile acids possess a 5β-hydrogen group and a cis-configuration along the plane of the fused A and B ring. Majority of the bile acids exist as glycine or taurine conjugates, and are thus referred to as “sodium salts”. Conjugated bile acids are negatively charged molecules with increased solubility under most physiological pH ranges. Bile acids usually have a few hydroxyl groups and the carboxyl group on one side of the carbon skeleton, which is the structural basis for bile acids to act as amphipathic detergent molecules in facilitating lipid absorption and cholesterol dissolution.
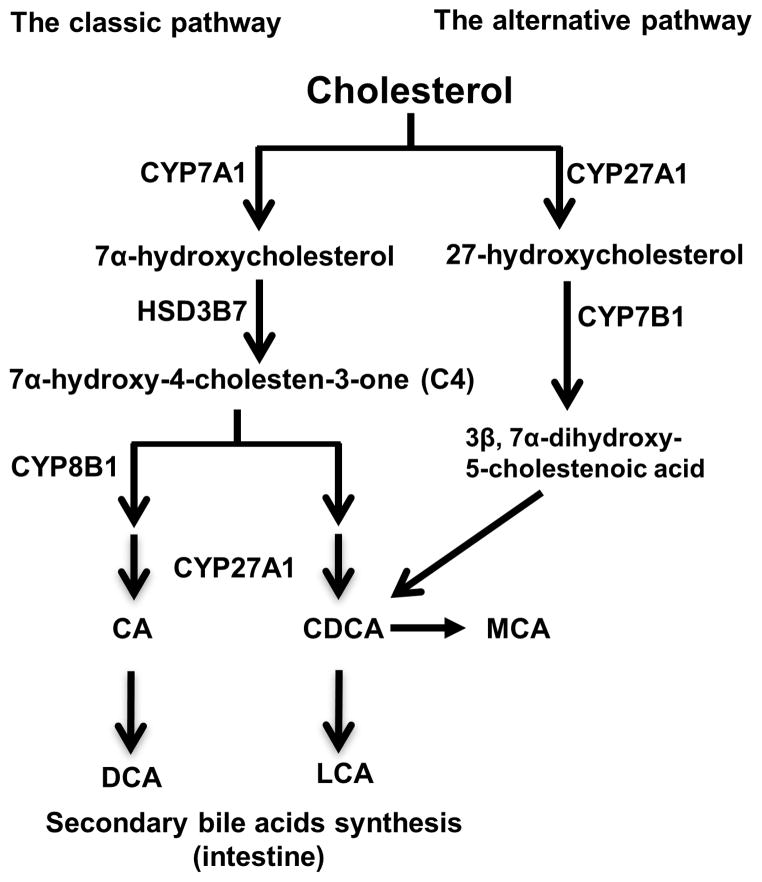
Figure 1. Bile acid synthetic pathways and bile acid structure.
Cholesterol is the common precursor for bile acid synthesis via two major bile acid biosynthetic pathways. In the classic pathway, the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1) in the endoplasmic reticulum converts cholesterol into 7α-hydroxycholesterol. The 3β-hydroxysteroid dehydrogenase (3βHSD, HSD3B7) converts 7α-hydroxycholesterol to 7α-hydroxy-4 cholesten-3-one (C4). C4 can be converted to cholic acid (CA) which requires the sterol 12α-hydroxylase (CYP8B1). Without 12α-hydroxylation by CYP8B1, C4 is eventually converted to chenodeoxycholic acid (CDCA). In the classic pathway, the mitochondrial sterol 27-hydroxylase (CYP27A1) catalyzes the steroid side-chain oxidation in both CA and CDCA synthesis. In the alternative pathway, CYP27A1 catalyzes the first step to convert cholesterol to 27-hydroxycholesterol. Oxysterol 7α-hydroxylase (CYP7B1) catalyzes hydroxylation of 27-hydroxycholesterol to 3β, 7α-dihydroxy-5-cholestenoic acid, which eventually is converted to CDCA. In the large intestine, bacterial 7α-dehydroxylase removes a hydroxyl group from C-7 and converts CA to deoxycholic acid (DCA) and CDCA to lithocholic acid (LCA). In mouse liver, most of CDCA is converted to α- and β-muricholic acid (MCA).
The classic pathway
The classic bile acid synthetic pathway is the major bile acid biosynthetic pathway in humans (Figure 1). It accounts for about 90% of total bile acid production in the liver. This pathway produces CA and CDCA in approximately equal amounts. The cholesterol 7α-hydroxylase (CYP7A1), a microsomal CYP enzyme, catalyzes the first and rate-limiting step to convert cholesterol to 7α-hydroxycholesterol (Chiang, 2009; Myant and Mitropoulos, 1977). The intermediate product 7α-hydroxy-4-cholestene-3-one (C4) is the common precursor for CA and CDCA. Another microsomal CYP enzyme sterol 12α-hydroxylase (CYP8B1) catalyzes the addition of a hydroxyl group at the C-12 position on C4, and the resulting intermediate is eventually converted to CA. The C4 that escapes from this 12-hydroxylation reaction catalyzed by CYP8B1 is converted to CDCA. The synthesis of all bile acids involves the steroid side-chain cleavage, which is catalyzed by the mitochondrial sterol 27-hydroxylase (CYP27A1). The hepatic bile acid synthesis rate is mainly controlled via the transcriptional regulation of the CYP7A1 gene (Chiang, 2009). Hepatic CYP8B1 activity may regulate the CA: CDCA ratio in the bile acid pool. However, other mechanisms such as intestine bile acid transformation and transport may also have a significant impact on the bile acid pool composition. Studies have shown that the plasma level of C4, the common precursor for CA and CDCA, correlates well with hepatic bile acid synthesis and thus has been used as a surrogate serum marker for the rate of hepatic bile acid synthesis in humans (Axelson et al., 1988).
The alternative pathway
This pathway is also called the acidic pathway due to the production of acidic intermediates (Figure 1). In this pathway, the mitochondrial CYP27A1 catalyzes the first hydroxylation reaction to convert cholesterol to 27-hydroxycholesterol. The hydroxylation at the C-7 position is catalyzed by oxysterol 7α-hydroxylase (CYP7B1). In contrast to the classic pathway, the alternative pathway only produces CDCA, but not CA. In addition to cholesterol, oxysterol intermediates formed in the peripheral tissues can enter the liver and feed into this pathway for bile acid production. In humans, the alternative pathway is considered a minor bile acid synthetic pathway because it produces less than 10% of the total bile acids under normal physiological conditions. However, the alternative pathway seemed to be more active in rodents and can be responsible for the synthesis of about 50% of bile acids in rodents (Li and Chiang, 2014; Russell and Setchell, 1992).
10.1.2. Bile acid biotransformation in the intestine
In the intestine, some bile acids undergo multi-step biotransformation that is usually catalyzed by bacterial enzymes in the intestinal tract (Ridlon et al., 2006). In the small and large intestine, a number of bacterial species have bile salt hydrolases. These are bile salt de-conjugating enzymes that convert some taurine- and glycine-conjugated bile acids to free bile acids. Free bile acids can passively diffuse through the plasma membrane, but this process accounts for a very small fraction of bile acid re-absorption in the small and large intestine. Probably more importantly, bile acid deconjugation by bile salt hydrolases is a required step for subsequent conversion of primary bile acids into secondary bile acids in the intestine. For example, in the large intestine, bacterial 7α-dehydroxylase catalyzes C-7 dehydroxylation reaction on primary bile acids, which converts CA and CDCA to deoxycholic acid (DCA) and lithocholic acid (LCA), respectively (Bjorkhem et al., 1989; Hylemon et al., 1991; Ridlon et al., 2006). These secondary bile acids are highly hydrophobic and toxic, and increased concentrations in the liver has been linked to inflammation, cholestasis, gallstone formation and carcinogenesis (Low-Beer and Nutter, 1978; Marcus and Heaton, 1988; Ridlon and Hylemon, 2006). Both elevated levels of DCA and LCA have been implicated in the promotion of colon cancer (Hussaini et al., 1995). Elevated LCA in cholestasis contributes to liver injury and inflammation (Staudinger et al., 2001). Unconjugated DCA is reabsorbed in the large intestine via passive absorption and transported back to the liver, where it can be converted to conjugated form (Trauner and Boyer, 2003). In normal conditions, most LCA is sulfated in the intestine and excreted into feces. The trace amount of LCA that is transported to the liver can be efficiently sulfated and secreted into the circulation for renal excretion. In the intestine, CYP3A and CYP2 family enzymes can also metabolize LCA to more soluble and thus less toxic hyocholic acid (HCA) and ursodeoxycholic acid (UDCA) in humans (Figure 2) (Araya and Wikvall, 1999; Teixeira and Gil, 1991).
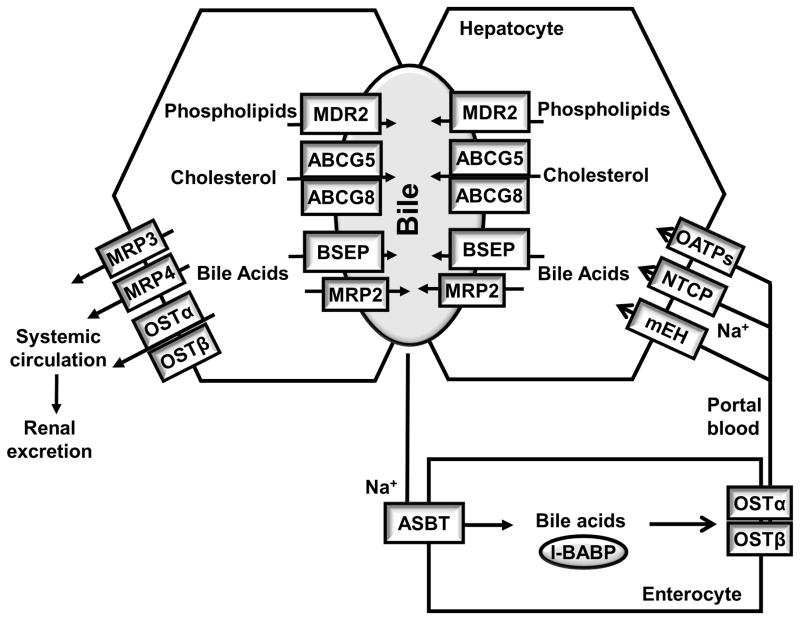
Figure 2. Bile acid transporters in the Enterohepatic circulation.
Bile acids, after synthesis, are secreted into the bile where bile acids, cholesterol and phospholipids form micelles. Food intake stimulates the gallbladder to release bile acids into the small intestine. Conjugated bile acids are efficiently re-absorbed in the ileum by active transport systems in the ileum, and a small amount of un-conjugated bile acids is reabsorbed by passive diffusion in the small and large intestine. Less than 5% of the bile acids is lost through fecal excretion, which is compensated by de novo synthesis in the liver. At the canalicular membrane of the hepatocytes, the bile salt export pump (BSEP) is the primary bile acid efflux transporter, while the multidrug resistance-associated protein-2 (MRP-2) can also secrete organic substrates including bile acids, bilirubin and glutathione. ABCG5 and ABCG8 form heterodimers and transport cholesterol into the bile, and multidrug resistance-2 (MDR2) is responsible for phospholipid secretion. At the basolateral membrane of the hepatocytes, The Na+-dependent taurocholate transporter (NTCP) is mainly responsible for Na+-dependent uptake of conjugated bile acids. The microsomal epoxide hydrolase (mEH) may also mediate Na+-dependent uptake of conjugated bile acids. The organic anion transporters (OATPs) show substrate specificity for unconjugated bile acids. At the basolateral membrane of the hepatocytes, organic solute transporters OSTα and OSTβ heterodimers, MRP3 and MRP4 secrete bile acids into the systemic circulation. In cholestasis, this pathway is induced and leads to subsequent renal bile acid excretion. In the intestine, the apical sodium-dependent bile acid transport (ASBT) mediates bile acid uptake in the ileum. Intracellular bile acids are bound to the intestinal bile acid binding protein (I-BABP) and are transported to the basolateral membrane where bile acids is secreted into the portal circulation by the OSTα and OSTβ heterodimer.
It should be noted that although rodents, especially mice and rats, have been widely used as a model for bile acid research, the bile acid pool composition and hydrophobicity in rats and mice are significantly different from that of humans. In mice and rats, the majority of CDCA is converted to more hydrophilic α-muricholic acid (α-MCA) and β-MCA in the liver, which deoes not occur in human livers. In humans, the highly hydrophobic bile acid pool consists of about 40% of CA, 40% of CDCA and 20% DCA. Mouse bile acid pool consists of about equal amount of CA and MCAs with relatively low levels of DCA and CDCA, and thus is more hydrophilic. In addition, rats do not have gallbladders, and they secrete bile directly into the small intestine.
