Abstract
B cells play a key role in generation of protective immunity against rotavirus infection, a major cause of gastroenteritis in children. Current RV vaccines are less effective in developing countries compared to developed countries. Commensals/probiotics influence mucosal immunity, but the role of early gut colonizing bacteria in modulating intestinal B cell responses to RV vaccines is largely unknown. We co-colonized neonatal gnotobiotic pigs, the only animal model susceptible to HRV diarrhea, with 2 dominant bacterial species present in the gut of breastfed infants, Lactobacillus rhamnosus strain GG and Bifidobacterium animalis lactis Bb12 to evaluate their impact on B cell responses to an attenuated (Att) human rotavirus (HRV) Wa strain vaccine. Following HRV challenge, probiotic-colonized, AttHRV vaccinated piglets had significantly lower fecal scores and reduced HRV shedding titers compared to uncolonized, AttHRV vaccinated pigs. The reduction in HRV diarrhea was significantly correlated with higher intestinal IgA HRV antibody titers and intestinal HRV-specific IgA antibody secreting cell (ASC) numbers in probiotic-colonized, AttHRV vaccinated pigs compared to uncolonized, vaccinated pigs. The significantly higher small intestinal HRV IgA antibody responses coincided with higher IL-6, IL-10 and APRIL responses of ileal mononuclear cells (MNCs) and the immunomodulatory effects of probiotics genomic DNA on TGF-β and IL-10 responses. However, serum RV IgG antibody titers and total IgG titers were significantly lower in probiotic-colonized, AttHRV vaccinated pigs compared to uncolonized, vaccinated pigs, both pre- and post-challenge. In summary, LGG and Bb12 beneficially modulated intestinal B cell responses to HRV vaccine.
Keywords: B cell responses, human rotavirus, neonatal diarrhea, probiotics, vaccine
Abbreviations
- APRIL
a proliferation-inducing ligand
- ASC
antibody secreting cell
- AttHRV
attenuated human rotavirus
- AUC
area under the curve
- Bb12
Bifidobacterium lactis Bb12
- FFU
fluorescent foci forming unit
- Gn
gnotobiotic
- HRV
human rotavirus
- LGG
Lactobacillus rhamnosus strain GG
- MNCs
mononuclear cells
- PCD
postchallenge day
- PID
postinoculation day
- Vac+Pro
vaccinated probiotic colonized group
- RAM
rat anti-mouse
- RV
rotavirus
- Vac
3XAttHRV Wa vaccinated only group
- VirHRV
virulent human rotavirus
- PBCD
post bacterial colonization day.
Introduction
The intestinal microbiota plays a significant role in maturation of mucosal immunity in neonates.1 Recent studies have shown that composition of the microbiota also influences gut immune responses2,3 and the immunomodulatory effects of commensals or probiotics are strain dependent.4 Mice monocolonized with L. johnsonii produced higher antigen-specific IgA antibody responses as compared to animals colonized with L. paracasei.5 Further, the colonization patterns of commensals also had significant effects on B cell responses in neonates.6 In infants, early colonization with Escherichia coli and Bifidobacteria was associated with increased memory B cells, but colonization with Staphylococcus aureus was negatively associated with number of memory B cells.7 Most of the previous studies mainly investigated the effects of specific commensals on total intestinal IgA responses.8,9 The impact of individual or a combination of early colonizing commensals in modulating intestinal B cell responses to enteric pathogens such as rotavirus, as well as enteric vaccines, is largely unexplored.
Rotavirus (RV) is a leading cause of diarrhea in children worldwide. Rotavirus infection causes ∼450,000 deaths in children <5 years of age annually worldwide. Approximately 85% of the rotavirus-associated deaths occur in developing countries.10,11 The efficacy of currently available RV vaccines is >80% in developed countries,12-14 but it is only ∼50% in developing countries.11 B cells play a significant role in generation of protective immunity against RV infection.15 Thus modulating adaptive B cell responses using beneficial commensals may be a cost effective strategy to prevent RV infection. Probiotic intervention is a potential strategy to modulate adaptive immunity to enteric infections/vaccines. Indeed, the composition of commensals in the small intestine significantly influenced antibody responses to enteric vaccines in children.16-20 Bifidobacteria and Lactobacilli are among the dominant members of the gut microbiota in breastfed infants21,22 and among the most commonly used probiotic bacterial species. Each species possesses various immunomodulatory properties.23-25 Supplementation of Bifidobacterium lactis Bb12 increased both total26 and poliovirus-specific fecal IgA levels in children.25 Interactions among probiotics affect their intestinal colonization patterns as well as their effects on gut immunity. An earlier study reported that initial Lactobacillus rhamnosus strain GG (LGG) colonization promotes subsequent colonization of Bifidobacterium in children.27 However, the effects of co-colonization by Bifidobacterium and Lactobacillus combined, on intestinal B cell responses to an oral HRV vaccine and on resolution of virulent HRV (VirHRV) infection is largely unknown.
Neonatal gnotobiotic (Gn) piglets are susceptible to HRV induced diarrhea and also their immune responses, gut physiology and milk diet are similar to that of infants.28 Thus Gn piglets are an important animal model to assess the impact of probiotics on a HRV vaccine or infection. In our earlier study, we demonstrated that dual colonization of L. acidophilus and L. reuteri had no impact on HRV specific immunity29 which may have been the result of a very short interval (2 days) between LAB colonization and HRV challenge of Gn piglets.29 In this study, we investigated the effect of co-colonization of Lactobacillus rhamnosus strain GG and Bifidobacterium lactis Bb12 on B cell responses to AttHRV Wa strain vaccine in a neonatal Gn pig model. We show that dual-colonization of Gn pigs with LGG and Bb12 significantly augmented small intestinal B cell responses including HRV specific IgA responses in vaccinated animals post-challenge. The enhanced intestinal HRV IgA antibody responses were inversely correlated with reduced fecal scores and also coincided with complete protection against virus shedding in probiotic colonized, vaccinated piglets post-challenge. In addition, LGG+Bb12 also enhanced intestinal as well as systemic total IgA responses.
