Abstract
Bacterioferritin from Rhodobacter capsulatus was crystallized and its structure was solved at 2.6 Å resolution. This first structure of a bacterioferritin from a photosynthetic organism is a spherical particle of 24 subunits displaying 432 point group symmetry like ferritin and bacterioferritin from Escherichia coli. Crystallized in the I422 space group, its structural analysis reveals for the first time, the non-symmetric heme molecule located on a two-fold crystallographic symmetry axis. Other hemes of the protomer are situated on two-fold noncrystallographic axes. Apparently both types of sites bind heme in two orientations, leading to an average structure consisting of a symmetric 50:50 mixture, thus satisfying the crystallographic and noncrystallographic symmetry of the crystal. Five water molecules are situated close to the heme which is bound in a hydrophobic pocket and axially coordinated by two crystallographic or noncrystallographically related methionine residues. Its ferroxidase center, in which Fe(II) is oxidized into Fe(III), is empty or fractionally occupied by metal ion. Two positions are observed for the coordinating Glu18 side chain instead of one in the E. coli enzyme in which the site is occupied. This result suggests that the orientation of the Glu18 side chain could be constrained by this interaction.
I- Introduction
Ferritins and bacterioferritins (Bfr) are enzymes found in all organisms : eukarya, bacteria and archae. These spherical particles of an inner 8 nm diameter are iron storage proteins that oxidise Fe2+ into Fe3+ (ferroxidase activity). Fe3+ is next stored as an inorganic complex in the cavity of the spherical shell. Ferritins and bacterioferritins are protomers of 24 subunits that display a 432 point group symmetry. Two protein chains called H and L are present in the protomer in vertebrate enzymes whereas only chain H (with the ferroxidase site) is present in bacterioferritin. In eukarya, ferritin stores Fe3+ less toxic than Fe2+, therefore reducing its availability to react with the superoxide ion, a reaction leading to the production of reactive hydroxyl radical. In bacteria, Bfr would act as anion detoxifier (1). Another difference between ferritins and Bfr is the presence in the bacterioferritins of an iron protoporphyrin IX or coproporphyrin whose role still remains unknown (2) (3). In the structure from the E. coli enzyme, the Fe atom of the heme group is axially coordinated to the sulfur atom of a methionine residue from each of two symmetrically related subunits (4). Although the percentage of sequence identity between these proteins is low (around 20 %), the overall fold of the subunit is similar and consists of a bundle of four antiparallel α helices enclosing the ferroxidase center (also called dinuclear center) that binds two metal ions (4). The ferroxidase center differs between the ferritins and bacterioferritins by two residues thought to be involved in the iron binding (5).
At this time, the three dimensional structure of bacterioferritin is known from only one species, E. coli. This structure is available at 2.9 Å resolution from two different space groups (4) (6). In both of these structures the twelve heme groups bound to the 24 subunits of the protomer are situated on 2-fold noncrystallographic axes and the ferroxidase center is occupied by 2 manganese ions from the crystallization medium.
In the present study, we report the structure of the Rhodobacter capsulatus bacterioferritin solved at 2.6 Å resolution in the I422 space group and compare it with the one from Escherichia coli. The structure was solved by the MIR method, independently from previous structures. The final steps of protein purification, crystallization, and data collection were carried out in the presence of the metal chelator EDTA, and the dinuclear site is empty or fractionally occupied. This Bfr structure is the first from a photosynthetic organism and also constitutes the first example of heme, a non-symmetric molecule, situated on a crystallographic axis.
II- Materials and methods
2-1 Purification and crystallization
Crystals of Rhodobacter capsulatus bacterioferritin were obtained while attempting to crystallize the cytochrome cbb3 cytochrome oxidase of this organism. A strain overproducing the cytochrome bc1 complex (pMTS1/MT-RBC1) was grown on MPYE medium as described (7). Membranes were obtained by passing the cells in 50 mM KPi pH 7.5 through a French pressure cell 2 times at 12000 lb/in2 pressure. After removing unbroken cells by centrifuging at 11,000 × g for 30 minutes, the membranes were pelleted (2 hr at 310,000 × g), washed once in the same buffer and then stored frozen until used. Membranes were thawed, resuspended to 10 g/l protein concentration (by the Lowry procedure, (8)) in 50 mM KPi pH 7.5 containing enough NaCl and dodecyl maltoside to give final concentrations of 260 mM and 10 g/l respectively. After stirring 30 minutes the mixture was centrifuged 30 minutes at 12,000 × g to remove undissolved material, and the supernatant was passed through a 2.5 × 100 cm column of DEAE-Sepharose CL6B to remove the cytochrome bc1 complex and other strongly anionic proteins. The unbound flowthrough was diluted 2-fold with 25 mM KPi (pH 7.5), 0.1 g/l DM, and re-applied to DEAE Sepharose 6B. This time the column was washed with 25 mM KPi, 130 mM NaCl, 0.1 g/l DM and eluted with 50 mM KPi, 260 mM NaCl, 0.1 g/l DM. Colored peaks were identified by taking spectra of several fractions before and after adding dithionite, and the peak containing oxidase (high potential cytochrome b reduced before dithionite, cytochrome c reduced by dithionite) was pooled and applied to a hydroxyapatite column, washed with 50 mM KPi, 0.1 g/l DM, and eluted with 200 mM KPi, 0.5 g/l DM.
A final gel filtration chromatography was performed on Sephacryl S-300 in 20 mM K-MOPS 7.5, 100 mM NaCl, 0.1 g/l dodecyl maltoside, and 0.5 mM EDTA. The colored peak containing oxidase was pooled and concentrated by ultrafiltration through Amicon (Pharmacia) YM-100 membranes. For crystallization 5 or 10 µl of the concentrated protein was supplemented with 20 g/l octyl glucoside, mixed with an equal volume of 0.1 M NaOAc pH 4.6, 4% PEG 4K (precipitant #37 of the “Screen Lite” screening kit from Hampton Research) and allowed to equilibrate by vapor diffusion with the same precipitant. Because the final chromatography buffer in which the protein was obtained had 0.5 mM EDTA, and the precipitant did not contain metal ions other than Na+ and K+, it can be assumed the concentration of di- and trivalent metal ions is quite low.
The major precipitation was fine and amorphous, but after several weeks tiny (< 0.1 mm) red crystals were observed that continued to grow slowly. In some cases we obtained small cubicshaped crystals, in other cases they were cross-shaped and dendritic. Because of the small size, it was not deemed practical to analyze the crystals by SDS-PAGE to ascertain their polypeptide content.
