Abstract
MicroRNA knockout by genome editing technologies is promising. In order to extend the application of the technology and to investigate the function of a specific miRNA, we used CRISPR/Cas9 to deplete human miR-93 from a cluster by targeting its 5’ region in HeLa cells. Various small indels were induced in the targeted region containing the Drosha processing site and seed sequences. Interestingly, we found that even a single nucleotide deletion led to complete knockout of the target miRNA with high specificity. Functional knockout was confirmed by phenotype analysis. Furthermore, de novo microRNAs were not found by RNA-seq. Nevertheless, expression of the pri-microRNAs was increased. When combined with structural analysis, the data indicated that biogenesis was impaired. Altogether, we showed that small indels in the 5’ region of a microRNA result in sequence depletion as well as Drosha processing retard.
Keywords: CRISPR/Cas9, microRNA depletion, microRNA biogenesis, miR-93, RNA-guided DNA cleavage
Abbreviations
- CRISPR
clustered, regularly interspaced, short palindromic repeats
- crRNA
CRISPR RNAs
- PAM
the protospacer adjacent motif
Introduction
MicroRNAs are a class of 22–25 nt endogenous non-coding RNAs that play pivotal roles in the regulation of gene expression.1 They are processed from pri-miRNAs, their precursors with stem-loop structure, to form ∼70 nt pre-miRNAs by the nuclear RNase III protein Drosha. The pre-miRNAs are then exported to the cytoplasm and cut by another RNase III family member, Dicer, to form a ∼23 bp miRNA/miRNA* duplex before one of the strands is incorporated into the RNA-induced silencing complex.2 MicroRNAs recognize their targets mainly through mechanisms mediated by the seed sequence (the 2-8 nt from the 5'end of the miRNA) and induce translational repression or mRNA de-adenylation and degradation.3
Loss-of-function methods have proved to be the best ways to delineate the biological roles of miRNAs in various model systems. Antisense technology has been widely used as a loss-of-function method in miRNA studies.4 The drawbacks of this method are incomplete masking of the miRNA function and off-target effects on miRNAs sharing similar sequences, especially in the seed region. Different gene-knockout techniques have also been used to understand the function of miRNAs in animal models.5-8 Homologous recombination-based miRNA knockout methodology has been limited due to its low efficiency and complicated procedure.9 Genome-editing technologies using engineered endonucleases have been shown to be useful for specific gene targeting such as zinc finger nuclease, and transcription activator-like effector nucleases (TALENs).10
The clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system provides a rapid and efficient technology that is quickly replacing TALENs and becoming the preferred platform for targeted genome editing.10-15 The Cas9 protein from the type II CRISPR/Cas system of Streptococcus pyogenes relies on small CRISPR RNAs (crRNAs) to target chromosomal DNA, thereby triggering an error-prone repair process and producing targeted mutagenesis of the genomic level in cells. A single RNA molecule that combines the transacting RNA with the crRNA, termed gRNA, directs site-specific DNA cleavage which occurs 3 base-pairs upstream of NGG, the protospacer adjacent motif (PAM) sequence of target genes.12
The miRNA database (miRBase release 20) has currently registered ≥2000 mature miRNAs in Homo sapiens.16 Among them, a significant number are expressed as miRNA clusters. During evolution, miRNA families were formed with miRNAs sharing the same 5’ seed region. Functional knockout of one such miRNA without affecting the expression of other cognate miRNAs is highly desirable. It has, however never been established how few nucleotides have to be altered in order to diminish the expression of one miRNA in such a scenario.
Functional knockout of a particular miRNA by genomic editing has been proved promising. However, little is known about the mechanism and consequences of the microRNA sequence impairment. MicroRNA knock-out by deleting fragments as large as possible is recommended,11 but 2 targeted cutting sites are required in such cases. Destroying the processing sites was an effective strategy using TALEN technique.17 In this study, we attempted to deplete a single microRNA by targeting its 5’ region, including the Drosha processing site and seed region using the CRISPR/Cas system as a novel tool. We choose miR-93, a critical onco-miRNA from a cluster, as the target. By establishing multiple cell lines carrying various mutants and exploring the consequences of genomic alteration, we showed that small indels in 5’ region of this miRNA leads to a successful and specific gene knockout due to sequence impairment and biogenesis of the microRNA was blocked as well.