Results
LGG and Bb12 colonized Gn piglets and the probiotics decreased fecal scores and virus shedding post-virulent HRV challenge
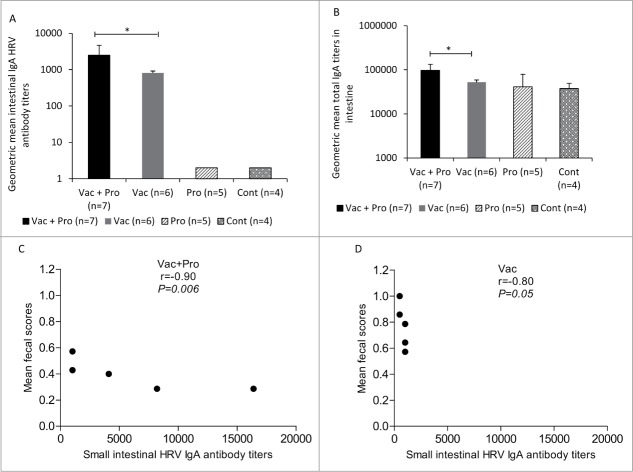
Co-colonization of LGG and Bb12 was confirmed by QPCR in rectal swab fluids from representative pigs from each group. Both LGG and Bb12 were recovered from fecal samples of all colonized Gn piglets throughout the study period,30 and the uncolonized piglets remained bacteriologically sterile. In addition, recovery of the probiotic bacteria from duodenum, jejunum, ileum, cecum and colon tissues from representative Pro and Vac+Pro piglets at PID34/PCD7 confirmed colonization of these probiotics in the intestinal tract of both groups of gnotobiotic piglets.30 Vac+Pro piglets had significantly (P < 0.05) lower mean cumulative fecal scores compared to Vac piglets.31 Fecal virus shedding was undetectable in Vac+Pro piglets but Vac piglets had higher incidence of fecal virus shedding.31 Further, a stronger significantly inverse correlation was observed between small intestinal HRV IgA titers and mean fecal scores in Vac+Pro piglets (Fig. 1C) compared with Vac piglets (Fig. 1D). This suggests that amelioration of RV severity in vaccinated probiotic colonized piglets may be partly mediated by probiotics-augmented intestinal B cell responses.
Figure 1.
Intestinal HRV specific-IgA antibody and total IgA responses and correlation between small intestinal HRV IgA antibody titers and mean diarrhea scores at post-challenge (PID34/PCD7). Geometric mean titers (GMT) of HRV specific-IgA (A) and total IgA (B) antibody titers in intestinal contents of Gn pigs vaccinated with AttHRV with or without LGG + Bb12 probiotic colonization at post-HRV challenge (PID34/PCD7)(one-way ANOVA followed by Duncan's multiple range test on log10 transferred titers, *P < 0.05). Correlations between SIC HRV IgA antibody titers and mean diarrhea scores (C and D) were determined using Spearman correlation coefficient. Error bars indicate SEM. PID-Post-inoculation day; PCD-Post-challenge day. SIC-Small intestinal contents; LIC-Large intestinal contents.
LGG+Bb12 probiotics significantly enhanced small intestinal HRV IgA antibody and total IgA responses in vaccinated piglets post-challenge
In the small intestine (Fig. 1A), Vac+Pro piglets had significantly (P < 0.05) higher HRV IgA antibody titers compared with Vac piglets post-challenge (PID34/PCD7). Thus LGG+Bb12 probiotics enhanced intestinal IgA antibody responses. Nonparametric Spearman correlation coefficient analyses revealed a significant negative correlation between small intestinal HRV IgA antibody titers and mean diarrhea scores that was higher in Vac+Pro (Fig 1C) compared to Vac piglets post-VirHRV challenge (Fig 1D). This finding suggests that the enhanced small intestinal HRV IgA antibody responses may play a role in ameliorating the severity of HRV infection in Vac + Pro compared to Vac piglets. Total IgA titers in the small intestine were also significantly higher in Vac+Pro piglets compared to Vac piglets post-challenge (PID34/PCD7) (Fig. 1B). These findings were further supported by the presence of higher total intestinal IgA IgSCs in Vac+Pro piglets compared to Vac piglets (Fig 2C). Thus LGG+Bb12 probiotics enhanced intestinal total IgA and HRV IgA antibody responses.
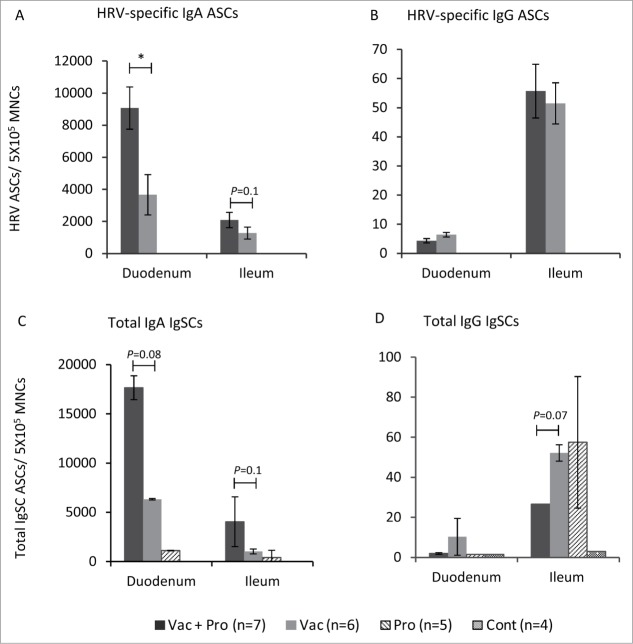
Figure 2.