2.2 Data collection phasing, and model refinement
Crystals were transferred via mixtures to a cryoprotectant solution containing 25% glycerol, 12% PEG 4K, 10 mM KMES 6.7, 3 mM azide, and 3 g/l OG for 10–30 minutes and then frozen in liquid nitrogen for data collection. X-ray fluorescence spectra taken at SSRL beamline 1–5 (unpublished results, H.D. Bellamy and E.A. Berry) showed the presence of iron and absence of copper. Data were collected at SSRL on beamline 7-1 using a wavelength of 1.08 Å. The best crystals diffracted to 2.6 Å.
The reciprocal space lattice was highly symmetrical, with primitive cell dimensions of a, b, c = 122.89, 122.02, 122.38 Å and α, β, γ = 109.06, 110.19, 109.25°. The diffraction could be accurately indexed on a trigonal, I-centered cubic (or tetragonal or orthorhombic), or F-centered orthorhombic lattice. Merging symmetry-related reflections ruled out the possibility of rhombohedral or cubic symmetry and confirmed tetragonal symmetry and the space group I422 (Table 1) with cell dimensions 142.37 × 142.37 × 140.88 Å.
Table 1.
statistics of X-ray diffraction data collection for the native crystal
| Mosaicity | 0.57 |
|---|---|
| Resolution (Å) | 21.5-2.6 (2.64-2.59) |
| Number of reflections | 260,696 |
| Unique reflections | 22,679 |
| Completeness (%) | 99.4 (94) |
| Rsym (%) | 7.7 (33.1) |
Values in parentheses refer to data in the highest resolution shell Rsym = □ |Ii -<I>|/ □<I> where i is the ith measurement and <I> is the weighted mean of I.
Initially it was assumed that the crystals contained the cytochrome cbb3 oxidase. Molecular replacement using subunit 1 of the Paracoccus denitrificans cytochrome aa3 cytochrome oxidase (PDB entry 1AR1) was unsuccessful, so further crystals were soaked in 5 mM solutions of several heavy atom reagents in the same cryoprotectant solution. Full datasets were collected for 4 of these, and these and the best native dataset were used for MIR phasing with the program SOLVE (9). SOLVE found useable heavy atom sites in three of the derivatives containing trimethyl lead, PCMB, and dimercury acetate. The resulting phases showed the asymmetric unit contained several hemes and at least 10 alpha helices of about the right length to be transmembraneous. The sites identified by SOLVE were further refined using SHARP (10). Statistics for the isomorphous phasing are presented in Table 2 and Table 3. The sites in the PCMB derivative refined to very low occupancy and did not contribute significantly to phasing. The density-modification program SOLOMON (11) was run using the script automatically generated by SHARP to further improve the phases. Maps generated from the phases from SHARP were not significantly better than those from SOLVE, however the density modification was very effective and the final map was quite interpretable.
Table 2.
phasing statistics for the heavy atom derivatives
| “Isomorphous” | “Anomalous” | |||||
|---|---|---|---|---|---|---|
| Compound | Sites | Resolution (Å) | Rcullis | PP | Rcullis | PP |
| Native | 0 | 2.6 | ||||
| Me3Pb | 6 | 2.8 | 0.87 | 1.64 | 0.80 | 1.55 |
| OAcHg2 | 4 | 2.6 | 0.77 | 1.74 | 0.92 | 1.06 |
| PCMB | 2 | 2.8 | 0.99 | 0.16 | 1.00 | 0.17 |
| Mersalyl | 0 | |||||
The Rcullis is defined here as Rcullis= (<phase-integrated lack of closure> / < | FPH - FP | >) and phasing power(PP) as PP= (< | FPcalc | / phase-integrated lack of closure >) from SHARP in shell 4.6–5.8 Å, for acentric reflections. FP and FPH represent respectively the structure factors for the protein and derivative.
Table 3.
figure of merit statistics for MIR phases from SHARP.
| Acentric reflections | Centric reflections | |||
|---|---|---|---|---|
| Dmin - Dmax (Å) | N | FOM | N | FOM |
| 37.94 - 7.20 | 835 | 0.84047 | 318 | 0.55480 |
| 7.20 - 5.14 | 1597 | 0.78699 | 325 | 0.66542 |
| 5.14 - 4.21 | 2107 | 0.64435 | 324 | 0.46472 |
| 4.21 - 3.65 | 2519 | 0.50549 | 329 | 0.45110 |
| 3.65 - 3.27 | 2873 | 0.40877 | 321 | 0.41385 |
| 3.27 - 2.98 | 3186 | 0.37257 | 325 | 0.43030 |
| 2.98 - 2.76 | 3485 | 0.31457 | 330 | 0.33725 |
| 2.76 - 2.59 | 3565 | 0.21986 | 301 | 0.20582 |
| Overall | 20167 | 0.43790 | 2573 | 0.44213 |
FOM: figure of merit for the quality of the centroid structure factor
N: number of reflections
At this point it becames clear that there was 3-fold noncrystallographic symmetry and the asymmetric unit contained three copies of a unit with only four long helices and 1.5 hemes, with the hemes ligated by the same residue in two different monomers and one of the hemes on a 2-fold axis ligated by a crystallographically related monomer (see Results and Discussion section). Solvent flattening and 3-fold NCS symmetry averaging (RAVE package (12)) further improved the maps allowing the chain to be completely traced and many side chains to be tentatively assigned. After several cycles of rebuilding, side chain assignment or modification, and refinement in CNS_SOLVE (13), the R and free-R factors (14) were 29 and 31%. The assigned sequence was then submitted in a BLAST search of the GenBank nonredundant database which showed the closest match to be bacterioferritin from Rhodobacter capsulatus (36% identity). Comparison with the structure of the E. coli homologue (PDB entries 1BCF (4) and 1BFR (6)) confirmed that our protein was a bacterioferritin. The sequence was thus corrected (15) and the structure was refined by energy minimization between 21.5 and 2.6 Å resolution, F/σF > 0 (22,526 independent reflections) using CNS_SOLVE and rebuilding using O (16). Bulk solvent correction was applied for all data during the refinement. Atoms of the heme group on a 2-fold crystallographic axis were given occupancy 0.5 and fixed during positional refinement (see Results and Discussion section). A noncrystallographic restraint of 300 kcal/mol was also applied between protein atoms of the three molecules in the asymmetric unit. The individual B-factor refinement was performed in the last stage of the refinement and 21 water molecules were included using the automated procedure implemented in CNS_SOLVE followed by a manual examination using O. After the inspection of the Fo-Fc electron density map, two conformations were imposed on the Glu18 side chain each with occupancy of 0.5. At the end of the refinement, the R and free-R factors were respectively 22.7 % and 24.7 % for all data (σF cutoff 0) between 21.5 and 2.6 Å resolution (Table 4). In order to refine the heme on the special position, the dataset was reduced again from unmerged intensities using I4 symmetry, and an asymmetric unit of 3 dimers enclosing three hemes was refined in this lower-symmetry spacegroup. After this refinement converged the symmetry was again returned to I422 and the trimer with 1.5 hemes was further refined. From these coordinates the trimeric structure with proper 3-fold symmetry (Figure 1b) was then generated and refined using CNS_SOLVE, without fixing the atoms of the heme situated on the crystallographic axis, and applying 3-fold NCS restraints to the protein. Interactions between the overlapping heme molecules were ignored during positional refinement. This resulted in the final submitted structure, with R and free-R factors of 22.5 and 24.2 %, respectively. Questions regarding symmetry of this heme will be discussed below.