Results and Discussion
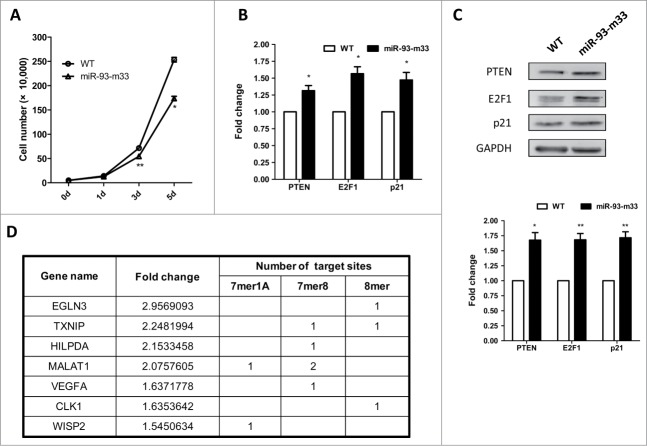
To knockout a particular single microRNA with the CRISPR/Cas system, we chose miR-93, a member of the miR-106b-25 cluster as the target. miR-106b-25 and miR-106a-363 clusters are paralogs of the miR-17-92 cluster (Fig. 1A).5,18-20 The miR-17 family members not only have identical sequences in the seed region but also share extensive similarities along the whole length of the mature miRNA, with only a few nucleotides differing from each other (Fig. 1B). Such a target gene provided a good example in which to demonstrate the feasibility of molecular ablation of one miRNA without affecting the expression and function of other miRNAs.
Figure 1.
Design of miR-93 depletion by CRISPR/Cas system. (A) Schematic representation of the miR-17 family and the miR-106b-25 cluster. Pre-miRNAs are shown as color-coded boxes and black boxes indicate mature miRNAs. Boxes of the same color represent miRNAs from the same miRNA family. (B) Sequences of the miR-17 family members. The seed sequences are underlined. (C) Schematic representation of the CRISPR strategy for miR-93 deletion. The miR-93 mature sequence is shown in bold and the seed sequence is underlined. The crRNA sequence is listed below. The box indicates the PAM sequence. The triangle indicates the possible cleavage site, which occurs in the middle of the seed sequence. (D) The T7EI assay was used to detect CRISPR/Cas-induced indels in human HeLa cells. Arrows indicate the position of the PCR product and the expected DNA bands cleaved by T7EI. Mutation frequencies were measured from the band intensities.
The 5′-end of microRNAs is determined by precise cleavage by Drosha and contains the seed region, which is critical for target recognition.1,3 We designed one gRNA targeting the PAM sequence at the 5′ region of miR-93. Indels therefore tended to occur around the Drosha cleavage site and in the seed region (Fig. 1C). After co-transfecting the expression constructs of Cas9 protein and gRNA into HeLa cells, we extracted whole genomic DNA for T7 endonuclease I (T7EI) assay21 to assess the mutation efficiency, and showed that the gRNA for mir-93 induced mutations at a frequency of 16% (Fig. 1D).
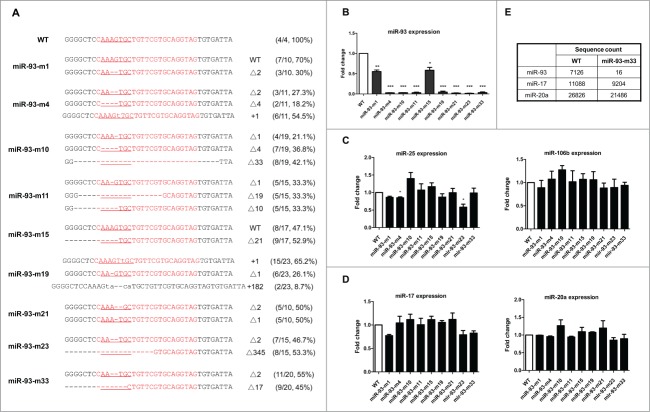
We cultured and expanded the HeLa cells containing mutated forms of miR-93. They were separated into single cells by flow cytometry and cultured for one week until single colonies were formed. Seventy clones were obtained and the genomic DNA was extracted. The miR-93 gene regions were individually amplified by PCR and sequenced. The clones with overlapping sequencing peaks were confirmed by sub-cloning and re-sequencing. As a result, 9 mutant cell clones were identified out of the 70. Among the 9, 7 clones had indels at the seed region of miR-93 in both alleles and 2 others had one mutant and one wild-type allele (Fig. 2A).
Figure 2.