Small intestinal HRV IgA and IgG ASC and total IgA IgG IgSC responses at post-challenge (PID34/PCD7). HRV-specific ASC (A and B) and total IgSC (C and D) responses in duodenum and ileum of Gn pigs vaccinated with AttHRV with or without LGG + Bb12 probiotics colonization post-HRV challenge (PID34/PCD7). Data represent the mean numbers of HRV-specific ASC and total IgSC per 5 × 105 MNC, respectively (Kruskal–Wallis Test, *P < 0.05). Error bars indicate SEM. PID-Post-inoculation day. PCD-Post-challenge day.
LGG+Bb12 probiotics significantly elevated duodenal HRV IgA ASC in vaccinated piglets post-challenge
HRV IgA and IgG ASC were determined in ileum and duodenum. Significantly (P < 0.05) higher duodenal HRV IgA ASC were observed in Vac+Pro piglets compared to Vac piglets post-challenge (Fig. 2A). A similar trend of higher HRV specific IgA ASC in Vac+Pro compared to Vac piglets was also observed in ileum (Fig. 2A). The enhanced small intestinal HRV IgA ASC (Fig. 2A) coincided with enhanced small intestinal HRV IgA antibody responses (Fig. 1A). However the Vac+Pro piglets had lower duodenal HRV IgG ASC (Fig. 2B) and HRV IgG antibody-responses (Fig. 4A) compared to Vac piglets post-challenge.
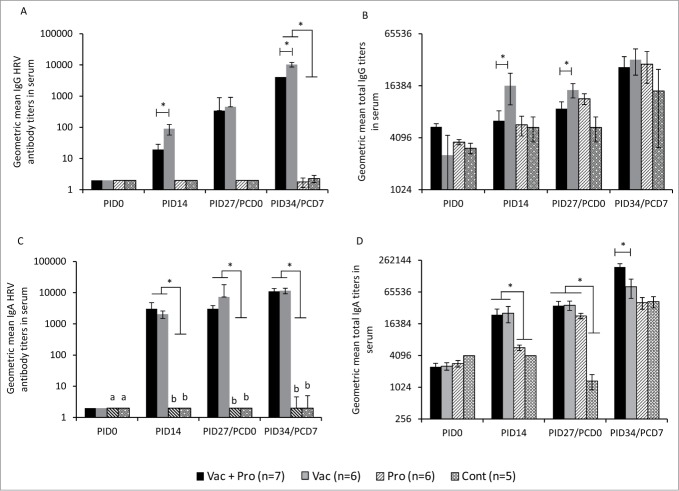
Figure 4.
LGG and Bb12 probiotic colonization modulates serum HRV IgG antibody responses, and total serum IgA and IgG responses. Geometric mean titers (GMT) of IgG (A) and IgA (C) antibody to HRV, total IgG (B) and IgA (D) gemometric mean titers in serum of Gn pigs vaccinated with AttHRV with or without LGG + Bb12 probiotics colonization at indicated PID/PCD (one-way ANOVA followed by Duncan's multiple range test on log10 transferred titers,*P < 0.05). PID-Post-inoculation day. PCD-Post-challenge day.
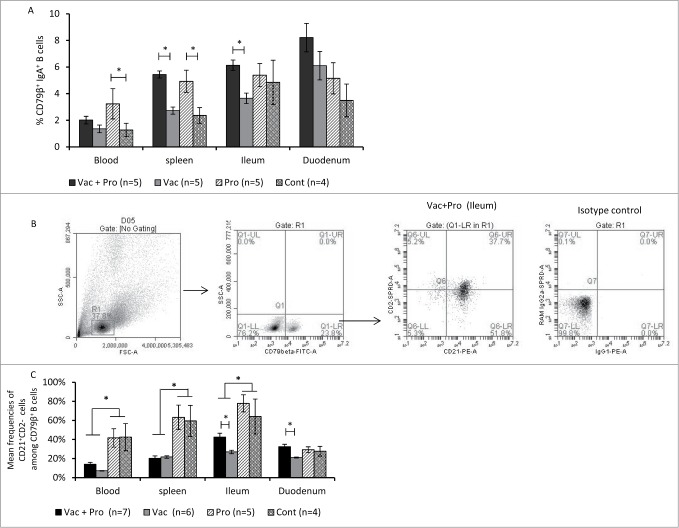
LGG+Bb12 probiotics enhanced the frequency of activated B cells as well as IgA+ B cells in the small intestine
Vac+Pro piglets had significantly higher frequencies of IgA+ B lymphocytes in spleen and ileum compared to that of Vac piglets. In addition, probiotic colonization alone resulted in significantly higher frequencies of IgA+ B lymphocytes in blood and spleen in comparison to unvaccinated, non-colonized control piglets (Fig. 3A). Significantly higher frequencies of activated CD21+CD2− B cells were observed in ileum and duodenum in Vac+Pro compared to Vac piglets (Fig. 3C) based on CD2 and CD21 expression (CD21+CD2− B cells are defined as activated B cells in pigs).32 The enhanced activation of B cells coincided with the higher frequencies of mature conventional dendritic cells (cDCs) as well as plasmacytoid DCs (pDC)30 and higher intestinal IgA responses in Vac+Pro piglets compared to Vac piglets. Further, significantly higher frequencies of activated B cells were observed in unvaccinated piglets compared to vaccinated piglets in ileum, blood and spleen post-challenge (Fig. 3C). RV is known to activate B cells33,34 and severe RV infection, as indicated by significantly higher RV shedding,31 coincided with the increased frequency of activated B cells in the unvaccinated piglets compared to vaccinated piglets.