Table 4.
refinement and model statistics
| Resolution range (Å) | 21.5-2.6 (2.69-2.60) |
|---|---|
| Number of reflections used for Rcryst calculation | 21,429 |
| Number of reflection used for the Rfree calculation | 1,097 |
| Data cutoff F/σ(F) | 0.0 |
| Rcrysta (%) | 22.5 (31.4) |
| Rfree (%) | 24.2 (39.8) |
| Number of non-hydrogen protein atoms | 3,804 |
| Number of water molecules | 21 |
| Mean B factor, protein main chain atoms (Å2) | 25.2 |
| Mean B factor, protein side chain atoms (Å2) | 27.1 |
| Mean B factor, solvent atoms (Å2) | 23.1 |
| Ramachandran plot | |
| Residues in most favored regions (%) | 96.9 |
| Residues in additionally allowed regions (%) | 3.1 |
| Rmsd from ideal geometry | |
| Bond length (Å) | 0.008 |
| Bond angle(°) | 1.2 |
values indicated in parentheses correspond to the highest resolution shell Rcryst= □ ||Fobs|-|Fcalc||/ □ |Fobs|. Rfree is the same as Rcryst but calculated for 4.9 % of the data omitted from the refinement.
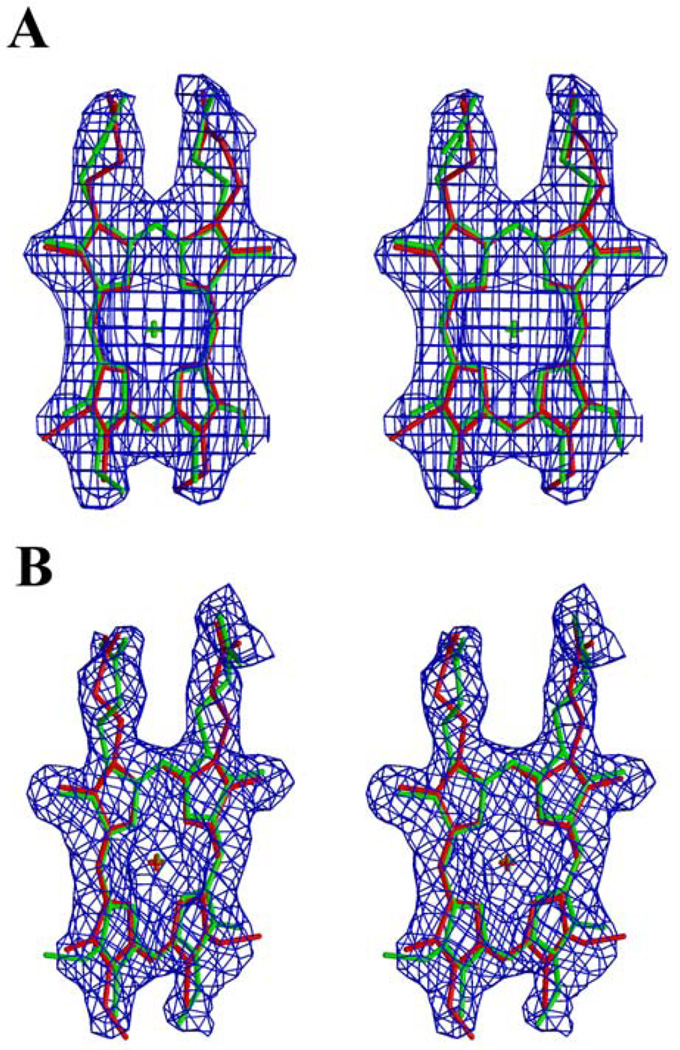
Figure 1.
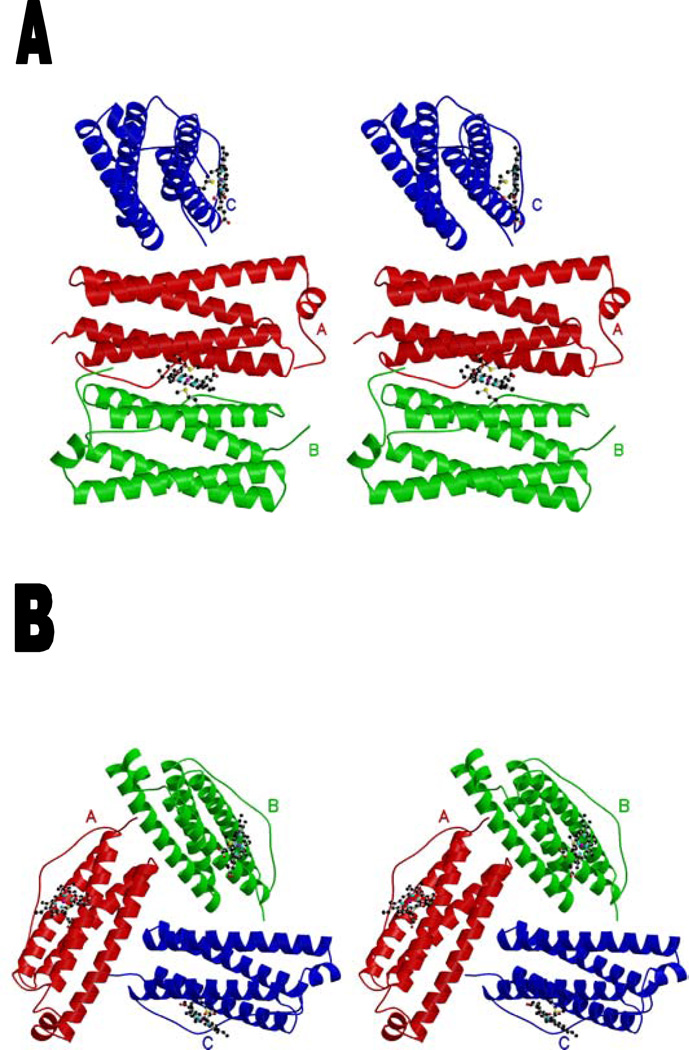
stereoview of the Rhodobacter capsulatus bacterioferritin subunits of the asymmetric unit schematized as a ribbon. The heme group is represented in ball-and-stick. Monomers related by a noncrystallographic 2-fold axis are colored in red (A) and green (B). The subunit (C) binding to the heme situated on the 2-fold crystallographic axis is colored in blue. (a) Trimeric structure including one heme with its enclosing noncrystallographic dimer and one heme on a crystallographic axis (at occupancy 0.5) with one monomer of its enclosing dimer. (b) Trimeric structure with proper 3-fold symmetry, including three heme moieties at 0.5 occupancy. The Met52 and heme are represented in ball-and-stick. The drawing was generated using Molscript (19) and Raster3D (20).