10.1.3. Genetic defects of cytochrome P450s in bile acid synthesis
Genetic mutations in CYP7A1 gene are very rare and only one case has been reported so far. Individuals of homozygous CYP7A1 mutation showed decreased bile acid levels, and developed hypercholesterolemia and premature gallstone disease, which is consistent with the key role of bile acid synthesis in overall cholesterol metabolism (Pullinger et al., 2002). Consistently, studies have linked CYP7A1 polymorphisms to risk of gallstone, plasma LDL cholesterol levels and cardiovascular risk (Hofman et al., 2005; Jiang et al., 2004; Kajinami et al., 2004). Mice lacking cyp7a1 showed decreased postnatal survival rate and fat and vitamin malabsorption which could be partially corrected with vitamin and bile acid supplements (Ishibashi et al., 1996). Later studies also showed that cyp7a1 knockout mice were hypercholesterolemia (Erickson et al., 2003). In contrast, CYP7A1 transgenic mice had increased hepatic cholesterol catabolism, and were resistant to diet-induced hypercholesterolemia and atherosclerosis (Miyake et al., 2001; Miyake et al., 2002) and metabolic disorders (Li et al., 2011; Li et al., 2010). CYP7A1 transgenic mice had ~2–3 fold increase in total bile acid pool size. Susceptibility to cholestatic liver injury has not been studied in these mice.
Humans with CYP8B1 mutations have not been reported so far. Cyp8b1 knockout mice lack CA production (Li-Hawkins et al., 2002). As mentioned early and also discussed in the later section, due to the efficient conversion of CDCA to MCAs, mice depend more on CA to inhibit cyp7a1 and to facilitate intestine cholesterol absorption because MCAs neither inhibit cyp7a1 nor facilitate cholesterol absorption. For these reasons, cyp8b1 knockout mice had increased hepatic CYP7A1 levels and larger bile acid pool, and decreased intestinal fractional cholesterol absorption (Slatis et al., 2010). Cyp8b1 knockout mice were also resistant to atherosclerosis development primarily due to decreased cholesterol absorption and increased hepatic CYP7A1 levels. In humans, CDCA is not converted to MCAs. Therefore, decreased CYP8B1 activity or defect in CA synthesis in humans may have different impact on hepatic bile acid synthesis and intestine cholesterol absorption compared to mice.
Because CYP27A1 mediates the sterol side chain cleavage in both classic and alternative bile acid synthesis pathways, CYP27A1 mutations in humans leads to decreased bile acid synthesis and increased levels of 7α-hydroxylated cholesterol metabolites, including C4 in the liver. Human mutations in CYP27A1 cause a rare lipid storage disorder called cerebrotendinous xanthomatosis (CTX) (Leitersdorf et al., 1994). CTX is characterized by the accumulation of cholesterol and cholestanol, a derivative of C4, in xanthomas and brains, leading to xanthomatosis, premature atherosclerosis and progressive neurological disorders. CTX patients are effectively treated with CDCA, which inhibits CYP7A1 and thus decrease the production of 7α-hydroxylated cholesterol metabolites and therefore cholestanol. CYP27A1 mutations in humans usually do not lead to hypercholesterolemia. Cyp27a1 knockout mice had decreased bile acid synthesis and up-regulation of hepatic CYP7A1 gene, but were absent of CTX-related phenotypes (Rosen et al., 1998).
10.1.4. Bile acid transport
Enterohepatic circulation of bile acids
Once synthesized, bile acids in the hepatocytes are efficiently secreted into the bile and stored in the gallbladder. Food intake stimulates the epithelial cells in the duodenum to secrete a peptide hormone called cholecystokinin (CCK), which stimulates gallbladder contraction to release bile acids into the intestinal tract (Otsuki, 2000). CCK also stimulates the pancreas to secrete digestive enzymes into the small intestine to facilitate digestion. Along the small intestine tract, concentrated bile acids form mixed micelles with dietary lipids, which is necessary for lipid digestion by pancreatic lipases. Bile acids are then reabsorbed in the ileum, and transported back to the liver via portal blood for re-secretion into the bile. The enterohepatic circulation of bile acids is a highly efficient process and only ~ 5% of the total bile acids is lost via excretion into the feces, which is replenished by the hepatic de novo synthesis of bile acid. The enterohepatic circulation of bile acids is illustrated in Figure 2, and the bile acid transporters involved in this process are discussed below.
Hepatic bile acid transport
Hepatocytes are polarized cells with basolateral (sinusoidal) and apical (canalicular) membrane domains. Bile acids in the portal circulation are taken up by hepatocytes across the basolateral membrane and then secreted into the bile canaliculi and subsequently drained into the bile. Hepatic bile acid uptake is highly efficient with about 90% first pass extraction rate for conjugated bile acids. Conjugated bile acids cannot diffuse through the membrane and thus require active transport systems for cellular uptake (Meier, 1995). The Na+-dependent taurocholate transporter (NTCP) is the major bile acid uptake transporter in the basolateral membrane of the hepatocytes (Figure 2). Early studies conducted in cultured hepatocytes or perfused livers support the role of NTCP as the basolateral Na+-dependent bile acid uptake transporter (Hagenbuch and Meier, 1994; Hagenbuch et al., 1991; Meier and Stieger, 2002). Recently, a human patient with loss-of-function mutation of the NTCP gene was reported to show significantly elevated circulating bile acids without clinical symptoms of cholestasis, consistent with impaired basolateral bile acid uptake into the hepatocytes (Vaz et al., 2014). This is further supported by a recent study of ntcp knockout mice (Slijepcevic et al., 2015). The microsomal epoxide hydrolase (mEH), a bi-functional enzyme that is involved in xenobiotic metabolism, is another candidate basolateral Na+-dependent bile acid transporter (Ananthanarayanan et al., 1988; Fretland and Omiecinski, 2000; Von Dippe et al., 1993; von Dippe et al., 1996). A human individual with significantly decreased mEH function due to a point mutation had hypercholanemia, a condition associated with highly elevated plasma bile acid levels in the absence of hepatocyte injury (Zhu et al., 2003). However, mEH knockout mice showed no apparent alteration in bile acid homeostasis (Miyata et al., 1999). Several organic anion transporter (OATP) isoforms, including human OATP1A2, OATP1B1 and OATP1B3, can mediate Na+-independent bile acid uptake at the basolateral membrane of the hepatocytes. This pathway is mainly responsible for the uptake of unconjugated bile acids, and accounts for about ~25% of bile acids uptake by the hepatocytes (Trauner and Boyer, 2003).
Bile acids, phospholipids and cholesterol are three major organic solutes in the bile. They form mixed micelles to increase cholesterol solubility and decrease bile acid toxicity in the bile. Canalicular bile acid secretion into the bile against the concentration gradient is the rate-limiting step in bile formation (Boyer, 2013). The bile salt export pump (BSEP, ABCB11), an ATP-binding cassette (ABC) transporter also known as the sister of P-glycoprotein (SPGP), is the major bile acid efflux transporter at the canalicular membrane (Childs et al., 1995). The multidrug resistance-associated protein-2 (MRP2, ABCC2) can also mediate the canalicular secretion of certain sulfated and conjugated bile acids into the bile. MRP2 also shows broad substrate specificity for bilirubin conjugates, glutathione and drugs. Hepatobiliary free cholesterol secretion into the bile is a major route for cholesterol elimination from the body. The ATP-binding cassette transporters ABCG5 and ABCG8 form a heterodimer, which mediates cholesterol secretion into the bile (Berge et al., 2000). Phosphatidylcholine is the major phospholipid in the bile and its secretion is mediated by the phospholipid flippase multi-drug resistant 3 (MDR3, ABCB4) (Smit et al., 1993).
Bile acids in the hepatocytes can also be secreted across the basolateral membrane back into the systemic circulation for subsequent renal excretion. This process is mediated by several transporters including the organic solute transporters OSTα and OSTβ heterodimer, MRP3 and MRP4 (Ballatori et al., 2005; Kullak-Ublick et al., 2000; Trauner and Boyer, 2003). This basolateral bile acid efflux route in the hepatocytes is much less significant than biliary bile acid secretion under normal physiology. However, in cholestasis when biliary bile acid secretion is impaired, basolateral bile acid efflux is often induced as a protective mechanism (Ballatori et al., 2005; Boyer et al., 2006; Cui et al., 2009). In the bile duct, some of the bile acids can be taken up by the cholangiocytes. Unconjugated bile acids can passively enter the cholangiocytes, and conjugated bile acids are taken up by the apical sodium-dependent bile salt transporter (ASBT, SLC10A1) (Xia et al., 2006). Bile acids in cholangiocytes are secreted into the peribiliary plexus via the OSTα/β heterodimer at the basolateral membrane. Cholangiocyte bile acid transport may play a role in bile formation. In addition, this process can also transport bile acids into the systemic circulation for renal elimination in cholestasis (Kullak-Ublick et al., 2000; Xia et al., 2006).
Intestinal bile acid transport
Intestine bile acid reabsorption mainly occurs at the terminal ileum where bile acid transporters are highly expressed. This is a highly efficient process and only about ~5% of the bile acids is loss via fecal excretion. At the apical side, the ASBT mediates ileal bile acid uptake into the enterocytes (Shneider et al., 1995). In the enterocytes, bile acids are transported to the basolateral membrane by the intestinal bile acid binding protein (I-BABP) (Gong et al., 1994). The OSTα and OSTβ form a functional heterodimer at the basolateral membrane and mediate bile acid efflux into the portal circulation (Ballatori et al., 2005; Rao et al., 2008). As most of the conjugated bile acids are efficiently reabsorbed in the small intestine via active transport systems, some unconjugated primary bile acids and secondary bile acids, mainly DCA and to a much less extent LCA can also be reabsorbed in the colon via passive diffusion and returned to the liver via portal circulation.
10.2. Regulation of bile acids toxicity and inflammation in cholestasis
10.2.1. Cholestatic liver diseases
Cholestasis is resulted from disrupted bile flow from the liver to the intestine tract, leading to accumulation of toxic bile acids and other metabolites in the liver, decreased bile acids in the intestine and increased bile acids in the systemic circulation. The accumulation of toxic bile acids in the hepatobiliary system damages bile duct epithelial cells and hepatocytes, causing liver injury and inflammation. Chronic cholestasis leads to fibrosis, cirrhosis and eventually liver failure or hepatocellular or cholangiocellular carcinomas. Mechanical obstruction of extrahepatic bile ducts or major intrahepatic bile ducts, such as by tumors or gallstones, genetic mutations of bile acid transporter genes, and acquired dysregulation of bile transport system by drugs, pregnancy and other pathophysiological conditions can cause either intra- or extra-hepatic cholestasis (Srivastava, 2014; Zollner and Trauner, 2008). Congenital or acquired defects in canalicular membrane transporters are the major cause of intrahepatic cholestasis (Zollner and Trauner, 2008). Congenital cholestasis usually occurs very early in life. These patients show progressive liver damage, jaundice, pruritus, slow growth due to malabsorption of fat-soluble vitamins. Progressive familial intrahepatic cholestasis and benign recurrent intrahepatic cholestasis are autosomal recessive diseases that are linked to genetic mutations in ATP8B1 (Type 1, PFIC1), BSEP (Type 2, PFIC2), and MDR3 (Type 3, PFIC3)(Srivastava, 2014). PFIC1, which is also known as Byler disease, is linked to mutations in the ATP8B1 gene. This gene encodes a P-type ATPase that functions as an aminophospholipid flippase. Mutations in the BSEP gene are associated with progressive familial intrahepatic cholestasis subtype 2 (PFIC-2). Defective BSEP function causes hepatic bile acid accumulation and cholestasis. These patients showed markedly elevated plasma bile acid levels and extremely low biliary bile acid concentration (Strautnieks et al., 1997). In addition, BSEP polymorphisms have been linked to intrahepatic cholestasis of pregnancy (ICP) (Noe et al., 2005; Pauli-Magnus et al., 2004) and drug-induced liver injury (Lang et al., 2007). ICP is a reversible form of intrahepatic cholestasis associated with adverse pregnancy outcomes. PFIC3 patients, with defective phospholipid transporter MDR3, have high levels of γ-glutamyl transpeptidase activity, progressive cholestasis, bile duct damage, and may require liver transplant. As mentioned early, phospholipids in bile are required for mixed micelle formation. If not incorporated into mixed micelles, bile acids will damage the canalicular membrane and cholangiocytes, causing liver injury and cholestasis. Genetic polymorphisms and heterozygote mutations of the PFIC1, PFIC2 and PFIC3 genes may increase susceptibility to acquired cholestasis in adults including ICP, drug-induced liver injury, primary biliary cirrhosis (PBC), and primary sclerosing cholangitis (PSC). MRP2 excretes conjugated bile acids, bilirubin and other organic anions into the bile. Mutations in the MRP2 gene have been linked to Dubin-Johnson syndrome, which is characterized by chronic hyperbilirubinemia (Keitel et al., 2003). These patients have elevated bile acids and cholestasis.