Figure 3.
Probiotics colonization enhanced frequencies of intestinal activated B cells and IgA+ B cells post-challenge (PID34/PCD7). Mean frequencies of CD79β+IgA+ B cells among lymphocytes (A), representative dot plot (B) and mean of frequencies (C) CD21+CD2− B cells among CD79β+ B cells in peripheral blood, spleen, ileum and duodenum of Gn pigs vaccinated with AttHRV with or without LGG + Bb12 probiotic colonization post-HRV challenge (PID34/PCD7). Data represent the mean frequencies of B cells ± SEM (Kruskal–Wallis Test, *P < 0.05). PID-Post-inoculation day. PCD-Post-challenge day.
LGG+Bb12 probiotics decreased serum HRV IgG antibody responses
Serum HRV IgG antibody titers were significantly (P < 0.05) lower in Vac+Pro piglets compared to Vac piglets at pre- (PID14) and post- (PID34/PCD7) challenge (Fig. 4A). The differential systemic HRV IgG antibody responses were associated with significant differences in systemic IL-4 cytokine concentrations between Vac+Pro and Vac piglets.31 However probiotics colonization had no effect on serum HRV IgA antibody responses in the vaccinated piglets (Fig. 4C). As expected, Vac piglets with or without probiotic colonization had significantly (P < 0.05) higher HRV IgG (Fig 4A) and IgA (Fig 4C) antibody titers compared to probiotic-colonized or control piglets at all PID or PCD time-points.
Systemic total Ig responses were modulated by LGG+Bb12 colonization
Total serum IgG titers also were consistently lower (Fig. 4B) post-probiotic colonization and were significantly lower at PID27/PCD0 (Fig 4B) in Vac+Pro piglets compared to Vac piglets. However, total serum IgA titers were significantly higher in Vac+Pro piglets compared to Vac piglets at PID34/PCD7 (Fig. 4D). In addition, the vaccinated piglets had significantly higher total serum IgA titers compared to unvaccinated piglets at PID14 irrespective of probiotic colonization (Fig. 4D). Pre-challenge (PID27/PCD0), the significantly higher (17-fold) total IgA titers in Pro piglets compared to Cont piglets (Fig. 4D) also indicates the marked effect of LGG+Bb12 probiotics on systemic total IgA antibody responses.
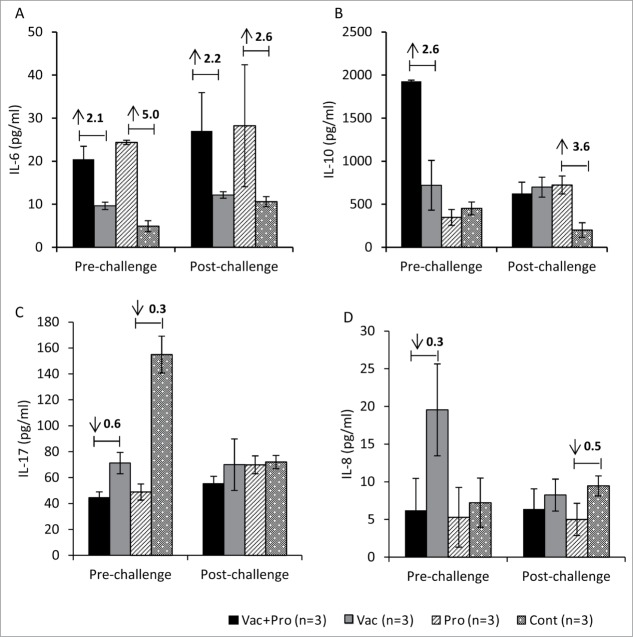
LGG+Bb12 probiotics increased IL-6 and IL-10 responses and decreased IL-8 and IL-17 responses in ileum
To assess immune responses at the induction site (ileum) of the small intestine,35 cytokine responses were measured in culture supernatants from HRV antigen re-stimulated ileal MNCs isolated from Vac+Pro, Vac, Pro and Cont piglets (Fig. 5). Changes (Vac+Pro vs Vac and Pro vs Cont) in cytokine responses were determined to assess effects of probiotics on induction of cytokines associated with IgA responses. Higher IL-6 responses were observed in probiotic colonized piglets compared to uncolonized piglets regardless of vaccination pre- and post-challenge (Fig. 5A). Pre-challenge, IL-10 levels were also higher in Vac+Pro piglets compared to Vac piglets (Fig. 5B). In contrast to IL-6 and IL-10 responses, a trend of lower IL-8 and IL-17 cytokine responses was observed in Vac+Pro piglets compared to Vac piglets pre-challenge (Fig. 5C and D). These results suggest that LGG+Bb12 colonization may enhance IL-6 and IL-10 cytokine responses which may in turn be involved in enhancing small intestinal RV specific IgA responses.
Figure 5.
Ileal MNCs from Gn pigs vaccinated with AttHRV with or without LGG + Bb12 probiotics colonization pre- and post-challenge were co-cultured with HRV antigen under in vitro condition to determine cytokine responses. Ileal MNCs were treated with HRV antigen (12 μg/ml) for 48 h at 37°C and culture supernatants were collected to quantify concentrations of IL-6, IL-10, IL-17 and IL-8 (A–D) cytokines. Differences in cytokine levels between groups (Vac+Pro vs Vac and Pro vs Cont) were expressed as fold changes (Fold change with values greater than 1 representing increased levels and values less than 1representing decreased levels) as indicated by values at top of bar diagram.