III- Results and discussion
3-1 Crystal contents and structure quality
The unit cell contains two 24-subunit protomers related by the crystallographic centering operator. The asymmetric unit contains 3 Bfr subunits of 161 residues each (A, B and C), 1.5 heme, and 21 water molecules. The C-terminal residue (E161) was not observed in the density and was omitted from the model. Despite presence of two detergents in the mother liquor and cryoprotectant, no detergent molecule was found in the structure. The rms deviation values between pairs of the molecules are 0.02 Å for all the atoms and 0.01 Å for the Cα atoms. Analysis of the model using CNS_SOLVE (13) and PROCHECK (17) shows a good stereochemistry with a rmsd of 0.008 Å for the bond lengths and 18° for the dihedral angles. No residue is observed in disallowed regions of the Ramachandran plot (Table 4).
3.2 Structure of the R. capsulatus Bfr
The R. capsulatus Bfr monomer is a bundle of 4 long antiparallel α helices (named A, B, C and D) and a small α helix (E) in the C-terminal part (Fig. 1a and 1b). These helices extend from Ala5 to Trp35 (helix A), from Ala38 to Phe64 (B), from Pro83 to Glu110 (C), and from Ile114 to Leu144 (D). Helix E is composed of residues from Pro146 to Leu152. The long loop connecting helix B to C (L loop (6), from residues Leu65 to Glu82) runs from one end of the bundle to the other. In doing so it loops out over the heme cleft, separating it from the external medium and providing hydrophobic residues 71 and 74 to the lining of the cavity which contains the heme ring and a cluster of 5 waters described below.
Bacterioferritin from R. capsulatus has 49.7 % sequence identity with the E. coli Bfr, whose structure is known (4). Except the C-terminal part of the subunit, which is longer and in a different orientation in R. capsulatus Bfr, the overall structure is similar. A subunit of the R. capsulatus bacterioferritin can be superimposed on the E. coli monomer (1BCF) with an rms deviation between Cα atoms of 0.78 Å. Dimers sharing a heme molecule (chains A+B) superimpose on the corresponding E. coli dimer with rms deviation 0.96 Å.
The choice of the asymmetric unit is somewhat arbitrary, but for the I422 R. capsulatus crystals it must contain 1/16 of the unit cell contents, i.e. 3 monomers and 1.5 hemes. For most of the refinement we have used the asymmetric unit shown in figure 1a, in which the three monomers are related by rotations of 180° (A and B), 120° (A and C), and 90° (B and C). Molecules A and B share one heme group in the asymmetric unit while molecule C binds heme with another molecule C related by a 2-fold crystallographic axis.
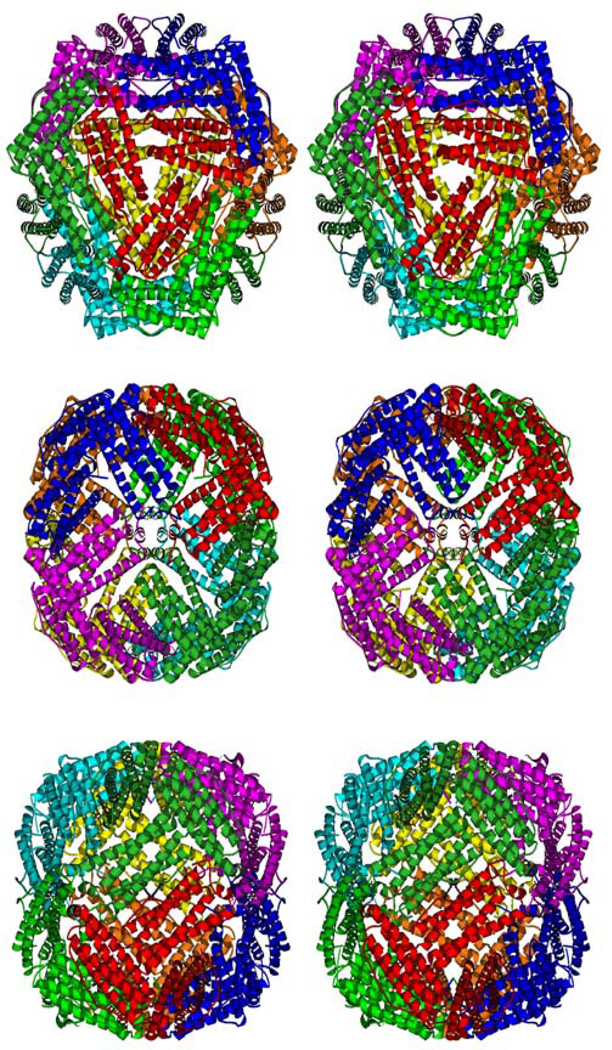
The protomer described as the biological form of bacterioferritin (4) contains 24 protein subunits and 12 heme molecules, i.e. 8 asymmetric units of the I422 crystal described above. Four 3-fold symmetry axes and three 4-fold axes are present in the protomer. Holes where these axes pass through the shell result in eight channels with 3-fold symmetry and six channels with 4-fold symmetry (Fig. 2a and 2b). There are six 2-fold axes (Fig 2c), but instead of passing through channels in the shell they pass through the 12 heme molecules, along the quasi-2-fold axis of heme as described (4). Correlation between this symmetry of the protomer and crystallographic/noncrystallographic symmetry in the different crystals will be discussed below.
Figure 2.
stereoview of the Bfr protomer from R. capsulatus along the 3-fold channel (a), along the 4-fold channel situated on the four-fold crystallographic axis (b) and along the 2-fold axis (c). The drawing was generated using Molscript (19).