Generation of cell lines carrying various indels in miR-93. (A) DNA sequences of the targeting sites in the miR-93-depleted cells are represented. The region of miRNA sequence is shown in red and the seed sequence is underlined. Lowercase letters show the DNA insertion. The number of mutated nucleotides and sequencing frequency are indicated at right. (B) miR-93 expression levels in the depleted cell lines were validated by qRT-PCR with stem-loop primers. The relative expression level of miR-93 was normalized to U6 small nuclear RNA and compared with the wild-type cell line. (C) The expression level of the miR-106b-25 cluster miRNAs in the deleted cell line was measured by qRT-PCR with stem-loop primers. The relative expression of microRNAs was normalized to U6 small nuclear RNA and compared with the wild-type cell line. (D) qRT-PCR measurement of the expression level of the miR-17 family members in the deleted cell lines. The relative levels of microRNAs were normalized to U6 small nuclear RNA and compared with the wild-type cell line. (E) RNA-seq showing that the sequence count of miR-93 was dramatically reduced compared with the other miRNAs in the same family (*P < 0.05, **P < 0.01, ***P < 0.001 compared to wild-type).
To evaluate whether these indels result in depletion of miR-93 in the mutated cells, we carried out quantitative reverse-transcription PCR (qRT-PCR) using stem-loop primers (Fig. 2B). Astonishingly, miR-93 was almost undetectable in all the 7 clones with disrupted seed sequence in both alleles, while in the 2 clones, miR-93-m1 and miR-93-m15, where one wild-type allele was still intact, the amount of miR-93 fell to half compared with the wild-type control. It was a surprise that even very small indels in the seed region (such as those in miR-93-m4 and miR-93-m21) had the ability to totally abolish the production of mature miR-93.
To assess whether the indels at miR-93 locus would affect the expression of miR-25 and miR-106b, the other 2 closely located miRNAs in the miR-106b-25 cluster, qRT-PCR was performed with the total RNA of the 9 mutant clones and it was found the expression of miR-25 and miR-106b did not change in the miR-93 knockouts, with the exception of miR-93-m23 (Fig. 2C). We noted that the indel in one allele of miR-93-m23 extended to miR-25 locus and disrupted this gene as well, and this is in good accordance to the fact that miR-25 showed ∼50% decrease. We further determined the expression of other miR-17 family members, which share the same seed sequence and highly conserved mature sequence with miR-93. According to the small-RNA-sequencing results, miR-17 and miR-20a are highly expressed in HeLa as well as miR-93 and miR-106b. We then performed qRT-PCR of these 2 microRNAs in the 9 mutant clones and found their expression was unaffected by miR-93 deletion (Fig. 2D). In brief, the CRISPR/Cas–mediated small indels in the 5’ region of a miRNA can serve as a very clean tool with which to dissect the functions of such miRNAs with high specificity.
In order to further confirm the depletion of miR-93, small-RNA-sequencing was performed in the representive miR-93-m33 clone. This cell line had a Δ2 deletion in one allele and a Δ17 deletion in the other, which contained small indels next to Drosha processing site (Δ2) and larger indels that completely deleted the Drosha cutting site (Δ17). As a result, we found that the sequence counts for this particular miRNA were 99.78% lower than in the wild-type control (from 7126 down to 16), representing a nearly complete depletion of miR-93. Meanwhile, we checked other miRNAs of this family expressed in Hela cells, and found that their counts did not change much (Fig. 2E). These results indicate that small indels generated by CRISPR/Cas system in the 5’ region are enough to eliminate the mature miRNA with high specificity.
miR-93 is a critical regulator of cell growth.22 To determine the functional consequence of miR-93 knockout by the CRISPR/Cas system, we determined cell growth by counting cell numbers of the miR-93-m33 cell line at different time points. We found that cell growth is slowed in the mutant cells, consistent with the function of miR-93 as an oncogene (Fig. 3A). To further analyze the consequence of these miR-93 indels on the targetome, we assessed the expression of PTEN, E2F1 and p21, 3 well-known targets of miR-93 responsible for cell proliferation18,20 in the miR-93-m33 cell line. We found that all the targets were upregulated at both mRNA and protein levels (Figs. 3B and C). In addition, we determined the molecular impact of miR-93 depletion on genome-wide gene expression using mRNA sequencing, followed by cuffdiff analysis. As a result, 140 genes were up-regulated ≥1.5-fold, 33 of which had miR-93 seed sequence in their 3'UTR, and 7 have been reported to play important role in cell proliferation (Fig. 3D).