LGG+Bb12 probiotics bacterial DNA induces TGF-β, IL-10 and IL-12 cytokines in ileal MNC
Soluble factors or microbe associated molecular patterns of beneficial bacteria such as CpG DNA of probiotic bacterial genomic DNA might mediate some of the observed immunomodulatory and enhanced IgA effects. Both Bifidobacteria and LGG have high GC percentage in their genomic DNA.36,37 Also, significantly higher small intestinal TLR9 expression was observed in vaccinated, probiotic colonized piglets compared to vaccinated, uncolonized piglets in our study.30 TLR9 recognizes bacterial CpG motifs38 and the higher expression of TLR9 of MNCs might have played a role in eliciting higher mucosal IgA responses in Vac+Pro piglets compared to Vac piglets. Treatment of ileal MNCs from naive Gn piglets (no prior exposure to any bacteria/virus) with probiotic bacterial genomic DNA (1:1 ratio of LGG and Bb12 genomic DNA) resulted in significantly higher levels of TGF-β (Fig. 6A), IL-10 (Fig. 6B) and IL-12 (Fig. 6C) cytokines under in vitro conditions. This indicates that probiotic bacterial genomic DNA may have induced a favorable microenvironment for enhanced IgA production in probiotic colonized, vaccinated piglets. In addition, pretreatment of MNCs with chloroquine, a known inhibitor of TLR9, resulted in complete inhibition of probiotic bacterial DNA induced TGF-β and IL-12 cytokine responses, but had no effect on IL-10 production. These results suggest that bacterial genomic DNA may modulate production of factors associated with IgA responses through a TLR9 dependent manner.
Figure 6.
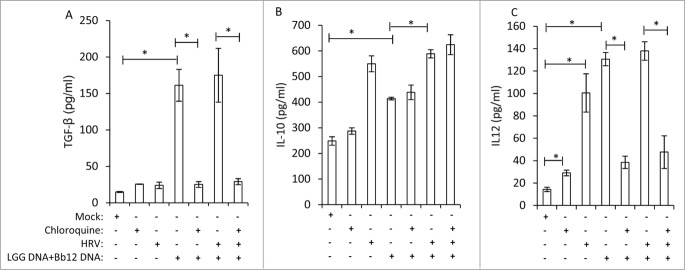
LGG and Bb12 probiotic bacterial DNA induces TGF-β, IL-10 and IL-12 cytokines in ileal MNC. Ileal MNCs from 28 day old control Gn piglets (no prior exposure to any bacteria/virus) were treated with a combination of LGG- (25 μg/ml) and Bb12-genomic bacterial DNA (25 μg/ml) in presence or absence of HRV antigen (12 μg/ml) for 48 h at 37°C, and supernatants were collected for TGF-β, IL-10 and IL-12 determination. When indicated, MNCs were pretreated with chloroquine (10 μM), a known inhibitor of TLR9, for 30 m prior to probiotic bacterial genomic DNA treatment. Results are mean ± SEM (n = 4, One-way ANOVA, *P < 0.05).
The increased small intestinal IgA antibody responses in probiotic-colonized piglets may also be related to induction of T-cell independent IgA inducing factors.39,40 To assess probiotics effect on T-cell independent IgA inducing factors such as APRIL and BAFF, ileal MNCS from control Gn piglets were treated with live LGG and Bb12 under in vitro condition. Probiotics treatment had no effect on BAFF expression in MNCs from naive piglets (data not shown). However, treatment of MNCs with live LGG and Bb12 probiotics or HRV antigen resulted in 15 (significant)- and 1.7-fold higher APRIL mRNA levels, respectively, compared to mock treatment (Supplementary Fig. 1A). Further, treatment of MNCs with a combination of the live probiotics and HRV antigen resulted in significantly higher APRIL mRNA (27-fold) compared to mock or LGG+Bb12 treatments (Supplementary Fig. 1A). This suggests a possible synergistic effect between LGG+Bb12 probiotics and HRV on induction of APRIL in ileum. Probiotic bacterial genomic DNA had no effect on APRIL and BAFF gene expression in MNCs under in vitro conditions (data not shown). Consistent with this in vitro finding, significantly higher levels of APRIL mRNA were also observed in ileal MNCs from Vac+Pro piglets compared to that of Vac piglets (Supplementary Fig. 1B). These findings suggest that APRIL may be involved in enhanced mucosal IgA production.
Discussion
Recent studies have shown a role of intestinal commensals in the development of mucosal immunity in neonates. In this study, we determined the specific impact of a combination of probiotics on B cell responses to RV vaccine post-VirHRV challenge. Our study demonstrates that co-colonization of LGG and Bb12 probiotics enhanced small intestinal HRV specific- and total IgA-responses and induced differential effects on systemic IgA- and IgG-antibody responses in Gn piglets.
Co-colonization of LGG+Bb12 probiotics enhanced both HRV specific- and total IgA responses in the small intestine. Although the immunomodulatory effects of either LGG or Bb12 on mucosal antibody responses were assessed in only a few previous studies,24,25 it is unknown whether these probiotics exert their effects directly on host immunity or act through modulating the gut microbiota. Results from our Gn piglet model system (devoid of microflora) revealed that the observed effects of LGG+Bb12 probiotics on intestinal IgA responses could be mediated by direct modulation of host immune responses by the probiotics.