The 4-fold channels are hydrophobic in R. capsulatus Bfr, being lined with 4 leucine residues (Leu151) (Fig. 3). This channel is also hydrophobic in the mammalian ferritins (1), however in the E. coli enzyme it was reported as hydrophylic (1), being defined by the Asn148 and Gln151 side chains (6). In R. capsulatus Bfr, these are replaced by glycine and leucine, respectively, resulting in a nonpolar channel. No positive electron density peak corresponding to an ion or water molecule, as described in the E. coli Bfr (6), was observed at the entrance of this channel in the R. capsulatus enzyme.
Figure 3.
stereoview of the 4-fold channel along the 4-fold crystallographic axis. The Leu151 residues situated in the channel are displayed in ball-and-stick. The drawing was generated using Molscript (19) and Raster3D (20).
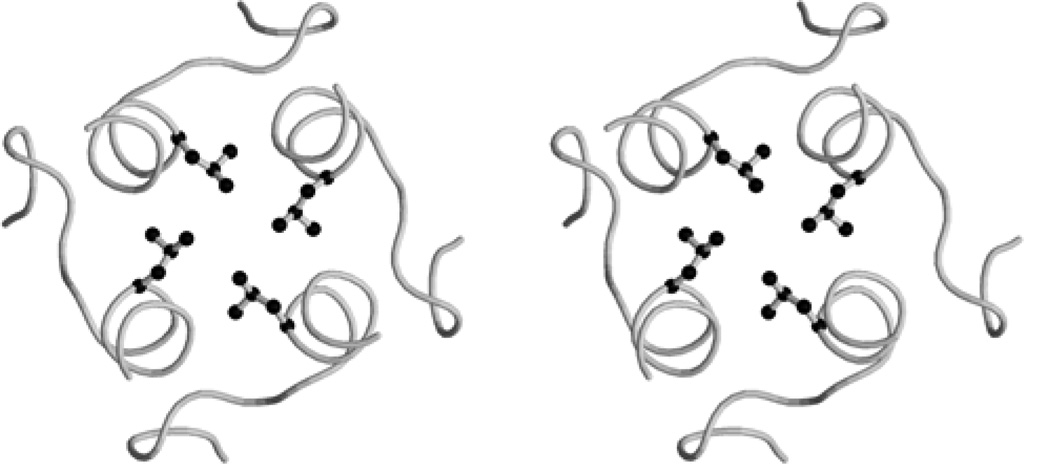
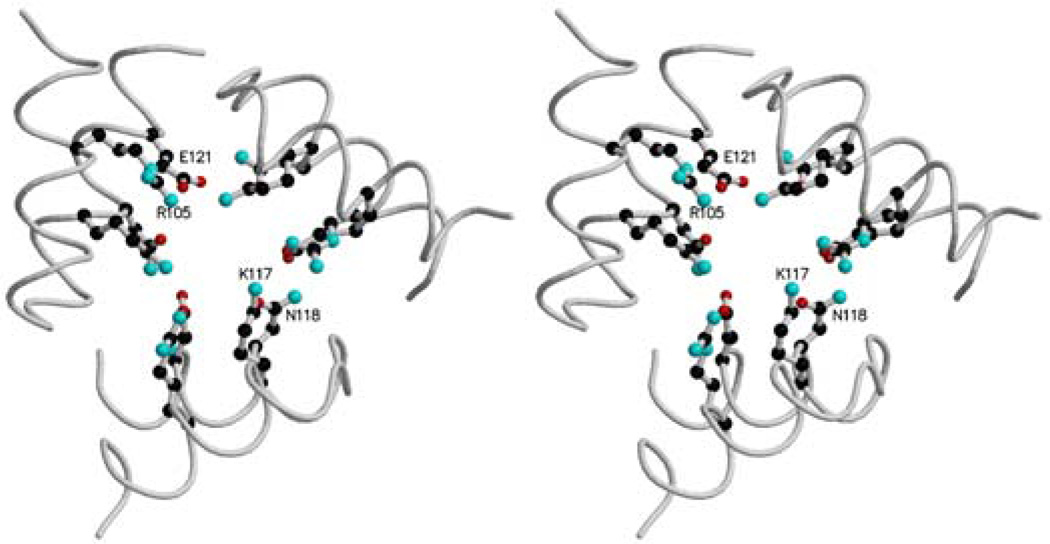
The 3-fold channels, through which Fe(II) ions are probably scavenged in ferritins and bacterioferritin (18), are hydrophilic as in mammalian ferritins and E. coli bacterioferritin (1). The charged side chains Arg105, Lys117, Glu121 and Asn118, which define this channel in the R. capsulatus protein (Fig. 4), are not highly conserved, but at least residues 117 and 118 are nearly always charged or polar giving this channel a hydrophylic lining. The 3-fold channel can bind different ions: Ca2+, Cd2+, Zn2+, Tb3+ (1).
Figure 4.
stereoview of the 3-fold channel. Arg105, Lys117, Asn118 and Glu119 are represented in ball-and-stick. The drawing was generated using Molscript (19) and rendered using Raster3D (20).