10.2.2. Bile acid-activated nuclear receptor regulation of bile acid metabolism and detoxification in cholestasis
Nuclear receptor
Nuclear receptors (NR) are ligand-activated transcription factors. They play important roles in embryogenesis, development and metabolism (Mangelsdorf and Evans, 1995; Mangelsdorf et al., 1995). A typical nuclear receptor consists of an N-terminal DNA binding domain (DBD) and a C-terminal ligand-binding domain (LBD) (Figure 3). The DBD is a highly conserved region containing two Zinc finger motifs that mediate the NR binding to a consensus DNA sequence called hormone response element (HRE). Some NRs bind to the HRE as a homodimer, while some NRs form a heterodimer with another nuclear receptor retinoid X receptor (RXR) to bind to DNA. A few NRs, such as the hepatocyte nuclear factor 4α (HNF4α) and the liver receptor homolog-1 (LRH-1), bind DNA as monomers. In the LBD, a number of α-helices form a hydrophobic ligand-binding cavity where lipophilic small molecules bind as ligands. The LBD also contains motifs for dimerization and recruitment of co-regulators (co-activators or co-repressors). In general, ligand binding causes a conformational change in the LBD of a NR, which allows the NRs to recruits the co-activators and displace the corepressors (Figure 3). This facilitates the assembly of general transcriptional machinery leading to transcriptional activation of the target genes. There are 48 nuclear receptor genes in the human genome and 49 in the mouse genome (Mangelsdorf et al., 1995). Among these NRs, the Farnesoid X receptor (FXR), Pregnane X receptor (PXR) and Vitamin D receptor (VDR) can be activated by bile acids (Chiang, 2003). Constitutive androstane receptor (CAR) is activated by drugs and xenobiotics, but not bile acids. However, CAR is activated in cholestasis likely by the accumulation of toxic metabolites. These NRs regulate a number of bile acid synthetic and metabolizing CYPs as well as bile acid conjugation enzymes and transporters. During cholestasis, activation of these NRs by bile acids and their metabolites play a protective role against injury by decreasing hepatic bile acid synthesis and uptake, promoting hepatic bile acid efflux, and inducing phase-I bile acid metabolizing and phase-II bile acid conjugation enzymes. The bile acid activated NRs and their target genes are summarized in Table 1. The role of major bile acid-activated receptors in the regulation of bile acid metabolism and detoxification are discussed below.
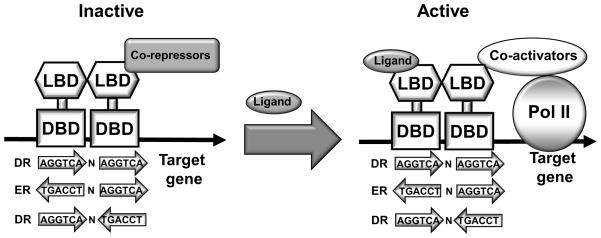
Figure 3. Nuclear receptors.
The domain structure of a typical nuclear receptor contains a DNA binding domain (DBD) and a ligand binding domain (LBD). Nuclear receptors recognize consensus DNA sequence AGGTCA half site arranged in direct repeat (DR), everted repeat (ER) and inverted repeat (IR). Ligand binding causes nuclear receptor LBD conformational change, which allows the nuclear receptor to recruit coactivators to replace corepressors. Co-activators facilitate chromatin remodeling and the assembly of general transcriptional machinery, leading to transactivation of the target gene.
Table 1.
Bile acid-activated nuclear receptor target genes in bile acid metabolism
| Receptors | Target genes | regulation | Function and pathways |
|---|---|---|---|
| FXR | CYP7A1 | Down | Enzyme in bile acid synthesis |
| CYP8B1 | Down | Enzyme in bile acid synthesis | |
| CYP27A1 | Down | Enzyme in bile acid synthesis | |
| SHP | Up | Negative regulator of bile acid synthesis | |
| FGF15/FGF19 | Up | Negative regulator of bile acid synthesis | |
| BSEP | Up | Canalicular bile acid secretion | |
| OSTα/OSTβ | Up | Basolateral bile acid secretion | |
| NTCP | Down | Basolateral bile acid uptake | |
| UGT2B4/UGT2B7 | Up | Bile acid conjugation | |
|
| |||
| PXR | CYP3A | Up | Phase-I bile acid/drug metabolism |
| CYP2B/CYP2C | Up | Phase-I bile acid/drug metabolism | |
| MRP1/MRP2/MRP3 | Up | Bile acid/drug transport | |
| UGT1A3 | Up | Bile acid conjugation | |
| SULT2A1 | Up | Bile acid conjugation | |
| OATP2 | Up | Basolateral bile acid/drug uptake | |
| CYP7A1 | Down | Enzyme in bile acid synthesis | |
|
| |||
| CAR | CYP3A | Up | Phase-I bile acid/drug metabolism |
| CYP2B, CYP2C | Up | Phase-I bile acid/drug metabolism | |
| UGT1A1 | Up | Bile acid conjugation | |
| MRP2 | Up | Bile acid/drug transport | |
| CYP7A1 | Down | Enzyme in bile acid synthesis | |
|
| |||
| VDR | CYP3A | Up | Phase-I bile acid/drug metabolism |
| CYP2B/CYP2C | Up | Phase-I bile acid/drug metabolism | |
| SULT2A1 | Up | Bile acid conjugation | |
| MRP3/MRP4 | Up | Bile acid/drug transport | |
| ASBT | Up | Canalicular bile acid uptake | |
| CYP7A1 | Down | Enzyme in bile acid synthesis | |
FXR regulation of bile acid feedback inhibition of bile acid synthesis
FXR is highly expressed in the hepatocytes and in the intestine, tissues that are exposed to high levels of bile acids. FXR can be activated by both free and conjugated-bile acids (Makishima et al., 1999; Parks et al., 1999). The hydrophobic bile acid CDCA is the most efficacious ligand of FXR (EC50 = ~10 μmol/L), followed by LCA, DCA, and CA The hydrophilic bile acid MCAs and UDCA do not activate FXR. In fact, a recent study showed that elevated concentration of MCA in the bile acid pool may act as FXR antagonists to inhibit FXR activity (Sayin et al., 2013).
Hepatic bile acid synthesis is tightly regulated by bile acids through negative feedback mechanisms, which maintains a constant bile acid pool size in normal physiology. In cholestasis, accumulation of bile acids in the liver represses hepatic bile acid synthesis, which is deemed as a protective mechanism to alleviate bile acid damage to the liver. The activity of hepatic bile acid synthetic cytochrome P450 enzymes, including CYP7A1, CYP8B1 and CYP27A1, are mainly controlled at the rate of gene transcription. Bile acid activated FXR has been shown to cause transcriptional repression of CYP7A1, CYP8B1 and CYP27A1, and thus plays a central role in mediating the negative feedback regulation of bile acid synthesis (Li and Chiang, 2014). The mechanisms of FXR regulation of the CYP7A1 gene have been extensively investigated in the past decades. FXR does not directly bind to the CYP7A1 gene promoter. Instead, two indirect mechanisms mediate the FXR repression of CYP7A1 gene (Figure 4). First, activation of FXR induces the transcription of a nuclear receptor small heterodimer partner (SHP). SHP is an atypical nuclear receptor without a DBD and thus does not bind to DNA. Instead, it often acts as a transcriptional repressor and inhibits the trans-activating activity of a number of nuclear receptors and transcriptional factors, leading to the inhibition of their target genes. On the CYP7A1 gene promoter, two nuclear receptors LRH-1 and HNF4α bind to their consensus DNA binding sites and activate the basal CYP7A1 gene transcription (Goodwin et al., 2000; Lu et al., 2000). Bile acid and FXR-induced SHP interacts with and inhibits LRH1 and HNF4α, leading to transcriptional repression of the CYP7A1 gene. In normal physiology, hepatic bile acids are efficiently excreted by canalicular transporters into the bile to maintain a relatively low level of intrahepatic bile acids. Food intake subsequently triggers gallbladder bile acid release into the small intestine. Indeed, studies in mice revealed that intestine is a major bile acid storage site that can retain about 60–80% of the bile acid pool in the body (Li et al., 2012; Li et al., 2010). Therefore, intestine bile acid sensing is very important in the bile acid pool maintenance. In 2005, Inagaki et al. reported that activation of FXR transcriptionally induces fibroblast growth factor 15 (FGF15) in mice. FGF15 then acts as an endocrine hormone, binds to the cell surface FGF receptor 4 (FGFR4) on the hepatocytes and inhibits hepatic CYP7A1 gene transcription (Inagaki et al., 2005). Studies have so far identified that β-Klotho, the SH2 domain containing protein tyrosine phosphatase (SHP-2) and FGF receptor substrate 2 (FRS2) are key components of the FGFR4 signaling complex at the plasma membrane, and that deletion of each of these signaling complex proteins led to increased hepatic bile acid synthesis and elevated bile acid pool size (Goetz et al., 2007; Ito et al., 2005; Li et al., 2014; Lin et al., 2007; Wang et al., 2014). Importantly, deletion of shp-2 resulted in a severe cholestasis phenotype in mice (Li et al., 2014). Human FGF19 shares ~51% amino acid sequence homology with mouse FGF15, and thus is the mouse FGF15 orthologue. Current evidence suggests that FGF19 also represses human CYP7A1 in an FGFR4-dependent signaling mechanism (Song et al., 2009). However, different from FGF15 that is not expressed in mouse hepatocytes, FGF19 mRNA is detectable in human livers and primary human hepatocytes, and its expression can be induced by FXR (Song et al., 2009). One study showed that circulating FGF19 levels increased while CYP7A1 expression decreased in human patients with obstructive cholestasis, indicating that human hepatocytes produce FGF19 (Schaap et al., 2009). The CYP8B1 and CYP27A1 genes have been shown to be repressed by bile acid/FXR/SHP cascade, but not by the bile acid/FXR/FGF15 signaling axis (Chen and Chiang, 2003; Zhang and Chiang, 2001). Therefore, FXR activation represses key genes in the biosynthetic pathway to decrease bile acid output.
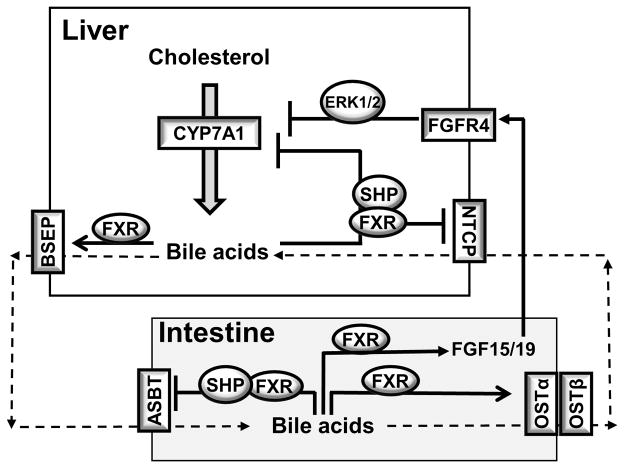
Figure 4. FXR regulation of bile acid feedback inhibition of bile acid synthesis and bile acid transport in the enterohepatic system.
In hepatocytes, bile acids-activated FXR induces the repressor SHP, which interacts with and represses the trans-activating action of HNF4α and LRH-1, leading to CYP7A1 inhibition. Bile acid/FXR induces SHP to repress NTCP. FXR binds to BSEP gene promoter and induced BSEP and canalicular bile acid secretion. In the intestine, FXR activation inhibits ASBT and induces OSTα and OSTβ, and thus decreases bile acid absorption and promotes basolateral bile acid secretion. Bile acid-activated FXR induces FGF15 (FGF19 in humans). FGF15 binds and activates FGFR4 on the hepatocytes, leading to the inhibition of CYP7A1 gene, a process that may involve ERK1/2 signaling. The downstream target of FGF15/19 has not been well characterized.