The potential mechanism(s) for the probiotic-enhanced intestinal IgA HRV antibody- and total IgA-responses or intestinal HRV IgA ASC may be through conditioning the intestinal microenvironment including modulating responses of dendritic cells to mediate increased IgA production. Surface MHC II expression on dendritic cells (DCs) is indicative of dentritic cell maturation41 and in our study, significantly higher frequencies of mature pDCs as well as cDCs were observed in ileum of LGG+Bb12 colonized, vaccinated piglets compared to uncolonized, vaccinated piglets.30 Further, comparison of maturation status between pDCs and cDCs revealed that nearly all the pDCs have a mature phenotype, but only 10% of cDCs possess such a phenotype. These results suggest that the probiotics indeed enhanced maturation of intestinal DCs and the probiotic induced maturational changes in pDCs may have played a role in enhancing IgA antibody responses in probiotic colonized piglets compared to uncolonized piglets.34,42
LGG+Bb12 colonized, vaccinated piglets had significantly higher TLR9 expression and lower TLR4 and TLR2 expression levels in small intestinal MNCs compared to that of uncolonized, vaccinated piglets in small intestine.30 Thus, probiotics-induced changes in expression of pattern recognition receptors may potentially modulate the IgA responses. Microbe associated molecular patterns such as bacterial nonmethylated CpG motifs can stimulate B cells and enhance antibody responses without BCR recognition.43 Genomic DNA of both Bifidobacteria and LGG probiotics possess CpG motifs with immunomodulatory effects on B cells.36,44 These motifs may have played a role in enhancing mucosal IgA production as observed in earlier studies.45,46 Further, the ability of genomic DNA from LGG and Bb12 to induce cytokines that mediate IgA antibody responses such as TGF-β and IL-1047 as shown in our study also suggest that these probiotics might possess immunomodulatory effects on IgA inducing factors in the small intestine. In addition, the complete inhibition of probiotic bacterial genomic DNA induced TGF-β response by chloroquine pretreatment in ileal MNCs also suggests possible involvement of TLR9 in induction of TGF-β. Further, colonization of various bifidobacterial strains from infants resulted in increased secretion of IL-10 and TGF-β cytokines in a Gn mouse model.48 Collectively, these probiotic induced changes in MNCs may be involved in augmenting intestinal IgA antibody responses.
In addition, the increased IgA production may have occurred through either T-cell dependent or T-cell independent processes or a combination of both processes. In duodenum, significantly higher CD4 T cells as well as activated CD4 T cells were observed in Vac+Pro piglets as compared to Vac piglets pre-HRV challenge.31 The higher frequency of activated CD4 T cells in LGG + Bb12 colonized, vaccinated piglets may act as a source of the cytokines that modulate IgA antibody responses. Others have shown that the cytokines TGF-β,49 IL-1047 and IL-650 are involved in stimulating intestinal IgA responses. Indeed, our finding of higher IL-6 and IL-10 cytokine responses of ileal MNCs from probiotic colonized, vaccinated piglets supports their potential role in mediating IgA induction in small intestine. The increased CD4 T cells along with higher IgA inducing cytokines such as IL-6 and IL-10 might have induced a favorable gut microenvironment for augmented intestinal IgA HRV antibody responses in vaccinated, probiotic colonized piglets.
T-cell independent IgA responses at mucosal sites also play a role in immunity against RV infection.51 BAFF and APRIL are secreted mainly by monocytes, dendritic cells, and intestinal epithelial cells52,53 and involved in generation of T-cell independent IgA responses.54 The higher APRIL responses in MNCs of Vac+Pro piglets compared to Vac piglets also suggest the possible involvement of those factors in augmenting HRV specific intestinal IgA antibody responses in the probiotic-colonized, vaccinated piglets post-HRV challenge. These results were further confirmed by our in vitro experiment in which live LGG+Bb12 probiotic treatment of ileal MNCs from naive Gn piglets induced significantly higher APRIL responses which were synergistically enhanced by co-treatment with HRV antigen. These findings and our recent observation of translocation of LGG to the mesenteric lymph nodes in LGG monnocolonized gnotobiotic piglets (unpublished) suggest a potential role of the probiotics in enhancing expression of APRIL in intestinal tissues. In addition, a higher percentage of intestinal B cells were IgA+ in probiotic-colonized piglets, which also indicates potential involvement of APRIL in augmenting intestinal IgA responses. These findings further coincided with the enhanced intestinal HRV IgA antibody responses.
Higher B cell activation in Vac+Pro compared to Vac piglets suggests that LGG+Bb12 probiotics played a role in activation of small intestinal B cells. Previous studies have shown that both genomic DNA of lactobacillus36 or commensal bacteria32 can activate B cells and the enhanced activation of B cells in Vac+Pro piglets may be induced by probiotic derived factors such as genomic DNA. Furthermore, a significantly higher frequency of activated B cells was observed post-challenge in the susceptible unvaccinated compared with the protected vaccinated piglets, regardless of probiotic colonization. The more severe rotavirus infection in unvaccinated piglets may have caused the increased activation of B cells as observed in a mouse model.55 Indeed, the presence of significantly higher IFN-α,30 a B cell activating factor that is induced during RV infection,34 coincided with the higher B cell activation that we observed in unvaccinated piglets compared to vaccinated piglets.