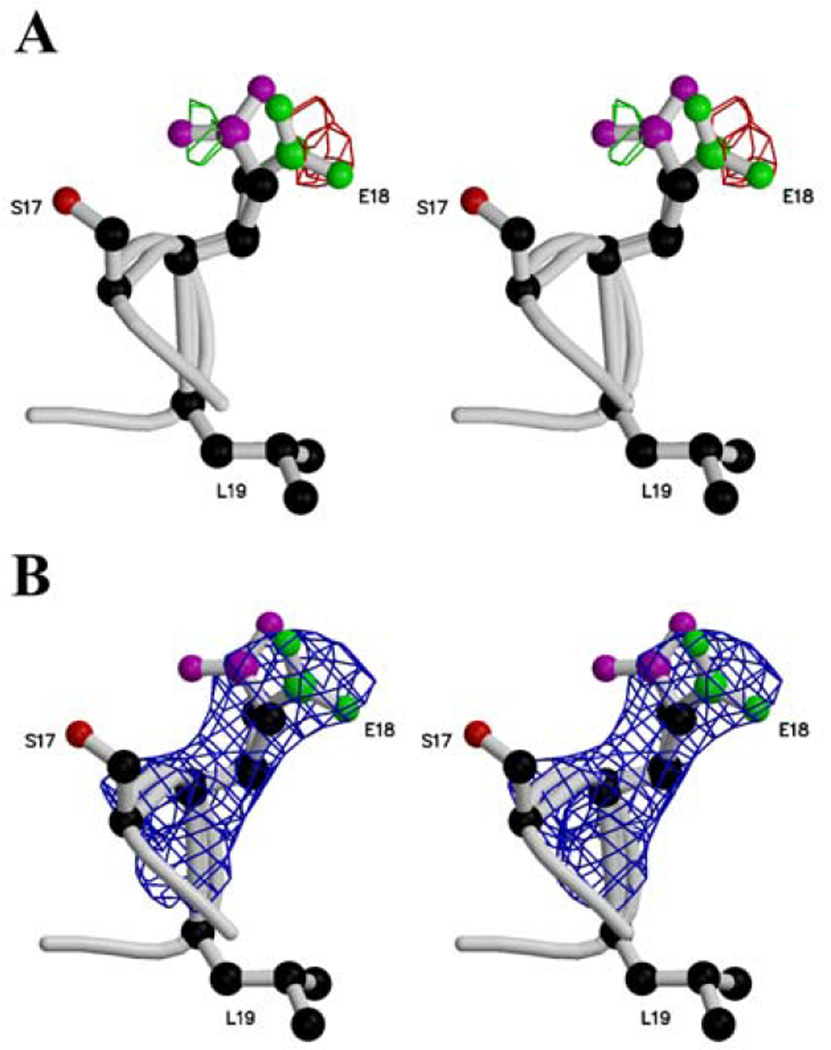
The two structures of E. coli bacterioferritin are from crystals grown in the presence of heavy metal ions, and both have metal bound at two adjacent sites comprising the “dinuclear site” at which Fe2+ is believed to be oxidized to Fe3+. In the deposited structures these are modeled as two manganese ions, mimicking probably the iron bound to the enzyme, and they are stabilized by glutamate and histidine residues (Glu18, Glu51, Glu94, Glu127, His54 and His130). The present crystals were grown and used without heavy metals and in the presence of the chelator EDTA, and the dinuclear site is either vacant or occupied to a small fractional extent. Analysis of the Fo-Fc electron density map reveals two positions for the Glu18 side chain (Fig. 5a). These two positions are also observed in the 3Fo-2Fc omit map calculated omitting this residue and the atoms around it in a sphere radius of 3.5 Å (Fig. 5b). Analysis of the Fo-Fc electron density map led us to attribute an occupancy of 0.5 to each position of the Glu18 side chain. Also, the B-factors and occupancy refinements starting with values around 26 Å2 and 1.0 for either positions of the side chain lead respectiveley to values around 50 Å2 and 0.5 for the Glu18 carboxylate group, reflecting the two positions or a partial disorder. The single Glu18 side chain orientation in the E. coli structure may be constrained by the presence of metal ions in the dinuclear center.
Figure 5.
stereoview of the two positions of the Glu18 side chain in a Fo-Fc electron density map (a) contoured at +3.5 σ (green) and −3.5 σ (red) and in a 3Fo-2Fc omit map (b) contoured at 1.2 σ and calculated at 2.6 Å resolution. The Ser17, Glu18 and Leu19 side chains are displayed in ball-and-stick. The two positions for the carboxylate group are displayed in different colors. The drawing was realized using Molscript (19) and O (16) and rendered using Raster3D (20).
A positive electron density peak in the Fo-Fc map higher than 3.5 σ in the dinuclear center suggests that the dinuclear center is partially occupied by a metal ion in R. capsulatus enzyme crystals even though no salt containing manganese ion was used to crystallize the protein. Distances from this peak to the oxygen atoms of each carboxylate group range between 4.06 and 1.95 Å. The closer contacts are too short for hydrogen bonds and thus not consistent with the peak being a water molecule. This peak was therefore interpreted as an iron ion with a low occupancy. Assignment of occupancy 0.33 and B-factor 40.0 in the three molecules of the asymmetric unit resulted in disappearance of the positive Fo-Fc peak. Refinement of the B-factors and the occupancies for these 3 Fe ions led to B-factors between 50.5 and 57.5 Å2 and occupancies between 0.28 and 0.44. This putative iron ion is in a position similar to that of Mn202 in the E. coli structure 1BFR and Mn601 in 1BCF; being ligated by Glu18, Glu51, His54 and Glu127. The presence of this iron ion at fractional occupancy could be responsible for the 2 positions observed for the Glu18 side chain.
A tetrahedron of four water molecules in the vicinity of the heme was found as described for the E. coli enzyme (4), and a fifth water molecule was observed situated at the center of the tetrahedron. The average B-factor for these five water molecules is 17.5 Å2. They are hydrogen bonded to the Ser23 hydroxyl group and to the carbonyl group of Leu19 and Leu71 in two monomers, as well as to each other. No functional role has so far been attributed to these water molecules.
There are 12 hemes in the protomer, located between pairs of monomers. The Fe atom in each heme is axially coordinated by the sulfur atoms of the Met52 side chains from two molecules related by the noncrystallographic or crystallographic symmetry. Axial ligation by methionine is also seen in class I type c cytochromes, however the chirality of the methionine sulfur atom here is the opposite of that in c cytochromes. This can be seen by comparing the structures visually or from the “improper” angle calculated for Sδ,Cγ,Cε,Fe which is −39 in cytochrome c (1YCC) but +19 in the R. capsulatus Bfr. This value is also positive in the E. coli structures, being +22 for entry 1BFR and +52 for 1BCF.
The tetrapyrol ring of the heme molecule is situated in a hydrophobic pocket constituted by Leu19, Ile22, Trp26, Ile49, Met52 and Leu71 of both monomers. The ring is perpendicular to the surface, with the edge bearing the vinyl groups (BC edge) directed outwards and the edge bearing the propionate groups (AD edge) inwards towards the internal cavity of the protomer. The vinly/methyl groups contact the cluster of 5 water molecules which is in turn covered by protein of the L linker loops, separating the water cluster and the heme from the external medium. The propionates actually extend into the solvent-filled volume inside the shell. This contributes to the highly polar and charged nature of the inner surface, which has in addition many side chains of Asp, Glu, and Arg protruding into the cavity.
The side chain of Arg45 in the two adjacent monomers approach the two heme propionates. As described below the use of two symmetry-related hemes at half occupancy in each site allowed the refinement program to fit two conformations for each propionate (atomic models of Fig 6a and 6b). In one conformation the propionate makes an ion pair with Arg45. The propionates are rather less ordered than the tetrapyrol ring or the Arg45 side chains and probably have two or more conformations, one making the ion pair directly with Arg45 and others hydrogen-bonding only with bulk water.