Figure 3.
Functional analysis of miR-93 depletion. (A) Cell growth in the miR-93-m33 cell line at different time points were monitored by cell number counts. (B) Relative mRNA expression levels of PTEN, E2F1, and p21 in the miR-93-m33 cell line were measured by qRT-PCR. The mRNA expression levels were normalized to GAPDH mRNA and compared with the wild-type cell line. (C) Upper lanel: protein expression detected by western blot. Lower lanel: relative expression levels measured from the band intensities normalized to GAPDH and compared with the wild-type cell line. (D) Seed sequences were found in the 3’-UTR of the upregulated and proliferation-related coding genes (or in the sequences of noncoding RNAs) (*P < 0.05, **P < 0.01, ***P < 0.001 compared to wild-type).
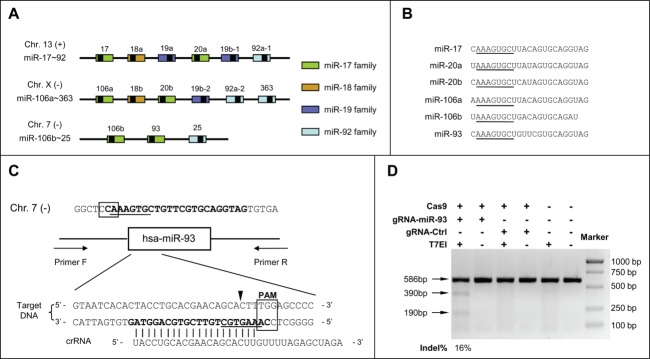
Next, we questioned whether these small indels could affect Drosha processing. We measured the expression level of pri-mir-93 using qRT-PCR with primers residing outside of the Drosha cleavage site (Fig. 4A). We assumed that the expression level of pri-miR-93 would increase if Drosha processing was disrupted and indeed, the expression level of pri-miR-93 increased markedly in the mutants with indels at both alleles but not in miR-93-m23, which had a Δ345 deletion disrupting the binding site of the PCR primer. As for miR-93-m1 and miR-93-m15, with a wild-type allele remaining, the level of pri-mir-93 only rose slightly compared to other mutants (Fig. 4B). We analyzed the secondary structure of the alleles with small indels in all the mutants and found the stem-loop was not totally disrupted, but the free energy value (ΔG) in the predicted structure increased, indicating the impaired thermodynamic stability of the pri-miRNA (one example is shown in Fig. 4C). As an example, we checked the small RNA sequencing data of miR-93-m33 to see whether there were newly generated miRNAs and only found one sequence related to the Δ2 allele with a sequence count of 1 (Fig. 4C). These results indicate that very small indels at the 5’ region of miRNA have the ability to disrupt Drosha processing.
Figure 4.
Retardance of Drosha processing by small indels around the cleavage site. (A) Design strategy of primers for pri-miR-93 detection. (B) pri-miR-93 expression levels in the depleted cell lines were validated by qRT-PCR. The relative expression level of pri-miR-93 was normalized to GAPDH mRNA and compared with the wild-type cell line. (C) Secondary structure of the miR-93 precursor was generated by Sfold (http://sfold.wadsworth.org/cgi-bin/srna.pl). Right: wild-type allele; the red circles indicate the seed sequence of miR-93. Left: the Δ2 allele of the miR-93-m33 cell line; the blue circles indicate the newly-generated miRNA with the sequence CAAUGCUGUUCGUGCAGGUAGUGU (*P < 0.05, **P < 0.01, ***P < 0.001 compared to wild-type).
In this study, we deplete single miRNA by introducing indels at the 5′ end of its mature sequence using the CRISPR/Cas system as a novel tool. We demonstrated that for a targeted miRNA, alteration of a single or a few nucleotides in the specific genomic sequence not only depletes the mature sequence, but also retards Drosha processing.