Co-colonizaton of LGG and Bb12 probiotics had differential immunomodulatory effects on systemic HRV IgG antibody and total IgG responses compared to IgA responses in the Gn piglet model. Serum HRV IgG antibody titers were lower in the LGG+Bb12 colonized, vaccinated piglets compared to uncolonized, vaccinated piglets at all-time points post-bacterial colonization. Furthermore, more marked significant reductions in HRV specific IgG responses (five-fold lower) were observed at PID14 in comparison to differences in the IgG responses between Vac+Pro and Vac piglets during later time points. These observations are consistent with our earlier finding in which Lactobacillus acidophilus colonized, AttHRV vaccinated piglets had lower spleen HRV IgG ASC compared to uncolonized, vaccinated piglets.56 These findings are also supported by an earlier study in which oral administration of Bifidobacteria to mice immunized with RV resulted in lower serum RV IgG antibody responses but enhanced serum RV specific IgA antibody responses as compared to immunized, non-probiotic colonized animals.57 Consistent with these findings, a recent study also demonstrated that administration of a probiotic mixture including Lactobacillus and Bifidobacteria decreased antigen specific serum IgG antibody responses in a murine autoimmune disease model.58 It appears that the Lactobacillus and Bifidobacterium probiotics may have an inherent capacity to induce systemic immune responses that favor systemic IgA- but not IgG-antibody responses. These results were further supported by presence of significantly higher probiotic specific IgA (supplementary Fig. 2A) - and lower probiotics specific IgG-responses (supplementary Fig. 2B) in serum of Vac+Pro piglets compared to Pro piglets at PID34/PCD7. One potential explanation is that the probiotic modulated systemic cytokine responses may be involved in the lower systemic IgG responses in the Vac+Pro piglets. In our study, serum IL-4 levels were significantly lower in Vac+Pro compared to Vac piglets post-challenge.31 An earlier study59 reported that IL-4 inhibits IL-21/CD40L mediated IgA isotype switching but enhances the IgG isotype switching process. Thus, lower systemic IL-4 levels in Vac+Pro piglets compared to Vac piglets might have caused the lower systemic IgG antibody responses. Further, dose dependent effects of IL-4 at lower concentrations on IgG responses (supplementary Fig. 3A and B) of RV antigen stimulated splenic B cells from AttHRV vaccinated pigs also suggest the potential role of IL-4 in differential systemic IgG antibody responses. Total IgG responses also followed a similar pattern in which LGG+Bb12 colonized, vaccinated piglets had lower total IgG titers as compared to uncolonized, vaccinated piglets. Lower serum HRV specific- and probiotic specific-IgG antibodies might have contributed to the lower serum total IgG responses in probiotic colonized, vaccinated piglets. Vac+Pro and Vac piglets had comparable serum total IgA titers pre-challenge (PID27/PCD0), but Vac+Pro piglets had higher serum total IgA titers at PID34/PCD7 indicating a potential synergistic interaction between the immunomodulatory effects of probiotics and VirHRV infection.
Experimental evidence in animal models has shown that B cell responses play a critical role in development of long-lasting protective immunity against RV infection.15,60 Our results suggest that LGG and Bb12 probiotics, which effectively colonized the Gn piglets, beneficially modulated B cell responses to HRV vaccine. One caveat of this study is that gnotobiotic piglets without other confounding microbiota were used to determine the specific beneficial effects of only LGG+Bb12 on rotavirus induced immunity. Whether similar effects will be observed using the selected probiotics in the presence of a complex gut microbiota remains to be determined. Colonization of beneficial probiotics prior to RV vaccination in children may be achieved directly through treatment of children or through maternal supplementation of probiotics for indirect treatment of infants. Schultz and co-workers,61 reported that maternal supplementation of LGG probiotics resulted in colonization of the LGG in infants for at least 12 months. In addition, maternal supplementation of LGG also promoted a bifidobacteria profile in infants without affecting gut microbiota diversity.62
Materials and Methods
Probiotic bacterial strains
The selected probiotic strains, Lactobacilli rhamnosus GG strain ATCC 53103 (ATCC, Manassas, VA) and Bifidobacterium animalis subsp. lactis Bb12 (Christian Hansen Ltd., Horsholm, Denmark) were used to colonize Gn piglets. Growth and enumeration of colony forming units (CFU) in bacterial cultures prior to feeding the probiotics to the piglets was done as described previously.29
Experimental design
All studies were approved by The Ohio State University Institutional Animal Care and Use Committee. Cesarean-derived Gn piglets from near-term sows were maintained in sterile isolators as described previously.63 The Gn piglets were assigned to one of the following 4 groups: Probiotic-colonized and 3XAttHRV Wa vaccinated (Vac+Pro, n = 7); 3XAttHRV Wa vaccinated and uncolonized (Vac, n = 6), unvaccinated and probiotic-colonized (Pro, n = 5), and unvaccinated and uncolonized negative controls (Cont, n = 4). For probiotic colonization, pigs were first inoculated at 3 days of age (post bacterial colonization day, PBCD0) with Bb12 at a dose of 105 CFU; subsequently, the Bb12 colonized piglets were inoculated at 5 days of age (PBCD2) with both LGG and Bb12 at a 1:1 ratio, and at a dose of 105 CFU of each bacteria per pig. For vaccination, cell culture adapted AttHRV Wa strain (G1P1A[8]) vaccine was given orally at a dose of 5 × 107 fluorescent-forming units (FFU) at 6- (Post inoculation day, PID0), 15- (PID9) and 26- (PID20) days of age. Serum samples were collected to assess HRV antibody responses at PID0, 14, 27 and 34. For VirHRV challenge, piglets were challenged with 105FFU VirHRV Wa strain at PID27 (Pre-challenge) and subsets euthanized at PID27/post-challenge day 0 (PCD0) and PID34/PCD7. To measure immune responses in intestinal and systemic lymphoid tissues, mononuclear cells (MNCs) were isolated from blood, spleen, duodenum, and ileum as previously described.31
Fecal probiotic counts, clinical signs and virus shedding
The total LGG and Bb12 colonies in rectal swab fluids and tissues and the specific colonization patterns of the probiotic bacteria were determined as described previously.29,31 VirHRV challenged piglets were examined daily from PCD0 to PCD7 to assess fecal consistency, duration of diarrhea60 and fecal virus shedding, all of which were determined as described previously.60
ELISA, ELISPOT and flow cytometry assays
HRV specific IgA and IgG antibody responses and total IgA and IgG antibody responses were measured by ELISA as described previously.60,64 LGG and Bb12 specific-IgA and -IgG responses were measured by ELISA as described previously.29 Enumeration of both isotype-specific HRV antibody secreting cells (ASC) and total immunoglobulin secreting cells (IgSC) were performed by enzyme-linked immunosorbent spot (ELISPOT) assay as previously described.60 Frequencies of IgA+ or IgG+ B lymphocytes were determined by identifying CD79β+IgA+ and CD79β+IgG+ B cell, respectively. Briefly, 1×106 cells were stained with monoclonal anti-porcine IgA (Clone K61 1B4, Serotec) or biotinylated anti-porcine IgG (Clone F007–1241, BD Biosciences) antibody for 15 min at 4°C. Subsequently, cells were washed and incubated with anti-mouse IgG1-Allophycocyanin (BD Biosciences, CA) or streptavidin PE-Cy7 (BD Biosciences) secondary antibodies. After washing, stained cells were permeabilized and then stained with porcine cross- reactive anti-mouse CD79β-FITC antibody (Clone AT1072, Serotec, NC). To determine activated B cells (CD21+CD2-), MNCs were stained with anti-porcine CD21-PE (Clone BB6–11C9.6, SouthernBiotech, AL) and anti-porcine-CD2 (Clone MSA4, VMRD, WA), followed by anti-mouse IgG2a-SPRD (Cat#1080–13, SouthernBiotech, AL). Subsequently, cells were stained with CD79β-FITC as described previously. Appropriate isotype matched control antibodies were included. Subsequently, 100,000 events were acquired per sample using BD Accuri C6 flow cytometer (BD Biosciences, CA) and data were analyzed using C6 flow sampler software.