Figure 6.
stereoview of the two positions of the heme situated on the 2-fold crystallographic axis (a) and on the 2-fold noncrystallographic axis (b) in a 3Fo-2Fc omit map calculated at 2.6 Å resolution and contoured at 1.2 □. The two orientations of the heme are showed in green and red colors. For (a) the map was calculated from data reduced in the space group I4 to avoid imposing symmetry, and in both figures the heme was omitted from the phasing model. The drawing was generated using Molscript (19) and O (16) and rendered using Raster3D (20).
3.3 Interaction of crystallographic and protomeric symmetry and heme quasi-symmetry
The symmetry of the bacterioferritin protomer can readily be visualized as the symmetry of a cube (point group 432): with three 4-fold axes (connecting midpoints of 3 pairs of opposite faces of the cube), four 3-fold axes (connecting 4 pairs of opposing corners), and six 2-folds (connecting midpoints of opposite edges). This symmetry results in 24 equivalent positions, implying that the “asymmetric unit” of the structure consists of one monomer and half a heme moiety.
The 12 hemes are located on the six 2-fold axes, so that each is shared between two identical monomers. The heme molecule is not 2-fold symmetric, but has a quasi-2-fold axis as described (4). The asymmetry involves the methyl and vinyl substituents on rings B and C of the tetrapyrol- rotation about the quasi-twofold brings the methyl of ring C onto the vinyl of ring B and vice versa. In bacterioferritin the quasi-2-fold axis is aligned with the 2-fold axes of the protomer. Thus the slight asymmetry of the heme breaks the symmetry of the protein structure which would otherwise have proper 432 symmetry. Or to put it another way the asymmetric heme is binding in a perfectly symmetrical binding site. Looking at a single binding site it must be assumed that the heme binds equally well in both orientations. It seems unlikely that the slight asymmetry of the heme in one binding site would introduce significant asymmetry in the next binding site, so we believe the orientations of heme in the twelve binding sites are all independent and random. If so then on the average, as seen by crystallography, the heme consists of two hemes in opposite orientations, each with occupancy 0.5. As discussed below, all the crystallographic evidence we could obtain was consistent with such a model.
E. coli bacterioferritin has been crystallized in space groups P42212 and P21. The P42212 form (4) has 4 protomers in the unit cell, and a half protomer (12 monomers and 6 hemes) in the asymmetric unit. This space group has 2-fold axes parallel to c and diagonally in the a-b plane. The 2-folds parallel to c and all the crystallographic screw axes pass between protomers. The 2-folds in the a-b plane pass through 4-fold axes of the protomer. Thus the protomeric 2-folds, on which the hemes lie, are all noncrystallographic 2-fold axes. In the P21 crystal form (6) all the symmetry of the protomer is noncrystallographic (the spacegroup has only a two-fold screw axis) so again the heme is not in a special position in terms of crystallographic symmetry.
In the I422 crystals from R. capsulatus Bfr described here, there is much closer matching of crystallographic and protomeric symmetry. The origin of the unit cell is at the center of a protomer (within an arbitrary choice of origin), with the 4-fold crystallographic axis along c passing through a protomeric 4-fold (defining the axial direction of the protomer in this lattice). The space group has 2-fold axes along a and b which pass through 2 equatorial 2-folds of the protomer, putting the 4 hemes on these 2-folds in special crystallographic positions. The space group also has 2-folds in the a-b plane along 1,1,0 and −1,1,0; these pass through the two equatorial 4-fold axes of the protomer. The only protomeric symmetry axes that do not fall on a crystallographic symmetry axis are the 3-fold axes. The translation (centering) operators then generate the second protomer at the center of the cell.
The asymmetric unit is three protein chains and 3/2 heme molecules. For the initial refinement we chose a heme on a non-crystallographic axis with its enclosing dimer (AB), plus an adjacent monomer C with its heme on a crystallographic axis, with that heme occupancy set to 0.5 (Fig. 1a). The heme on the noncrystallographic axis was refined positionally, while the atoms of the heme on the crystallographic axis had to be fixed during positional refinement due to special position and overlap with symmetry-related heme atoms. In order to refine positionally the heme on the crystallographic axis, we refined a hexameric asymmetric unit (consisting of the trimeric asymmetric unit of figure 1a together with the trimer related by the two-fold axis passing through the heme on monomer c) against the dataset reduced in space group I4, which lacks this 2-fold axis.
By using data reduced in space group I4 from unmerged intensities, we were also able to test whether the heme on the crystallographic axis conforms to the higher symmetry of I422, which would imply equal binding of heme in two orientations. Ordinarily the space group I422 would be chosen over I4 based on the similar Rsym values (7.7 and 7.4 %) obtained in the two space groups. The asymmetry involves only two methyl groups, which might not contribute enough to the scatterring to give a high Rsym in I422, however asymmetric binding of the heme could only be explained by an asymmetry in the protein shell which presumably would result in a high Rsym in I422. Nonetheless, to test this question refinement was carried out with this heme in either of its two orientations, and we looked at positively and negatively contoured Fo-Fc maps. If the heme is actually predominantly in one orientation, then when the wrong orientation is used there should be strong positive peaks on the two vinyl side chains (because they are actually methyls), and negative peaks at the two methyl groups which are actually vinyl. When the correct orientation is used, positive and negative peaks should be much less significant. This was not observed, actually the fit was about the same with either orientation. The analysis of the 3Fo-2Fc omit map calculated in the I4 space group (Fig. 6a) did not clearly favor one orientation, although it was not perfectly symmetrical. So, we conclude that two orientations of the heme situated on the crystallographic axis are present (as modeled in Fig. 6a) and the high symmetry space group (I422) is justified.
Frolow et al. (4) considered the possibility that the heme preferentially binds in one orientation about its quasi-2-fold axis in the apparently symmetrical binding site. Since all of their hemes were on local and not crystallographic 2-folds this could not be excluded, and in fact they did report some indication of asymmetry in their 2.9 Å structure.
In our case, for the heme situated on the noncrystallographic axis between monomers A and B, a similar approach to the one described before was used. This time our original trimeric asymmetric unit (Fig. 1a) was refined against data reduced in space group I422. Each heme orientation was refined independently with occupancy of 1.0, leading to similar values for the free-R and R factors. Fo-Fc maps did not show a strong preference for one orientation. Again, the 3Fo-2Fc omit map did show some asymmetry (Fig. 6b), but mainly in the position of the propionate side chains which are not well ordered (see above). No significant asymmetry is found in the vinyl and methyl groups on the other side of the ring which are responsible for the intrinsic asymmetry of heme. Thus we conclude that a model in which all binding sites are occupied by a symmetric 0.5:0.5 mixture of heme in two orientations is more realistic than one in which heme binds with a particular orientation, the space group I422 is justified for the crystal, and the protomeric point group symmetry of 432 is appropriate for the biological entity including the heme when the ensemble of a large number of molecules is considered. If there is some non-random pattern to the heme orientation in the protomer, it does not affect crystal contacts and so is lost due to random packing of the protomer into the lattice.