FXR regulation of bile acid transport
In addition to inhibiting hepatic bile acid synthesis, FXR activation by bile acids also inhibits hepatic bile acid uptake and promotes biliary bile acid secretion by regulating the expression of hepatic bile acid transporters, and thus prevents bile acid accumulation in the hepatocytes. FXR induces the apical bile acid efflux transporters BSEP (Ananthanarayanan et al., 2001), MRP2 (Kast et al., 2002), and the phospholipid transporter ABCB4 (Liu et al., 2003). At the basolateral membrane of the hepatocytes, FXR activation limits bile acid uptake by inhibiting the expression of bile acid uptake transporter NTCP via induction of SHP that represses the transactivation of NTCP by nuclear receptors such as retinoic acid receptor (RAR) and HNF4α (Denson et al., 2001). In conditions associated with hepatic bile acid accumulation, there is usually a compensatory induction of transporters including OSTs and MRPs at the basolateral membrane of the hepatocytes to efflux bile acids into the systemic blood circulation for subsequent renal excretion (Figure 2), resulting in elevated plasma bile acid concentration in cholestasis (Ballatori et al., 2005; Boyer et al., 2006; Cui et al., 2009). The OSTα and OSTβ genes are directly induced by bile acid activated FXR, while MRP1, MRP3 and MRP4 are induced by bile acid-activated PXR during cholestasis, which will be discussed in the next section. In the intestine, FXR induces OSTα and OSTβ genes (Frankenberg et al., 2006) and represses ASBT gene (Chen et al., 2003; Neimark et al., 2004). Therefore, similar to the role of FXR in the hepatocytes, FXR activation in the intestine also inhibits bile acid uptake and promote basolateral bile acid secretion, which decreases cellular bile acid accumulation.
Bile acid/xenobiotic receptors in bile acid metabolism and detoxification
Like drugs and xenobiotics, bile acids accumulation during cholestasis feedforward induces genes that are involved in phase I and phase II bile acid metabolism and detoxification. In the liver, CYP3A4 is the major enzyme that catalyzes the hydroxylation of both primary and secondary bile acids at various positions, converting bile acids into more hydrophilic and less toxic molecules (Araya and Wikvall, 1999; Bodin et al., 2005; Chen et al., 2014). However, unlike bile acid transporters, genetic variation of CYP3A4 has not been associated with the risk of cholestasis. Bile acids also undergo phase II glucuronidation and sulfoconjugation reactions before biliary or renal excretion (Belanger et al., 2003; Kauffman, 2004). Significantly elevated levels of sulfonated and glucoronidated bile acids are often seen in patients with cholestasis (Takikawa et al., 1983). Sulfotransferase 2A isoforms (SULT2As) are major enzymes involved in the sulfoconjugation reactions of bile acids (Weinshilboum et al., 1997), which usually lead to the formation of 3-alpha-sulfated bile acids. As mentioned earlier, LCA in the liver can be efficiently sulfated and subsequently excretion in the kidney. The protective role of SULT enzymes against bile acid toxicity has been demonstrated in LCA-induced liver toxicity in mice (Kitada et al., 2003). Glucuronide conjugation of bile acids are catalyzed by UDP-glucuronosyltransferase (UGT) isoforms UGT2B4, UGT2B7 and UGT1A3 (Gall et al., 1999; Pillot et al., 1993; Trottier et al., 2006). These enzymes mediate the glucuronide conjugation of hydroxylated bile acids at different positions. For example, UGT2B4 is involved in the glucoronidation of 6α-hydroxylated bile acids, while UGT2B7 can mediate the glucoronidation of both 3α-hydroxylated bile acids and 6α-hydroxylated bile acids. Although some studies have shown that bile acid-activated FXR can regulate a few of the above mentioned genes (Barbier et al., 2003; Lu et al., 2005), extensive studies suggest that the drug and bile acid/xenobiotic-activated nuclear receptors PXR, CAR and VDR play a predominant role in inducing the phase I and phase II bile acid metabolizing genes in cholestasis.
Pregnane X receptor
PXR can be activated by a wide range of xenobiotics, endobiotics and clinical drugs, and in turn induces a number of CYP3A and CYP2 family enzymes, conjugation enzymes and transporters in drug metabolism (Kliewer et al., 2002). In cholestasis, high levels of primary bile acids and secondary bile acids and some of the bile acid intermediate metabolites can activate PXR. Upon ligand activation, PXR usually induces gene transcription via binding to the xenobiotic response elements in its target gene promoter or enhancer regions. In phase I bile acid metabolism, PXR induces CYP3A4 and CYP2B genes (Staudinger et al., 2001). Hydroxylation of bile acids by CYP3A4 not only detoxifies bile acids, but also increases their conjugation and subsequent excretion. Specifically, CYP3A4 can convert CDCA to hyocholic acid and 3α, 7α-dihydroxy-3-oxo-5β-cholanoic acid, and CA to 3-dehydro-CA. CYP3A4 can convert the highly toxic and carcinogenic DCA into 3-dehydro-DCA and 1β, 3α, 12α-trihydroxy-5β-cholanoic acid, and LCA into 3-dehydro-LCA, hyodeoxycholic acid and 1β-hydroxy-LCA. In phase II and phase III bile acid metabolism, PXR induces the bile acid conjugation enzymes, SULT2A1 and UGTs, the canalicular transporter MRP2 and the basolateral transporter OATP2 (Kliewer and Willson, 2002). These PXR target genes are induced in mice treated with LCA or subjected to bile duct ligation, while these adaptive responses were impaired in pxr knockout mice. PCN, a mouse PXR agonist, repressed hepatic CYP7A1 mRNA expression, enzyme activity and biliary bile acid secretion in rodents (Mason and Boyd, 1978; Stahlberg, 1995; Staudinger et al., 2001), suggesting that PXR may regulate bile acid synthesis. Rifampicin-activated PXR was also shown to repress human CYP7A1 gene transcription (Bhalla et al., 2004; Li and Chiang, 2005). PXR does not directly bind to the CYP7A1 gene promoter. Instead, it interacts with and represses the transactivation activity of HNF4α that binds the CYP7A1 gene promoter. In addition, activation of PXR in the intestine was shown to induce FGF15 or FGF19 expression, and a PXR response element was identified in the promoter of the FGF19 (Wang et al., 2011a; Wistuba et al., 2007). Mice lacking PXR were more susceptible to hepatotoxicity caused by LCA treatment or bile duct ligation (Staudinger et al., 2001; Stedman et al., 2005). Studies have also shown that pharmacological activation of PXR protected against bile acid-induced liver injury in experimental cholestasis models (Stedman et al., 2005). Rifampicin, the human PXR agonist, has been used to reduce pruritus associated with cholestasis in humans (Hofmann, 2002). The effectiveness of rifampicin in treating pruritus varied among individuals (Hofmann, 2002).
Constitutive androstane receptor
CAR is another key regulator of drug and bile acid metabolizing genes in the liver (Stanley et al., 2006). CAR and PXR can bind to the same xenobiotic response elements in the target gene promoters and thus regulate an overlapping set of target genes including CYP3A and CYP2Bs. Phenobarbital and TCPOBOP are frequently used as CAR agonists in various experimental settings. Bile acids do not directly activate CAR, but CAR may be activated by toxic metabolites in cholestasis. CAR agonists have been shown to repress CYP7A1 gene in hepatocytes (Miao et al., 2006). Studies have shown that activation of CAR was beneficial for protecting against bile acid toxicity during cholestasis in mice (Beilke et al., 2009; Guo et al., 2003; Saini et al., 2004; Stedman et al., 2005). The car knockout mice had higher degree of liver injury than wild type mice upon LCA treatment or bile duct ligation (Stedman et al., 2005). CAR may play an important role in inducing sulfation of LCA because car transgenic mice had increased levels of sulfated LCA and were resistant to LCA toxicity (Saini et al., 2004). Treatment with CAR ligands phenobarbital or TCPOBOP in fxr/pxr double knockout mice protected these mice from CA feeding-induced bile acid toxicity, which was attributed to the induction of CAR target genes Cpy2b, Cyp3a, Mrp2, Ugt1a1 and Gsta (Guo et al., 2003).
Vitamin D3 receptor
VDR is highly expressed in the intestine, but is expressed at very low levels in human hepatocytes, and is not expressed in mouse liver. The role of VDR in the regulation of intestine bile acid metabolism is well documented (Makishima et al., 2002), but its role in hepatic bile acid metabolism is less clear. Among all bile acids, LCA and its metabolite 3 keto-LCA are the most potent activators of VDR. VDR acts as a bile acid sensor in the intestine to protect the gut from bile acid toxicity (Makishima et al., 2002; Nagpal et al., 2005). VDR can recognize the same xenobiotic response elements as PXR and CAR, and activation of VDR by 1α, 25- dihydroxyvitamin D3 induces CYP3A4, CYP2B and CYP2C in drug and bile acid metabolism (Drocourt et al., 2002; Schmiedlin-Ren et al., 2001; Thummel et al., 2001). Activation of VDR also induced SULT2A1 and thus can stimulate bile acid sulfoconjugation (Chatterjee et al., 2005). Furthermore, VDR induced two bile acid transporters MRP3 and ASBT in the intestine (Chen et al., 2006; McCarthy et al., 2005). It has been suggested that high levels of LCA not only causes cholestasis, but may also be involved in the promotion of colon cancer (Ajouz et al., 2014). When LCA levels increase in the gut, VDR may be activated as an adaptive response to convert LCA to less toxic intermediates for excretion (Makishima et al., 2002).
Unlike PXR and CAR, VDR seemed to be expressed at very low levels in primary human hepatocytes (Han et al., 2010). Treatment of 1α, 25-dihydroxyvitamin D3 induced CYP3A, CYP2B, and CYP2C in primary human hepatocytes (Drocourt et al., 2002). During cholestasis, LCA levels may increase significantly in the liver, which leads to VDR activation. A few studies have investigated the role of VDR in cholestasis in mice. These studies used 1α, 25-dihydroxyvitamin D3 as a VDR agonist but not bile acids which also activate PXR and FXR. One study showed that 1α, 25-dihydroxyvitamin D3 treatment did not affect hepatic or plasma bile acid levels in bile duct ligated mice, suggesting a minimal role of VDR in modulating bile acid metabolism in cholestasis (Ogura et al., 2009). However, VDR activation by 1α, 25-dihydroxyvitamin D3 treatment in bile duct ligated mice reduced pro-inflammatory cytokine expression, suggesting the anti-inflammatory properties of VDR may provide certain benefits during cholestasis (Nagpal et al., 2005). Another study showed that 1α, 25-dihydroxyvitamin D3 treatment increased renal MRP2, MRP3 and MRP4 mRNA expression and increased renal bile acid secretion (Nishida et al., 2009).
Some studies have shown that VDR agonists affected hepatic bile acid synthesis and bile acid pool size, but both the effects and the mechanisms are still somewhat controversial. Treating primary human hepatocytes with 1α, 25-dihydroxyvitamin D3 repressed CYP7A1 mRNA expression, suggesting VDR may inhibit bile acid synthesis in cholestasis (Han et al., 2010). A recent study showed that vdr knockout mice showed higher hepatic cyp7a1 gene expression and a larger bile acid pool size, while 1α, 25-dihydroxyvitamin D3 treatment decreased hepatic cyp7a1 gene expression in mice (Schmidt et al., 2010). VDR is not expressed in mouse hepatocytes. Instead, it was suggested that VDR may affect hepatic bile acid synthesis via regulation of intestine FGF15 transcription (Schmidt et al., 2010). In contrast, another study showed that injecting mice with 1α, 25-dihydroxyvitamin D3 increased hepatic CYP7A1 mRNA and lowered cholesterol levels (Chow et al., 2013). This was associated with decreased SHP expression in the liver upon VDR activation. These effects of VDR activation on bile acid synthesis are likely mediated by extrahepatic mechanisms.
10.2.3. Bile acid modulation of hepatic inflammation and cholestatic liver injury
Both CDCA and ursodeoxycholic acid (UDCA) have been used for effective gallstone dissolution in human patients for many years (Lioudaki et al., 2011). While CDCA can cause mild hepatotoxicity in some patients, UDCA is highly soluble and was found to be generally non-toxic to humans. UDCA (Ursodiol™) has also been approved by FDA for treating PBC, and has been shown to significantly improve liver tests and prolong the time needed for liver transplantation in these patients (Dyson et al., 2015). In contrast, UDCA is not effective in treating patient with PSC. Current evidence suggest that UDCA can provide multiple benefits including decreased hydrophobicity of the bile acid pool, increased hepatobiliary secretion, reduced inflammation and cell death. Nor-ursodeoxycholic acid (norUDCA) is a side chain-shortened C23 homologue of UDCA (Yeh et al., 1997; Yoon et al., 1986). It cannot be conjugated and after administration it is secreted into the bile, reabsorbed by cholangiocytes and returned to the liver. It has been shown that norUDCA increased bicarbonate in bile and thus hypercholeresis. It was shown that norUDCA improved sclerosing cholangitis in the Mdr2−/− model of cholangiopathy (Halilbasic et al., 2009).