Of note, although mature miR-93 was undetectable in all mutant cells, the expression level of pri-miR-93 was not lowered, but increased. Analysis of the secondary structure indicate that very small indels lead to a new stem-loop structure that has the potential to be processed to a de novo miRNA, but small-RNA-sequencing failed to detect such novel small RNAs. A possible explanation is that Drosha cleavage was blocked by the structural alterations induced by the small indels. It is thought that Drosha processing was determined by the terminal loop as well as the stem-single-stranded RNA junction.23-25 Our study showed that indels around the Drosha processing site inhibit its cleavage ability. This may be explained by the altered stem length which is critical for recognition of a miRNA by Drosha, as shown in a previous study.25
In summary, we for the first time disrupt a single microRNA from a cluster and an miRNA family by introducing small indels around its 5′ region to disrupt seed sequence and impair Drosha processing using the CRISPR/Cas system, providing a strategy to knockout microRNAs, particularly those residing in or overlapping within functional genes, and the members of a tight cluster or from one miRNA family. Importantly, this approach enhances our understanding of genome-editing technology for miRNAs.
Materials and Methods
Cas9 and gRNA design
The Cas9 and gRNA expression vectors were made as previously described.14 In brief, we chose the PAM sequence nearest to the miR-93 seed sequence and the 20-bp sequence upstream as the targeting sequence of gRNA (Fig. 1C).
Cell culture and transfection
Human cervical carcinoma cells (HeLa) were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone) containing 10% fetal bovine serum (Hyclone) and 1% penicillin-streptomycin (Gibco) in 5% CO2 at 37℃. The HeLa cells were seeded into 6-well plates (Corning) and after 24h, the cells were transfected with the 2 plasmids expressing Cas9 and gRNA respectively using Lipofectamine 2000 (Invitrogen) with 2.5μg plasmid each per well. Forty-eight hours later, the cells were harvested for genomic DNA extraction.
T7 Endonuclease I (T7EI) assay
Targeted miR-93 genome DNA fragments were amplified with the primers: F 5′-GGCCAGTCCCGGAGTT-3′, R 5′-CCTATACTTCTGCCCGGACC-3′. The PCR products were purified, denatured, and annealed. Then T7EI (NEB) was added to the annealed PCR products and incubated at 37 for 60min. The reaction mixture was resolved on 2% agarose gel.
Generation of the miR-93-deleted cell lines
The transfected HeLa cells were cultured on 60-mm dish. After washing and trypsinization, the cells were re-suspended in DMEM. Single cells were picked by flow cytometry (BD FACSCalibur) and then seeded in 96-well plates. After the colonies formed, the genomic DNA of each clone was extracted for target segment amplification and DNA sequencing.
Cell proliferation assay
Cells were seeded in 6-well plates at 50,000 cells per well. After 1, 3, and 5 d, the medium was discarded and the numbers of cells were counted after trypsinization. Independent experiments were performed 3 times.
RNA extraction and quantitative reverse-transcription PCR
Total RNA was isolated from cells in culture using TRIzol regant (Sigma). For miRNA detection, 500 ng total RNA was reverse-transcribed using specific stem-loop primers (RiboBio Co., Ltd) and SYBR Green-based qPCR was carried out using a specific forward primer and a universal reverse primer (RiboBio Co., Ltd). Independent experiments were performed 3 times.
For pri-miR-93 and target mRNA qRT-PCR, 500 ng of total RNA digested with DNase I was reverse-transcribed with a random primer (TransGen Biotech) for pri-miR-93 or an oligo(dT) primer (TransGen Biotech) for target mRNA, and gene-specific primers (Table 1) were used for SYBR Green-based qPCR. Independent experiments were performed 3 times.
Table 1.
Primers for qRT-PCR of pri-miR-93, PTEN, E2F1, and p21
| Primer name | Sequence (5′-3′) |
|---|---|
| pri-miR-93-RT-F | TCCTTTCTGTCCTCCCGTCT |
| pri-miR-93-RT-R | TTGGCAGAGAGAACGTGTCC |
| PTEN-RT-F | GTTTACCGGCAGCATCAAAT |
| PTEN-RT-R | CCCCCACTTTAGTGCACAGT |
| E2F1-RT-F | GCCACTGATCTGCCACCATAG |
| E2F1-RT-R | CTGCCCATCCGGGACAAC |
| p21-RT-F | TTAGCAGCGGAACAAGGAGT |
| p21-RT-R | AGCCGAGAGAAAACAGTCCA |
| GAPDH-RT-F | TTCACCACCATGGAGAAGGC |
| GAPDH-RT-R | GGCATGGACTGTGGTCATGA |
Western blot
Cell lysates were prepared by incubation in lysis buffer (Sigma).Fifty micrograms of protein from each lysate was separated by SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad laboratories). Primary antibodies were used to detect PTEN, E2F1, p21 (Cell Signaling Technology, Inc..) and GAPDH (EASYBIO). Anti-rabbit (for PTEN, E2F1, and p21) or anti-mouse (for GAPDH) secondary antibodies were used and visualized with the ECL substrate (CWBIO).