This situation cannot be represented by our original asymmetric unit depicted in Figure 1a unless the heme molecule between monomers A and B is allowed two orientations at half occupancy, in which case the asymmetric unit of Figure 1b would be more intuitive, with three monomers and three hemes at half occupancy, all related by proper 3-fold noncrystallographic symmetry. This is the asymmetric unit used in our final refinement and submitted to the PDB.
Just as this model accomodates the asymmetry of the methyl and vinyl sidechains on the B and C pyrol rings in a symmetrical structure, it allows the propionates on the A and D rings (which are symmetrical in covalent structure) to take on different conformations without violating the crystallographic and noncrystallographic symmetry. The refinement process took advantage of this freedom to refine two different conformations for the propionates on the A and D ring of both the crystallographic and non-crystallographic hemes (Figure 6). This does not violate the symmetry of the crystallographically averaged structure, in which the crystallographic or crystallographic plus noncrystallographic symmetry superimposes the D ring of one heme with the A ring of another. Also in any heme binding site of a single molecule, with one heme bound in a particular orientation, it is not expected that the two propionates will be different. Rather both propionates will take on each conformation for a fraction of the time, resulting in an essentially symmetric structure. This is quite different from the case of the methyl and vinyl substituents on the B and C rings, which are asymmetric in covalent structure and will of course be asymmetric, showing either one or the other of two orientations, in any single heme binding site. Only in the averaged structure seen by crystallography is the heme symmetrical with respect to these substituents.
It may be noticed in Figures 6a and especially 6b that the electron density for the propionates does appear asymmetric, contrary to the above description. We cannot think of any cause for this asymmetry, and attribute it to noise in the map, which is contoured at a relatively low level in order to show density for the relatively disordered propionates. In any case this has no bearing on the symmetric orientation of the heme which is our main point here, as the propionates of heme are symmetric as far as the covalent structure is concerned and so give no information about the orientation.
IV- Conclusion
The structure of Rhodobacter capsulatus bacterioferritin presented here is the first from a photosynthetic organism, or from any organism other than E. coli. The resolution (2.6 Å) is significantly higher than that of previous bacterioferritin structures. We believe this is the only structure in the database in which the quasi-symmetric protoheme molecule is located on a two-fold crystallographic axis. Combining different crystallographic approaches we conclude that this does not break the symmetry of the space group because the heme is bound in a symmetric mixture of two orientations. Other hemes in the structure, situated on a two-fold noncrystallographic axis, probably also exist as a mixture of two orientations, and we can also suppose a similar characteristic for the E. coli Bfr, not shown in previous work.
The dinuclear metal-binding site is empty or fractionally occupied, probably due to presence of EDTA in the purification and crystallization solutions. One of the residues making up this site, Glu18, displayed two side chain orientations in the ferroxidase center probably resulting from the partial occupancy of the heavy atom which it ligates. The lining of the 4-fold channel in the bacterioferritin shell is nonpolar in the R. capsulatus enzyme, unlike that of the E. coli enzyme.
The fact that bacterioferritin was obtained while purifying the cytochrome cbb3 oxidase is suggestive of a functional association between these proteins, however a coincidental copurification cannot be ruled out. This question is currently being looked into.
Aknowledgments
We would like to acknowledge the contribution of Henry Bellamy and the staff at the Stanford Synchrotron Radiation Laboratory (SSRL) for performing the XAFS scan and assistance with data collection. This work was supported by NIH R01 grants DK44842 and GM62563, and by the US Department of Energy contract DOE-FG0291ER20052 to F.D and DE-AC03-76SF00098 to LBNL. Crystallographic data collection was carried out at SSRL which is operated by the Department of Energy, Office of Basic Energy Sciences. The SSRL Biotechnology Program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and by the Department of Energy, Office of Biological and Environmental Research.
The atomic coordinates have been deposited in the Protein Data Bank with the accession code 1JGC.
References
- 1.Harrison PM, Arosio P. Biochim. Biophys. Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Le Brun N, Barynin V, Thomson AJ, Moore GR, Guest JR, Harrison PM. J. Biol. Chem. 1995;270:23268–23274. doi: 10.1074/jbc.270.40.23268. [DOI] [PubMed] [Google Scholar]
- 3.Romao CV, Louro R, Timkovich R, Lubben M, Liu MY, LeGall J, Xavier AV, Teixeira M. FEBS Lett. 2000;480:213–216. doi: 10.1016/s0014-5793(00)01939-6. [DOI] [PubMed] [Google Scholar]
- 4.Frolow F, Gilboa AJK, Yariv J. Nat. Struc. Biol. 1994;1:453–460. doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Le Brun NE, Thomson AJ, Moore GR, Chasteen ND. Biochemistry. 2000;39:4915–4923. doi: 10.1021/bi992631f. [DOI] [PubMed] [Google Scholar]
- 6.Dautant A, Meyer JB, Yariv J, Precigoux G, Sweet RM, Kalb AJ, Frolow F. Acta Crystallogr. 1998;D54:16–24. doi: 10.1107/s0907444997006811. [DOI] [PubMed] [Google Scholar]
- 7.Gray KA, Dutton PL, Daldal F. Biochemistry. 1994;33:723–733. doi: 10.1021/bi00169a014. [DOI] [PubMed] [Google Scholar]
- 8.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 9.Terwilliger TC, Berendzen J. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La Fortelle Ed, Irwin JJ, Bricogne G. Crystallographic Computing. 1997:7. [Google Scholar]
- 11.Abrahams JP, Leslie AGW. Acta Crystallogr. 1996;D52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 12.Kleywegt GJ, Jones TA. In: From First Map to Final Model. Bailey S, Hubbard R, Waller DA, editors. Daresbury: SERC Daresbury Lab; 1994. pp. 59–66. [Google Scholar]
- 13.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Brunger AT. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 15.Penfold CN, Ringeling PL, Davy SL, Moore GR, McEwan AG, Spiro S. FEMS Microbiol. Lett. 1996;139:143–148. doi: 10.1111/j.1574-6968.1996.tb08194.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones TA, Zhou J-Y, Cowan SW, Kjeldgaard M. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Laskowsky RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Chasteen ND, Harrison PM. J. Struct. Biol. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- 19.Kraulis PJ. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 20.Merrit EA, Murphy MEP. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]