As discussed earlier, activation of FXR provides multiple benefits in alleviating cholestatic liver injury. Based on these rationales, a potent FXR agonist obeticholic acid (OCA) has been tested for treating cholestasis in both experimental animal models and in humans (Ali et al., 2015). OCA is a 6α-ethyl CDCA derivative that selectively activates FXR with a ~ 100-fold higher potency than CDCA (Pellicciari et al., 2004; Pellicciari et al., 2002). In animal models of cholestasis, OCA effectively protected against cholestatic liver injury and inflammation (Fiorucci et al., 2005; Pellicciari et al., 2002). Recent clinical trials also showed that OCA significantly improved liver tests in patients with PBC (Hirschfield et al., 2015). In addition to decreasing bile acid synthesis, increasing bile flow and promoting bile acid detoxification, FXR has recently been shown to directly modulate immune response in both hepatic and extrahepatic tissues. Fxr knockout mice showed increased liver inflammation, while FXR activation decreased lipopolysaccharide (LPS)-induced hepatic inflammation (Wang et al., 2008). Consistently, FXR activation protected against liver injury in Mdr2 knockout mouse model of chronic cholangiopathy (Baghdasaryan et al., 2011). FXR has also been shown to play an anti-inflammatory role in extrahepatic tissues. For example, FXR modulates intestine immunity and FXR activation was shown to reduce inflammation in inflammatory bowel disease (Gadaleta et al., 2011; Vavassori et al., 2009). FXR is expressed in vascular smooth muscle cells (VSMC) and FXR agonists have been shown to inhibit inflammation in VSMC and slow the progression of atherosclerosis by decreasing inflammation of the vasculature (Bishop-Bailey et al., 2004; Hanniman et al., 2005; Zhang et al., 2008a). The underlying molecular mechanism by which FXR modulates immune response is still not fully clear. FXR activation may antagonize nuclear factor κB (NF-κB) signaling to decrease pro-inflammatory cytokine production in the liver (Wang et al., 2008). Some studies have reported that FXR was expressed in macrophages and activation of FXR repressed LPS-induced pro-inflammatory cytokine expression, an effect that was abolished in fxr−/− macrophages (Mencarelli et al., 2009). In VSMC, FXR may induce SHP to inhibit the expression of cyclooxygenase 2 (COX-2) and inducible nitric oxide synthase (iNOS), which are involved in vascular inflammation and VSMC migration. It is noted that beside cholestasis, FXR agonist OCA has also shown promise in treating nonalchoholic steatohepatitis (NASH) based on both animal studies and clinical trials (Ali et al., 2015; Neuschwander-Tetri et al., 2015). OCA improved lipid and glucose homeostasis, liver enzyme tests and insulin sensitivity, which may be attributed to the role of FXR in the regulation of lipid and glucose homeostasis, inflammation, insulin sensitivity and bile acid metabolism (Ali et al., 2015).
The G protein-coupled receptor TGR5 is a bile acid-activated membrane receptor (Kawamata et al., 2003; Maruyama et al., 2002). TGR5 activation stimulates adenylate cyclase, intracellular cAMP production and PKA activation. Among all bile acids, LCA and 3-keto-LCA are the most potent TGR5 agonists with an EC50 of less than 1 μM. DCA, CDCA and CA also activate TGR5 with an EC50 of ~1.0, 4.4 and 7.7 μM, respectively. Despite the liver being a major bile acid target organ, TGR5 is not expressed in hepatocytes. However, TGR5 is expressed in the liver sinusoidal endothelial cells (Keitel et al., 2007), gallbladder epithelial cells and Kupffer cells (Keitel et al., 2008). TGR5 is highly expressed in the ileum and colon (Kawamata et al., 2003) and in non-traditional bile acid target organs including white and brown adipose, spleen, kidney, pancreas, lung, macrophages and the central nervous system (Kawamata et al., 2003). Activation of TGR5 in adipose, muscle and intestine has been shown to regulate lipid, glucose and energy metabolism and thus improve metabolic homeostasis (Li and Chiang, 2014). TGR5 may be a potential therapeutic target for the treatment of diabetes and cardiovascular diseases. The metabolic regulation by TGR5 signaling will not be further discussed here.
How TGR5 regulates bile acid synthesis and metabolism under normal physiology is currently not very clear. However, it was reported that mice lacking TGR5 had reduced bile acid pool size (Maruyama et al., 2006), a more hydrophobic bile acid composition and showed more severe liver injury upon bile acid feeding or bile duct ligation (Pean et al., 2013). Studies have shown that pharmacological activation of TGR5 in macrophages may play an anti-inflammatory role in the immune system, which is supported by recent studies demonstrating a protective role of TGR5 activation in cholestasis and NASH (Kawamata et al., 2003; Keitel et al., 2008; McMahan et al., 2013; Pean et al., 2013). Activation of TGR5 reduced LPS-stimulated pro-inflammatory cytokine production (Keitel et al., 2008). Tgr5 knockout mice challenged with LPS had higher plasma liver enzymes and elevated cytokine expression, while the selective TGR5 agonist 23(S)-mCDCA antagonized LPS-induced cytokine expression in mouse liver (Wang et al., 2011b). In the vasculature, TGR5 activation by 6-EMCA or INT-777 attenuated atherosclerosis in mice. Importantly, it was shown that INT-777 did not attenuate atherosclerosis in mice transplanted with bone marrow of tgr5 knockout mice, proving the anti-inflammatory and anti-atherogenic role of macrophage TGR5. In the intestine where TGR5 is highly expressed, a TGR5 selective agonist protected the integrity of intestinal barrier function, immune response, and pro-inflammatory cytokine production in experimental colitis models (Cipriani et al., 2011; Yoneno et al., 2013). Pruritus is commonly associated with cholestasis and treatment with bile acid derivatives. A recent study suggests that TGR5 mediates bile acid-induced itch and analgesia (Alemi et al., 2013). Bile acids activate TGR5 on sensory nerves and stimulate the release of neuropeptides in the spinal cord that transmits itch and analgesia.
10.3. Role of bile acids in liver injury, regeneration and cancer
Patients with chronic and advanced stage cholestasis including PBC and PSC may be at higher risk of developing HCC and bile duct cancer (Eaton et al., 2013; Tomiyama et al., 2013). One of the unique characteristics of liver is its ability to regenerate after liver injury or surgical resection (Michalopoulos, 2013). Proper liver regeneration is an important determinant of final outcome after toxin or drug-induced liver injury (Mehendale, 2005). In cholestasis, both biliary epithelial cells and hepatocytes proliferate to compensate for liver cell death. On one hand, cholestasis is associated with reduced regenerating capability (Yokoyama et al., 2007); On the other hand, repeated injury and repair cycles during chronic cholestasis exacerbate liver fibrosis, inflammation, cirrhosis and cancer. During cholestasis, bile acids not only act as toxins to cause chronic inflammation and cell death, bile acids also activate cellular mitogenic signaling pathways to regulate cell proliferation (Fan et al., 2015). Studies indicate that bile acids are tumor promoters and involved in the pathogenesis of hepatocellular carcinoma (HCC) (Kim et al., 2007; Yang et al., 2007).
Liver can regenerate upon surgical resection. This is modeled using partial hepatectomy (PHX) procedures where about two-thirds of the liver was surgically removed in rodents to allow the study of the regeneration process. Studies in this model have obtains important information on the role and regulation of bile acids metabolism and signaling in the regulation of hepatocyte proliferation. In rats and mice, hepatic CYP7A1 enzyme activity and mRNA expression were significantly decreased immediately following PHX (Huang et al., 2006; Maeda et al., 2005; Nakano et al., 1995). This may reflect an increased bile acid influx and activation of the FXR signaling in the regenerating lobe. Overexpression of CYP7A1 in the liver impaired liver regeneration in mice, suggesting that repression of CYP7A1 and hepatic bile acid synthesis may be necessary for normal regeneration (Zhang et al., 2009). In addition, deletion of fxr gene in mice resulted in high mortality coupled with inhibited liver regeneration following PHX (Huang et al., 2006). Later studies also revealed a similar effect of FXR deletion in liver regeneration after CCl4 and APAP-induced liver injury (Meng et al., 2010). Global deletion of FXR resulted in significantly induced CYP7A1, which caused enlarged bile acid pool and higher plasma bile acid levels in mice. Indeed, later studies showed that hepatocyte-specific fxr knockout mice displayed only a moderate delay in liver regeneration after PHX without any liver injury or necrosis in the regenerating lobes (Borude et al., 2012; Zhang et al., 2012). Hepatocyte-specific fxr knockout mice have intact intestine bile acids/FXR/FGF15 axis that limits the CYP7A1 expression and the expansion of bile acid pool. In addition, bile acid-induced FGF15 was recently shown to act as a secondary or auxiliary mitogen for hepatocytes and may enhance promitogenic effects of primary mitogens such as hepatocyte growth factors (HGF) and epidermal growth factor (EGF) (Limaye et al., 2008). Both fgf15 knockout mice and fgfr4 knockout mice had impaired liver regeneration and increased mortality after PHX (Chen et al., 2014; Uriarte et al., 2013). Current evidence suggests that intestine bile acid/FXR/FGF15 signaling axis is required to promote normal liver regeneration, while hepatic bile acid overload in cholestasis impairs normal liver regeneration.
The initial studies on the role of bile acids and liver cancers, specifically hepatocellular carcinoma, performed between 1970s and early 2000s showed that plasma bile acid concentrations are higher in HCC patients as compared to normal healthy subjects (Changbumrung et al., 1990; El-Mir et al., 2001; Hirayama and Irisa, 1976). These studies indicated that bile acid homeostasis was disturbed during HCC and cholangiocarcinoma development and could be potentially used as a biomarker. The first causative link between higher bile acids and HCC pathogenesis came in mid 2000s when two groups independently demonstrated that the fxr knockout mice develop spontaneous liver tumors (Kim et al., 2007; Yang et al., 2007). Further studies showed that genetic deletion of shp gene in mice also led to spontaneous HCC (Anakk et al., 2011; Katzenellenbogen et al., 2007; Zhang et al., 2008b). Deletion of Mdr2 gene resulted in portal regurgitation of bile, extensive cholestasis resulting in a condition similar to progressive familial intrahepatic cholestasis (PFIC). The Mdr2−/− mice developed biliary fibrosis and HCC (Katzenellenbogen et al., 2006). Similar condition was observed in humans where chronic cholestasis led to the development of HCC in many cases (Eaton et al., 2013; Tomiyama et al., 2013). In cholestasis, high levels of bile acids can lead to the generation of reactive oxygen species, disruption of cell membrane, impairment of mitochondrial function and induction of DNA damage and mutation. Toxic bile acids-induced chronic inflammation and injury-repair response in the liver likely contributes to tumor promotion. In addition, bile acids, specifically the highly hydrophobic and highly toxic DCA, are known to activate cellular signaling pathways such as MAPK, STAT-3 and NF-κB to induce cytokines such as tumor necrosis factor α (TNFα), interleukin 6 (IL-6). It is also known that bile acids can activate pro-inflammatory molecules such as Egr1 via an EGFR-dependent, FXR-independent mechanism (Allen et al., 2011). More recent studies revealed that conjugated bile acids can activate the sphingosine 1-phosphate receptor 2 that activates intracellular ERK1/2 and AKT signaling to promote the invasive growth of cholangiocarcinoma, which is commonly associated with chronic cholestasis (Liu et al., 2014; Studer et al., 2012). In summary, current studies suggest that dysregulation of hepatic bile acid synthetic and metabolizing enzymes and bile acid-activated receptors in cholestatic liver injury have an impact on hepatocyte proliferation, regeneration and tumorigenesis.
Conclusion
Bile acids are not only physiological detergent molecules that facilitate hepatic excretion of endogenous metabolites, xenobiotic and drugs and intestine fat and nutrient absorption, but also act as signaling molecules that regulate various cellular processes involved in metabolism, immune response and cell growth. Bile acids are highly toxic and accumulation of bile acids in cholestasis leads to tissue inflammation and injury, and increases the risk of liver cancer. Cytochrome P450 enzymes are involved in bile acid synthesis and detoxification. Bile acids, like drugs, undergo phase II conjugation and Phase III excretion. Bile acid synthesis and transport are tightly regulated by bile acid and drug/xenobiotic sensing nuclear receptors to maintain bile acid homeostasis. These receptors regulate genes in phase I, phase II and phase II bile acid metabolism and play a critical role in the detoxification of bile acids. Knowledge on the mechanisms of the regulation of bile acid metabolism and signaling provide important molecular basis for the development of novel therapeutic approaches for the treatment of cholestasis and inflammation-related liver diseases.
Acknowledgments
The American Diabetes Association Junior Faculty Award (T.L.), NIH grant 1R01DK102487-01 (T.L), the National Center for Research Resources (5P20RR021940-07) and the National Institute of General Medical Sciences (8 P20 GM103549-07) of the National Institutes of Health (TL). NIH grant 1R01DK098414 (UA). The American Association for the Study of Liver Diseases/American Liver Foundation Liver Scholar Award (UA).