RNA sequencing
Total RNA from miR-93-m33 and wild-type controls were isolated in cultured cells using TRIzol regant (Sigma) and treated with DNase I (NEB). Library construction and sequencing for mRNA and small RNA were both carried out at the high-throughput sequencing center of the Biodynamic Optical Imaging Center, Peking University. As for mRNA sequencing, the RNA-seq reads were aligned to the human reference genome (hg19) using TopHat with default parameters. The bam files were used as input of Cufflink and Cuffdiff to detect differentially-expressed genes. And the small RNA sequencing data were analyzed by BGI Tech Solutions Co., Ltd.
Statistical Analysis
All data are shown as mean ± SEM. Statistical analysis was performed by t-test to evaluate single-factor differences between 2 sets of data. P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Chuanyun Li and Dr. Peng Yu for their kind help in bioinformatics analysis.
Funding
This study was supported by the National High-tech R&D Program of China (2012AA022501), and the National Natural Science Foundation of China (81273422 and 81170097).
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 2. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597-610; PMID:20661255 [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012; 3:1; PMID:22230293; http://dx.doi.org/ 10.1186/1758-907X-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17∼92 Family of miRNA Clusters. Cell 2008; 132:875-86; PMID:18329372; http://dx.doi.org/ 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell 2007; 129:303-17; PMID:17397913; http://dx.doi.org/ 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- 7. Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev 2010; 24:1173-85; PMID:20466812; http://dx.doi.org/ 10.1101/gad.1915510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callis TE, Pandya K, Seok HY, Tang R-H, Tatsuguchi M, Huang Z-P, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 2009; 119:2772-86; PMID:19726871; http://dx.doi.org/ 10.1172/JCI36154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Z. miRNA Knockout Technology. In: Wang Z, ed. MicroRNA Interference Technologies: Springer; Berlin Heidelberg, 2009:175-82. [Google Scholar]
- 10. Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013; 31:397-405; PMID:23664777; http://dx.doi.org/ 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR Cas in zebrafish. Nucleic Acids Res 2013; 41:e141; PMID:23748566; http://dx.doi.org/ 10.1093/nar/gkt464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012; 337:816-21; PMID:22745249; http://dx.doi.org/ 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31:230-2; PMID:23360966; http://dx.doi.org/ 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 14. Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res 2013; 23:465-72; PMID:23528705; http://dx.doi.org/ 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu X, Chang N, Wang X, Zhou F, Zhou X, Zhu X, Xiong JW. Heritable gene-targeting with gRNA Cas9 in rats. Cell Res 2013; 23:1322-5; PMID:24145754; http://dx.doi.org/ 10.1038/cr.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011; 39:D152-7; PMID:21037258; http://dx.doi.org/ 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim YK, Wee G, Park J, Kim J, Baek D, Kim JS, Kim VN. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol 2013; 20:1458-64; PMID:24213537; http://dx.doi.org/ 10.1038/nsmb.2701 [DOI] [PubMed] [Google Scholar]
- 18. Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 2010; 3:ra29; PMID:20388916; http://dx.doi.org/ 10.1126/scisignal.2000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olive V, Li Q, He L. mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev 2013; 253:158-66; PMID:23550645; http://dx.doi.org/ 10.1111/imr.12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Tan W, Neo TWL, Aung MO, Wasser S, Lim SG, Tan TM. Role of themiR-106b-25microRNA cluster in hepatocellular carcinoma. Cancer Sci 2009; 100:1234-42; PMID:19486339; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01164.x [DOI] [PubMed] [Google Scholar]
- 21. Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9 RNA-mediated gene targeting. Cell Res 2013; 23:720-3; PMID:23545779; http://dx.doi.org/ 10.1038/cr.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene 2010; 30:806-21; PMID:20956944; http://dx.doi.org/ 10.1038/onc.2010.465 [DOI] [PubMed] [Google Scholar]
- 23. Ma H, Wu Y, Choi JG, Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc Natl Acad Sci U S A 2013; 110:20687-92; PMID:24297910; http://dx.doi.org/ 10.1073/pnas.1311639110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006; 125:887-901; PMID:16751099; http://dx.doi.org/ 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 25. Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 2005; 24:138-48; PMID:15565168; http://dx.doi.org/ 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]