Abbreviations
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- CYP7A1
cholesterol 7α-hydroxylase
- CYP8B1
sterol 12α-hydroxylase
- CYP27A1
sterol 27-hydroxylase (CYP27A1)
- CYP7B1
oxysterol 7α-hydroxylase
- LCA
lithocholic acid
- MCA
muricholic acid
- HCA
hyocholic acid
- UDCA
ursodeoxycholic acid
- CCK
cholecystokinin
- NTCP
Na+-dependent taurocholate transporter
- mEH
microsomal epoxide hydrolase (mEH)
- OATP
organic anion transporter
- BSEP
bile salt export pump
- SPGP
the sister of P-glycoprotein
- PFIC
progressive familial intrahepatic cholestasis
- MRP
multidrug resistance-associated protein
- ABC
ATP-binding cassette transporter
- OST
organic solute transporter
- ASBT
apical sodium-dependent bile salt transporter
- I-BABP
intestinal bile acid binding protein
- ICP
intrahepatic cholestasis of pregnancy
- PBC
primary biliary cirrhosis
- PSC
primary sclerosing cholangitis
- NR
Nuclear receptors
- RXR
receptor retinoid X receptor
- HNF4α
hepatocyte nuclear factor 4α
- LRH-1
liver receptor homolog-1
- FXR
Farnesoid X receptor (FXR)
- PXR
Pregnane X receptor
- VDR
vitamin D receptor
- SHP
small heterodimer partner
- FGF15
fibroblast growth factor 15
- FGFR4
FGF receptor 4
- FRS2
FGF receptor substrate 2
- RAR
retinoic acid receptor
- norUDCA
Nor-ursodeoxycholic acid
- OCA
obeticholic acid
- LPS
lipopolysaccharide
- VSMC
vascular smooth muscle cells
- NF-κB
nuclear factor κB
- COX-2
cyclooxygenase 2
- iNOS
inducible nitric oxide synthase
- PHX
partial hepatectomy
- APAP
acetaminophen
- HCC
hepatocellular carcinoma
- HGF
hepatocyte growth factors
- EGF
epidermal growth factor
- TNFα
tumor necrosis factor α
- IL-6
interleukin 6
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Tiangang Li, Email: tli@kumc.edu.
Udayan Apte, Email: uapte@kumc.edu.
References
- Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World journal of surgical oncology. 2014;12:164. doi: 10.1186/1477-7819-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemi F, Kwon E, Poole DP, Lieu T, Lyo V, Cattaruzza F, Cevikbas F, Steinhoff M, Nassini R, Materazzi S, Guerrero-Alba R, Valdez-Morales E, Cottrell GS, Schoonjans K, Geppetti P, Vanner SJ, Bunnett NW, Corvera CU. The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest. 2013;123:1513–1530. doi: 10.1172/JCI64551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AH, Carey EJ, Lindor KD. Recent advances in the development of farnesoid X receptor agonists. Annals of translational medicine. 2015;3:5. doi: 10.3978/j.issn.2305-5839.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175–186. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anakk S, Watanabe M, Ochsner SA, McKenna NJ, Finegold MJ, Moore DD. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J Clin Invest. 2011;121:86–95. doi: 10.1172/JCI42846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan M, von Dippe P, Levy D. Identification of the hepatocyte Na+-dependent bile acid transport protein using monoclonal antibodies. J Biol Chem. 1988;263:8338–8343. [PubMed] [Google Scholar]
- Araya Z, Wikvall K. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by CYP3A4 in human liver microsomes. Biochim Biophys Acta. 1999;1438:47–54. doi: 10.1016/s1388-1981(99)00031-1. [DOI] [PubMed] [Google Scholar]
- Axelson M, Mork B, Sjovall J. Occurrence of 3 beta-hydroxy-5-cholestenoic acid, 3 beta,7 alpha-dihydroxy-5-cholestenoic acid, and 7 alpha-hydroxy-3-oxo-4-cholestenoic acid as normal constituents in human blood. J Lipid Res. 1988;29:629–641. [PubMed] [Google Scholar]
- Baghdasaryan A, Claudel T, Gumhold J, Silbert D, Adorini L, Roda A, Vecchiotti S, Gonzalez FJ, Schoonjans K, Strazzabosco M, Fickert P, Trauner M. Dual farnesoid X receptor/TGR5 agonist INT-767 reduces liver injury in the Mdr2−/− (Abcb4−/−) mouse cholangiopathy model by promoting biliary HCO(−)(3) output. Hepatology. 2011;54:1303–1312. doi: 10.1002/hep.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. OSTalpha-OSTbeta: a major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology. 2005;42:1270–1279. doi: 10.1002/hep.20961. [DOI] [PubMed] [Google Scholar]
- Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003;124:1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- Beilke LD, Aleksunes LM, Holland RD, Besselsen DG, Beger RD, Klaassen CD, Cherrington NJ. Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metab Dispos. 2009;37:1035–1045. doi: 10.1124/dmd.108.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab. 2003;14:473–479. doi: 10.1016/j.tem.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated PXR interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha : Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004 doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- Bishop-Bailey D, Walsh DT, Warner TD. Expression and activation of the farnesoid X receptor in the vasculature. Proc Natl Acad Sci U S A. 2004;101:3668–3673. doi: 10.1073/pnas.0400046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkhem I, Einarsson K, Melone P, Hylemon P. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: evidence for a 3-oxo-delta 4-steroid intermediate. J Lipid Res. 1989;30:1033–1039. [PubMed] [Google Scholar]
- Bodin K, Lindbom U, Diczfalusy U. Novel pathways of bile acid metabolism involving CYP3A4. Biochim Biophys Acta. 2005;1687:84–93. doi: 10.1016/j.bbalip.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Borude P, Edwards G, Walesky C, Li F, Ma X, Kong B, Guo GL, Apte U. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology. 2012;56:2344–2352. doi: 10.1002/hep.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JL. Bile formation and secretion. Comprehensive Physiology. 2013;3:1035–1078. doi: 10.1002/cphy.c120027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, Zollner G, Lee JY, Ballatori N. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1124–1130. doi: 10.1152/ajpgi.00539.2005. [DOI] [PubMed] [Google Scholar]
- Changbumrung S, Tungtrongchitr R, Migasena P, Chamroenngan S. Serum unconjugated primary and secondary bile acids in patients with cholangiocarcinoma and hepatocellular carcinoma. Journal of the Medical Association of Thailand = Chotmaihet thangphaet. 1990;73:81–90. [PubMed] [Google Scholar]
- Chatterjee B, Echchgadda I, Song CS. Vitamin D receptor regulation of the steroid/bile acid sulfotransferase SULT2A1. Methods in enzymology. 2005;400:165–191. doi: 10.1016/S0076-6879(05)00010-8. [DOI] [PubMed] [Google Scholar]
- Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Breslow J, Ananthanarayanan M, Shneider BL. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem. 2003;278:19909–19916. doi: 10.1074/jbc.M207903200. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhao KN, Chen C. The role of CYP3A4 in the biotransformation of bile acids and therapeutic implication for cholestasis. Annals of translational medicine. 2014;2:7. doi: 10.3978/j.issn.2305-5839.2013.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chiang JY. Regulation of human sterol 27-hydroxylase gene (CYP27A1) by bile acids and hepatocyte nuclear factor 4alpha (HNF4alpha) Gene. 2003;313:71–82. doi: 10.1016/s0378-1119(03)00631-0. [DOI] [PubMed] [Google Scholar]
- Chen X, Chen F, Liu S, Glaeser H, Dawson PA, Hofmann AF, Kim RB, Shneider BL, Pang KS. Transactivation of rat apical sodium-dependent bile acid transporter and increased bile acid transport by 1alpha,25-dihydroxyvitamin D3 via the vitamin D receptor. Mol Pharmacol. 2006;69:1913–1923. doi: 10.1124/mol.105.020792. [DOI] [PubMed] [Google Scholar]
- Chiang JY. Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003;284:G349–356. doi: 10.1152/ajpgi.00417.2002. [DOI] [PubMed] [Google Scholar]
- Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs S, Yeh RL, Georges E, Ling V. Identification of a sister gene to P-glycoprotein. Cancer Res. 1995;55:2029–2034. [PubMed] [Google Scholar]
- Chow EC, Magomedova L, Quach HP, Patel RH, Durk MR, Fan J, Maeng HJ, Irondi K, Anakk S, Moore DD, Cummins CL, Pang KS. Vitamin D Receptor Activation Down-regulates Small Heterodimer Partner and Increases CYP7A1 to Lower Cholesterol. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Cipriani S, Mencarelli A, Chini MG, Distrutti E, Renga B, Bifulco G, Baldelli F, Donini A, Fiorucci S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Aleksunes LM, Tanaka Y, Goedken MJ, Klaassen CD. Compensatory induction of liver efflux transporters in response to ANIT-induced liver injury is impaired in FXR-null mice. Toxicological sciences : an official journal of the Society of Toxicology. 2009;110:47–60. doi: 10.1093/toxsci/kfp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J Biol Chem. 2002;277:25125–25132. doi: 10.1074/jbc.M201323200. [DOI] [PubMed] [Google Scholar]
- Dyson JK, Hirschfield GM, Adams DH, Beuers U, Mann DA, Lindor KD, Jones DE. Novel therapeutic targets in primary biliary cirrhosis. Nature reviews Gastroenterology & hepatology. 2015 doi: 10.1038/nrgastro.2015.12. [DOI] [PubMed] [Google Scholar]
- Eaton JE, Talwalkar JA, Lazaridis KN, Gores GJ, Lindor KD. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology. 2013;145:521–536. doi: 10.1053/j.gastro.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mir MY, Badia MD, Luengo N, Monte MJ, Marin JJ. Increased levels of typically fetal bile acid species in patients with hepatocellular carcinoma. Clinical science. 2001;100:499–508. [PubMed] [Google Scholar]
- Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, Nguyen L, Bollineni JS, Shefer S, Hyogo H, Cohen DE, Shneider B, Sehayek E, Ananthanarayanan M, Balasubramaniyan N, Suchy FJ, Batta AK, Salen G. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J Lipid Res. 2003;44:1001–1009. doi: 10.1194/jlr.M200489-JLR200. [DOI] [PubMed] [Google Scholar]
- Fan M, Wang X, Xu G, Yan Q, Huang W. Bile acid signaling and liver regeneration. Biochim Biophys Acta. 2015;1849:196–200. doi: 10.1016/j.bbagrm.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Clerici C, Antonelli E, Orlandi S, Goodwin B, Sadeghpour BM, Sabatino G, Russo G, Castellani D, Willson TM, Pruzanski M, Pellicciari R, Morelli A. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor ligand, in estrogen-induced cholestasis. J Pharmacol Exp Ther. 2005;313:604–612. doi: 10.1124/jpet.104.079665. [DOI] [PubMed] [Google Scholar]
- Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA. Regulation of the mouse organic solute transporter alpha-beta, Ostalpha-Ostbeta, by bile acids. Am J Physiol Gastrointest Liver Physiol. 2006;290:G912–922. doi: 10.1152/ajpgi.00479.2005. [DOI] [PubMed] [Google Scholar]
- Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chemico-biological interactions. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, Penna G, Laverny G, Adorini L, Moschetta A, van Mil SW. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, Mackenzie PI, Radominska-Pandya A. Differential glucuronidation of bile acids, androgens and estrogens by human UGT1A3 and 2B7. J Steroid Biochem Mol Biol. 1999;70:101–108. doi: 10.1016/s0960-0760(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ, Yu X, White KE, Inagaki T, Kliewer SA, Yamamoto M, Kurosu H, Ogawa Y, Kuro-o M, Lanske B, Razzaque MS, Mohammadi M. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol. 2007;27:3417–3428. doi: 10.1128/MCB.02249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YZ, Everett ET, Schwartz DA, Norris JS, Wilson FA. Molecular cloning, tissue distribution, and expression of a 14-kDa bile acid-binding protein from rat ileal cytosol. Proc Natl Acad Sci U S A. 1994;91:4741–4745. doi: 10.1073/pnas.91.11.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest. 1994;93:1326–1331. doi: 10.1172/JCI117091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuch B, Stieger B, Foguet M, Lubbert H, Meier PJ. Functional expression cloning and characterization of the hepatocyte Na+/bile acid cotransport system. Proc Natl Acad Sci U S A. 1991;88:10629–10633. doi: 10.1073/pnas.88.23.10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halilbasic E, Fiorotto R, Fickert P, Marschall HU, Moustafa T, Spirli C, Fuchsbichler A, Gumhold J, Silbert D, Zatloukal K, Langner C, Maitra U, Denk H, Hofmann AF, Strazzabosco M, Trauner M. Side chain structure determines unique physiologic and therapeutic properties of norursodeoxycholic acid in Mdr2−/− mice. Hepatology. 2009;49:1972–1981. doi: 10.1002/hep.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Li T, Ellis E, Strom S, Chiang JY. A novel bile acid-activated vitamin D receptor signaling in human hepatocytes. Mol Endocrinol. 2010;24:1151–1164. doi: 10.1210/me.2009-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanniman EA, Lambert G, McCarthy TC, Sinal CJ. Loss of functional farnesoid X receptor increases atherosclerotic lesions in apolipoprotein E-deficient mice. J Lipid Res. 2005;46:2595–2604. doi: 10.1194/jlr.M500390-JLR200. [DOI] [PubMed] [Google Scholar]
- Hirayama C, Irisa T. Serum cholesterol and bile acid in primary hepatoma. Clinica chimica acta; international journal of clinical chemistry. 1976;71:21–25. doi: 10.1016/0009-8981(76)90270-9. [DOI] [PubMed] [Google Scholar]
- Hirschfield GM, Mason A, Luketic V, Lindor K, Gordon SC, Mayo M, Kowdley KV, Vincent C, Bodhenheimer HC, Jr, Pares A, Trauner M, Marschall HU, Adorini L, Sciacca C, Beecher-Jones T, Castelloe E, Bohm O, Shapiro D. Efficacy of obeticholic Acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic Acid. Gastroenterology. 2015;148:751–761. e758. doi: 10.1053/j.gastro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Hofman MK, Princen HM, Zwinderman AH, Jukema JW. Genetic variation in the rate-limiting enzyme in cholesterol catabolism (cholesterol 7alpha-hydroxylase) influences the progression of atherosclerosis and risk of new clinical events. Clinical science. 2005;108:539–545. doi: 10.1042/CS20040339. [DOI] [PubMed] [Google Scholar]
- Hofmann AF. Rifampicin and treatment of cholestatic pruritus. Gut. 2002;51:756–757. doi: 10.1136/gut.51.5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- Hussaini SH, Pereira SP, Murphy GM, Dowling RH. Deoxycholic acid influences cholesterol solubilization and microcrystal nucleation time in gallbladder bile. Hepatology. 1995;22:1735–1744. [PubMed] [Google Scholar]
- Hylemon PB, Melone PD, Franklund CV, Lund E, Bjorkhem I. Mechanism of intestinal 7 alpha-dehydroxylation of cholic acid: evidence that allo-deoxycholic acid is an inducible side-product. J Lipid Res. 1991;32:89–96. [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW. Disruption of cholesterol 7alpha-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. J Biol Chem. 1996;271:18017–18023. doi: 10.1074/jbc.271.30.18017. [DOI] [PubMed] [Google Scholar]
- Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZY, Han TQ, Suo GJ, Feng DX, Chen S, Cai XX, Jiang ZH, Shang J, Zhang Y, Jiang Y, Zhang SD. Polymorphisms at cholesterol 7alpha-hydroxylase, apolipoproteins B and E and low density lipoprotein receptor genes in patients with gallbladder stone disease. World J Gastroenterol. 2004;10:1508–1512. doi: 10.3748/wjg.v10.i10.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. Interactions between common genetic polymorphisms in ABCG5/G8 and CYP7A1 on LDL cholesterol-lowering response to atorvastatin. Atherosclerosis. 2004;175:287–293. doi: 10.1016/j.atherosclerosis.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–2915. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen M, Mizrahi L, Pappo O, Klopstock N, Olam D, Jacob-Hirsch J, Amariglio N, Rechavi G, Domany E, Galun E, Goldenberg D. Molecular mechanisms of liver carcinogenesis in the mdr2-knockout mice. Molecular cancer research : MCR. 2007;5:1159–1170. doi: 10.1158/1541-7786.MCR-07-0172. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen M, Pappo O, Barash H, Klopstock N, Mizrahi L, Olam D, Jacob-Hirsch J, Amariglio N, Rechavi G, Mitchell LA, Kohen R, Domany E, Galun E, Goldenberg D. Multiple adaptive mechanisms to chronic liver disease revealed at early stages of liver carcinogenesis in the Mdr2-knockout mice. Cancer Res. 2006;66:4001–4010. doi: 10.1158/0008-5472.CAN-05-2937. [DOI] [PubMed] [Google Scholar]
- Kauffman FC. Sulfonation in pharmacology and toxicology. Drug Metab Rev. 2004;36:823–843. doi: 10.1081/dmr-200033496. [DOI] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- Keitel V, Nies AT, Brom M, Hummel-Eisenbeiss J, Spring H, Keppler D. A common Dubin-Johnson syndrome mutation impairs protein maturation and transport activity of MRP2 (ABCC2) Am J Physiol Gastrointest Liver Physiol. 2003;284:G165–174. doi: 10.1152/ajpgi.00362.2002. [DOI] [PubMed] [Google Scholar]
- Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, Haussinger D, Kubitz R. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem. 2003;278:17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev. 2002;23:687–702. doi: 10.1210/er.2001-0038. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res. 2002;43:359–364. [PubMed] [Google Scholar]
- Kullak-Ublick GA, Stieger B, Hagenbuch B, Meier PJ. Hepatic transport of bile salts. Semin Liver Dis. 2000;20:273–292. doi: 10.1055/s-2000-9426. [DOI] [PubMed] [Google Scholar]
- Lang C, Meier Y, Stieger B, Beuers U, Lang T, Kerb R, Kullak-Ublick GA, Meier PJ, Pauli-Magnus C. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenetics and genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- Leitersdorf E, Safadi R, Meiner V, Reshef A, Bjorkhem I, Friedlander Y, Morkos S, Berginer VM. Cerebrotendinous xanthomatosis in the Israeli Druze: molecular genetics and phenotypic characteristics. Am J Hum Genet. 1994;55:907–915. [PMC free article] [PubMed] [Google Scholar]
- Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, Bjorkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 2002;110:1191–1200. doi: 10.1172/JCI16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hsu DD, Li B, Luo X, Alderson N, Qiao L, Ma L, Zhu HH, He Z, Suino-Powell K, Ji K, Li J, Shao J, Xu HE, Li T, Feng GS. Cytoplasmic tyrosine phosphatase Shp2 coordinates hepatic regulation of bile acid and FGF15/19 signaling to repress bile acid synthesis. Cell Metab. 2014;20:320–332. doi: 10.1016/j.cmet.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- Li T, Chiang JY. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, Chiang JY. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Owsley E, Matozel M, Hsu P, Novak CM, Chiang JY. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye PB, Bowen WC, Orr AV, Luo J, Tseng GC, Michalopoulos GK. Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium. Hepatology. 2008;47:1702–1713. doi: 10.1002/hep.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem. 2007;282:27277–27284. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- Lioudaki E, Ganotakis ES, Mikhailidis DP. Lipid lowering drugs and gallstones: a therapeutic option? Curr Pharm Des. 2011;17:3622–3631. doi: 10.2174/138161211798220909. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhao R, Zhou X, Liang X, Campbell DJ, Zhang X, Zhang L, Shi R, Wang G, Pandak WM, Sirica AE, Hylemon PB, Zhou H. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology. 2014;60:908–918. doi: 10.1002/hep.27085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Binz J, Numerick MJ, Dennis S, Luo G, Desai B, MacKenzie KI, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Hepatoprotection by the farnesoid X receptor agonist GW4064 in rat models of intra- and extrahepatic cholestasis. J Clin Invest. 2003;112:1678–1687. doi: 10.1172/JCI18945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Beer TS, Nutter S. Colonic bacterial activity, biliary cholesterol saturation, and pathogenesis of gallstones. Lancet. 1978;2:1063–1065. doi: 10.1016/s0140-6736(78)91800-7. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Lu Y, Heydel JM, Li X, Bratton S, Lindblom T, Radominska-Pandya A. Lithocholic acid decreases expression of UGT2B7 in Caco-2 cells: a potential role for a negative farnesoid X receptor response element. Drug Metab Dispos. 2005;33:937–946. doi: 10.1124/dmd.104.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Nagatomo H, Kuroki N, Nagatomo J, Uchiyama F, Shimozono K, Kohno Y, Chijiiwa K. Bile acid biosynthesis during liver regeneration: enzyme activities of cholesterol 7alpha-hydroxylase and 3beta-hydroxy-delta5-C27-steroid dehydrogenase in rats. Annals of clinical and laboratory science. 2005;35:323–328. [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SN, Heaton KW. Deoxycholic acid and the pathogenesis of gall stones. Gut. 1988;29:522–533. doi: 10.1136/gut.29.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197–205. doi: 10.1677/joe.1.06546. [DOI] [PubMed] [Google Scholar]
- Mason JI, Boyd GS. The suppressive effect of the catatoxic steroid, pregnenolone-16alpha-carbonitrile, on liver microsomal cholesterol-7alpha-hydroxlyase. Steroids. 1978;31:849–854. doi: 10.1016/s0039-128x(78)80048-8. [DOI] [PubMed] [Google Scholar]
- McCarthy TC, Li X, Sinal CJ. Vitamin D receptor-dependent regulation of colon multidrug resistance-associated protein 3 gene expression by bile acids. J Biol Chem. 2005;280:23232–23242. doi: 10.1074/jbc.M411520200. [DOI] [PubMed] [Google Scholar]
- McMahan RH, Wang XX, Cheng LL, Krisko T, Smith M, El Kasmi K, Pruzanski M, Adorini L, Golden-Mason L, Levi M, Rosen HR. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J Biol Chem. 2013;288:11761–11770. doi: 10.1074/jbc.M112.446575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehendale HM. Tissue repair: an important determinant of final outcome of toxicant-induced injury. Toxicol Pathol. 2005;33:41–51. doi: 10.1080/01926230590881808. [DOI] [PubMed] [Google Scholar]
- Meier PJ. Molecular mechanisms of hepatic bile salt transport from sinusoidal blood into bile. Am J Physiol. 1995;269:G801–812. doi: 10.1152/ajpgi.1995.269.6.G801. [DOI] [PubMed] [Google Scholar]
- Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002;64:635–661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- Mencarelli A, Renga B, Distrutti E, Fiorucci S. Antiatherosclerotic effect of farnesoid X receptor. American journal of physiology Heart and circulatory physiology. 2009;296:H272–281. doi: 10.1152/ajpheart.01075.2008. [DOI] [PubMed] [Google Scholar]
- Meng Z, Wang Y, Wang L, Jin W, Liu N, Pan H, Liu L, Wagman L, Forman BM, Huang W. FXR regulates liver repair after CCl4-induced toxic injury. Mol Endocrinol. 2010;24:886–897. doi: 10.1210/me.2009-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Comprehensive Physiology. 2013;3:485–513. doi: 10.1002/cphy.c120014. [DOI] [PubMed] [Google Scholar]
- Miyake JH, Doung XD, Strauss W, Moore GL, Castellani LW, Curtiss LK, Taylor JM, Davis RA. Increased production of apolipoprotein B-containing lipoproteins in the absence of hyperlipidemia in transgenic mice expressing cholesterol 7alpha-hydroxylase. J Biol Chem. 2001;276:23304–23311. doi: 10.1074/jbc.M101853200. [DOI] [PubMed] [Google Scholar]
- Miyake JH, Duong-Polk XT, Taylor JM, Du EZ, Castellani LW, Lusis AJ, Davis RA. Transgenic expression of cholesterol-7-alpha-hydroxylase prevents atherosclerosis in C57BL/6J mice. Arterioscler Thromb Vasc Biol. 2002;22:121–126. doi: 10.1161/hq0102.102588. [DOI] [PubMed] [Google Scholar]
- Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J Biol Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- Myant NB, Mitropoulos KA. Cholesterol 7 alpha-hydroxylase. J Lipid Res. 1977;18:135–153. [PubMed] [Google Scholar]
- Nagpal S, Na S, Rathnachalam R. Noncalcemic actions of vitamin D receptor ligands. Endocr Rev. 2005;26:662–687. doi: 10.1210/er.2004-0002. [DOI] [PubMed] [Google Scholar]
- Nakano K, Chijiiwa K, Okamoto S, Yamashita H, Kuroki S, Tanaka M. Hepatic cholesterol 7 alpha-hydroxylase activity and serum 7 alpha-hydroxy-cholesterol level during liver regeneration after partial hepatectomy in rats. European surgical research Europaische chirurgische Forschung Recherches chirurgicales europeennes. 1995;27:389–395. doi: 10.1159/000129425. [DOI] [PubMed] [Google Scholar]
- Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–156. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E, Network NCR. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida S, Ozeki J, Makishima M. Modulation of bile acid metabolism by 1alpha-hydroxyvitamin D3 administration in mice. Drug Metab Dispos. 2009;37:2037–2044. doi: 10.1124/dmd.109.027334. [DOI] [PubMed] [Google Scholar]
- Noe J, Kullak-Ublick GA, Jochum W, Stieger B, Kerb R, Haberl M, Mullhaupt B, Meier PJ, Pauli-Magnus C. Impaired expression and function of the bile salt export pump due to three novel ABCB11 mutations in intrahepatic cholestasis. J Hepatol. 2005;43:536–543. doi: 10.1016/j.jhep.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Ogura M, Nishida S, Ishizawa M, Sakurai K, Shimizu M, Matsuo S, Amano S, Uno S, Makishima M. Vitamin D3 modulates the expression of bile acid regulatory genes and represses inflammation in bile duct-ligated mice. J Pharmacol Exp Ther. 2009;328:564–570. doi: 10.1124/jpet.108.145987. [DOI] [PubMed] [Google Scholar]
- Otsuki M. Pathophysiological role of cholecystokinin in humans. Journal of gastroenterology and hepatology. 2000;15(Suppl):D71–83. doi: 10.1046/j.1440-1746.2000.02178.x. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pauli-Magnus C, Lang T, Meier Y, Zodan-Marin T, Jung D, Breymann C, Zimmermann R, Kenngott S, Beuers U, Reichel C, Kerb R, Penger A, Meier PJ, Kullak-Ublick GA. Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics. 2004;14:91–102. doi: 10.1097/00008571-200402000-00003. [DOI] [PubMed] [Google Scholar]
- Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, Schoonjans K, Rainteau D, Tordjmann T. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58:1451–1460. doi: 10.1002/hep.26463. [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Costantino G, Camaioni E, Sadeghpour BM, Entrena A, Willson TM, Fiorucci S, Clerici C, Gioiello A. Bile acid derivatives as ligands of the farnesoid X receptor. Synthesis, evaluation, and structure-activity relationship of a series of body and side chain modified analogues of chenodeoxycholic acid. J Med Chem. 2004;47:4559–4569. doi: 10.1021/jm049904b. [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45:3569–3572. doi: 10.1021/jm025529g. [DOI] [PubMed] [Google Scholar]
- Pillot T, Ouzzine M, Fournel-Gigleux S, Lafaurie C, Radominska A, Burchell B, Siest G, Magdalou J. Glucuronidation of hyodeoxycholic acid in human liver. Evidence for a selective role of UDP-glucuronosyltransferase 2B4. J Biol Chem. 1993;268:25636–25642. [PubMed] [Google Scholar]
- Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, Verhagen A, Rivera CR, Mulvihill SJ, Malloy MJ, Kane JP. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110:109–117. doi: 10.1172/JCI15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci U S A. 2008;105:3891–3896. doi: 10.1073/pnas.0712328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Hylemon PB. A potential role for resistant starch fermentation in modulating colonic bacterial metabolism and colon cancer risk. Cancer biology & therapy. 2006;5:273–274. doi: 10.4161/cbt.5.3.2728. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Bjorkhem I, Leitersdorf E. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- Russell DW. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu Rev Biochem. 2003;72:1370174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Russell DW, Setchell KD. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49:1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem. 2010;285:14486–14494. doi: 10.1074/jbc.M110.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos. 2001;29:1446–1453. [PubMed] [Google Scholar]
- Shneider BL, Dawson PA, Christie DM, Hardikar W, Wong MH, Suchy FJ. Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J Clin Invest. 1995;95:745–754. doi: 10.1172/JCI117722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatis K, Gafvels M, Kannisto K, Ovchinnikova O, Paulsson-Berne G, Parini P, Jiang ZY, Eggertsen G. Abolished synthesis of cholic acid reduces atherosclerotic development in apolipoprotein E knockout mice. J Lipid Res. 2010;51:3289–3298. doi: 10.1194/jlr.M009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijepcevic D, Kaufman C, Wichers CG, Gilglioni EH, Lempp FA, Duijst S, de Waart DR, Oude Elferink RP, Mier W, Stieger B, Beuers U, Urban S, van de Graaf SF. Impaired uptake of conjugated bile acids and Hepatitis B Virus preS1-binding in Na -taurocholate cotransporting polypeptide knockout mice. Hepatology. 2015 doi: 10.1002/hep.27694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, Mol CA, Ottenhoff R, van der Lugt NM, van Roon MA, et al. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell. 1993;75:451–462. doi: 10.1016/0092-8674(93)90380-9. [DOI] [PubMed] [Google Scholar]
- Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. Progressive familial intrahepatic cholestasis. Journal of clinical and experimental hepatology. 2014;4:25–36. doi: 10.1016/j.jceh.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg D. Effects of pregnenolone-16 alpha-carbonitrile on the metabolism of cholesterol in rat liver microsomes. Lipids. 1995;30:361–364. doi: 10.1007/BF02536046. [DOI] [PubMed] [Google Scholar]
- Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. PXR and CAR: nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab Rev. 2006;38:515–597. doi: 10.1080/03602530600786232. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman CA, Liddle C, Coulter SA, Sonoda J, Alvarez JG, Moore DD, Evans RM, Downes M. Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci U S A. 2005;102:2063–2068. doi: 10.1073/pnas.0409794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strautnieks SS, Kagalwalla AF, Tanner MS, Knisely AS, Bull L, Freimer N, Kocoshis SA, Gardiner RM, Thompson RJ. Identification of a locus for progressive familial intrahepatic cholestasis PFIC2 on chromosome 2q24. Am J Hum Genet. 1997;61:630–633. doi: 10.1086/515501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, Dent P, Spiegel S, Shi R, Xu W, Liu X, Bohdan P, Zhang L, Zhou H, Hylemon PB. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takikawa H, Otsuka H, Beppu T, Seyama Y, Yamakawa T. Serum concentrations of bile acid glucuronides in hepatobiliary diseases. Digestion. 1983;27:189–195. doi: 10.1159/000198952. [DOI] [PubMed] [Google Scholar]
- Teixeira J, Gil G. Cloning, expression, and regulation of lithocholic acid 6 beta-hydroxylase. J Biol Chem. 1991;266:21030–21036. [PubMed] [Google Scholar]
- Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol Pharmacol. 2001;60:1399–1406. doi: 10.1124/mol.60.6.1399. [DOI] [PubMed] [Google Scholar]
- Tomiyama Y, Takenaka K, Kodama T, Kawanaka M, Sasaki K, Nishina S, Yoshioka N, Hara Y, Hino K. Risk factors for survival and the development of hepatocellular carcinoma in patients with primary biliary cirrhosis. Intern Med. 2013;52:1553–1559. doi: 10.2169/internalmedicine.52.0010. [DOI] [PubMed] [Google Scholar]
- Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- Trottier J, Verreault M, Grepper S, Monte D, Belanger J, Kaeding J, Caron P, Inaba TT, Barbier O. Human UDP-glucuronosyltransferase (UGT)1A3 enzyme conjugates chenodeoxycholic acid in the liver. Hepatology. 2006;44:1158–1170. doi: 10.1002/hep.21362. [DOI] [PubMed] [Google Scholar]
- Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HC, Carotti S, Vespasiani-Gentilucci U, Morini S, Vicente E, Concepcion AR, Medina JF, Marin JJ, Berasain C, Prieto J, Avila MA. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. [DOI] [PubMed] [Google Scholar]
- Vacca M, Degirolamo C, Massafra V, Polimeno L, Mariani-Costantini R, Palasciano G, Moschetta A. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol Cell Endocrinol. 2013;368:108–119. doi: 10.1016/j.mce.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. Journal of immunology. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- Vaz FM, Paulusma CC, Huidekoper H, de Ru M, Lim C, Koster J, Ho-Mok K, Bootsma AH, Groen AK, Schaap FG, Oude Elferink RP, Waterham HR, Wanders RJ. Sodium taurocholate cotransporting polypeptide (SLC10A1) deficiency: Conjugated hypercholanemia without a clear clinical phenotype. Hepatology. 2014 doi: 10.1002/hep.27240. [DOI] [PubMed] [Google Scholar]
- Von Dippe P, Amoui M, Alves C, Levy D. Na(+)-dependent bile acid transport by hepatocytes is mediated by a protein similar to microsomal epoxide hydrolase. Am J Physiol. 1993;264:G528–534. doi: 10.1152/ajpgi.1993.264.3.G528. [DOI] [PubMed] [Google Scholar]
- von Dippe P, Amoui M, Stellwagen RH, Levy D. The functional expression of sodium-dependent bile acid transport in Madin-Darby canine kidney cells transfected with the cDNA for microsomal epoxide hydrolase. J Biol Chem. 1996;271:18176–18180. doi: 10.1074/jbc.271.30.18176. [DOI] [PubMed] [Google Scholar]
- Wang C, Yang C, Chang JY, You P, Li Y, Jin C, Luo Y, Li X, McKeehan WL, Wang F. Hepatocyte FRS2alpha is Essential for the Endocrine Fibroblast Growth Factor to Limit the Amplitude of Bile Acid Production Induced by Prandial Activity. Curr Mol Med. 2014;14:703–711. doi: 10.2174/1566524014666140724095112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Venkatesh M, Li H, Goetz R, Mukherjee S, Biswas A, Zhu L, Kaubisch A, Wang L, Pullman J, Whitney K, Kuro-o M, Roig AI, Shay JW, Mohammadi M, Mani S. Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J Clin Invest. 2011a;121:3220–3232. doi: 10.1172/JCI41514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-Protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011b;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB. Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11:3–14. [PubMed] [Google Scholar]
- Wistuba W, Gnewuch C, Liebisch G, Schmitz G, Langmann T. Lithocholic acid induction of the FGF19 promoter in intestinal cells is mediated by PXR. World J Gastroenterol. 2007;13:4230–4235. doi: 10.3748/wjg.v13.i31.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Francis H, Glaser S, Alpini G, LeSage G. Bile acid interactions with cholangiocytes. World J Gastroenterol. 2006;12:3553–3563. doi: 10.3748/wjg.v12.i22.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- Yeh HZ, Schteingart CD, Hagey LR, Ton-Nu HT, Bolder U, Gavrilkina MA, Steinbach JH, Hofmann AF. Effect of side chain length on biotransformation, hepatic transport, and choleretic properties of chenodeoxycholyl homologues in the rodent: studies with dinorchenodeoxycholic acid, norchenodeoxycholic acid, and chenodeoxycholic acid. Hepatology. 1997;26:374–385. doi: 10.1002/hep.510260218. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Nagino M, Nimura Y. Mechanism of impaired hepatic regeneration in cholestatic liver. Journal of hepato-biliary-pancreatic surgery. 2007;14:159–166. doi: 10.1007/s00534-006-1125-1. [DOI] [PubMed] [Google Scholar]
- Yoneno K, Hisamatsu T, Shimamura K, Kamada N, Ichikawa R, Kitazume MT, Mori M, Uo M, Namikawa Y, Matsuoka K, Sato T, Koganei K, Sugita A, Kanai T, Hibi T. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology. 2013;139:19–29. doi: 10.1111/imm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YB, Hagey LR, Hofmann AF, Gurantz D, Michelotti EL, Steinbach JH. Effect of side-chain shortening on the physiologic properties of bile acids: hepatic transport and effect on biliary secretion of 23-nor-ursodeoxycholate in rodents. Gastroenterology. 1986;90:837–852. doi: 10.1016/0016-5085(86)90859-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang X, Meng Z, Dong B, Shiah S, Moore DD, Huang W. Significance and mechanism of CYP7a1 gene regulation during the acute phase of liver regeneration. Mol Endocrinol. 2009;23:137–145. doi: 10.1210/me.2008-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang YD, Chen WD, Wang X, Lou G, Liu N, Lin M, Forman BM, Huang W. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine. Hepatology. 2012 doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12a-hydroxylase gene (CYP8B1): Roles of hepatocyte nuclear factor 4a (HNF4a) in mediating bile acid repression. J Biol Chem. 2001;276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- Zhang Q, He F, Kuruba R, Gao X, Wilson A, Li J, Billiar TR, Pitt BR, Xie W, Li S. FXR-mediated regulation of angiotensin type 2 receptor expression in vascular smooth muscle cells. Cardiovascular research. 2008a;77:560–569. doi: 10.1093/cvr/cvm068. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008b;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QS, Xing W, Qian B, von Dippe P, Shneider BL, Fox VL, Levy D. Inhibition of human m-epoxide hydrolase gene expression in a case of hypercholanemia. Biochim Biophys Acta. 2003;1638:208–216. doi: 10.1016/s0925-4439(03)00085-1. [DOI] [PubMed] [Google Scholar]
- Zollner G, Trauner M. Mechanisms of cholestasis. Clin Liver Dis. 2008;12:1–26. vii. doi: 10.1016/j.cld.2007.11.010. [DOI] [PubMed] [Google Scholar]