In vitro stimulation with probiotics, probiotic bacterial genomic DNA and HRV antigen
Effects of probiotics or probiotics genomic DNA on induction of cytokine- as well as T-cell independent IgA inducing factor-responses were assessed in freshly isolated ileal MNCs from naive Gn piglets (no prior exposure to any bacteria/viruses). Isolated Ileal MNCs (2.5 × 105 cells/well in 48 well cell culture plate) from 28 day old naive Gn piglets were treated with live LGG and Bb12 bacteria at ratio of 10:1 (bacteria : MNCs) in the presence or absence of purified inactivated HRV antigen (12 μg/ml) for 24 h in RPMI containing 8% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 20 mM HEPES, and antibiotics (E-RPMI). Subsequently, total RNA was isolated from the cells using RNeasy mini kit (Qiagen, MD) to determine changes in expression of B cell activation factor of the TNF family (BAFF) and a proliferation-inducing ligand (APRIL). Genomic DNA from LGG and Bb12 probiotics was isolated using GenElute Bacterial Genomic DNA Kit (Sigma, MO) to determine probiotic bacterial genomic DNA-induced cytokine responses. Ileal MNCs from control Gn piglets were co-cultured with a combination of LGG (25 μg/ml) - and Bb12 (25 μg/ml)-genomic DNA with or without HRV antigen (12 μg/ml). When indicated, cells were pretreated with chloroquine (10 μM) (InvivoGen, CA), a known inhibitor of TLR9, for 30 min. Culture supernatants were collected for determination of TGF-β, IL-10 and IL-12 cytokines by ELISA as previously described.31,65
Cytokine responses of ileal MNCs from Vac+Pro, Vac, Pro and Cont piglets stimulated with HRV antigen under in vitro condition were also determined. Briefly, freshly isolated MNCs were treated with inactivated purified HRV antigen (12 μg/ml) for 48 h at 37°C in E-RPMI. Subsequently, culture supernatants were collected to quantify IL-6, IL-10, IL-17 and IL-8 cytokine concentrations by ELISA as previously described.31,65
Quantitative real-time RT-PCR (QPCR)
The MNCs from the ileum of pigs vaccinated with AttHRV with or without LGG+Bb12 probiotic colonization were extracted from euthanized pigs on PID34/PCD7 (Post-HRV challenge). Subsequently, total RNA was isolated from the isolated MNCs using RNeasy Mini kit for QPCR. QPCR was performed for BAFF, APRIL, and β-actin genes using gene-specific primers (Table 1). The QPCR was performed using a QuantiTect SYBR Green RT-PCR Kit (Qiagen, USA) as instructed by the manufacturer. QPCR data were analyzed by the 2−ΔΔCT method.66
Table 1.
Details of primer sequences used for QPCR experiments
| Gene | Forward and reverse primer sequences (5′→3′) | Amplicon length (bp) | Accession number |
|---|---|---|---|
| β-actin | 5′- CAGGTCATCACCA TCGGCAACG -3′ 5′- GACAGCACCGTGTTGGCGTAGAGGT -3′ |
164 | DQ845171 |
| APRIL | 5′- CAGCCTCATCTCCTTCCTTG -3′ 5′- TTTGCAGCTCTGTTTGTTGG -3′ |
162 | NM_001112690 |
| BAFF | 5′- CAGCTCCATTCAAAGCAACA -3′ 5′- CCGTTTCTTTGACCACGATT -3′ |
203 | NM_001097498 |
Statistical analysis
Log10 transformed isotype-specific ELISA antibody titers were analyzed using one-way ANOVA followed by Duncan's multiple range test. The HRV-specific ASC, total IgSC and frequencies of CD79β+IgA+ B cells were compared among groups using the Kruskal–Wallis rank sum test. All statistical analyses were performed using SAS program (SAS Institute, NC) or GraphPad Prism version 5 (San Diego, CA). Differences were considered significant at P < 0.05. Error bars indicate the standard error of the mean (SEM).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We gratefully acknowledge the technical assistance of Dr. Juliette Hanson, Rich McCormick, Lindsey Good, Ozkan Timurkan, Joshua Amimo, and Kyle T. Scheuer.
Funding
This work was supported by a grant from the NCCAM at NIH (grant # R21 AT004716 to LJS), NIAID at NIH (grant # R01 A1099451 to LJS) and federal funds appropriated to the Ohio Agricultural Research and Development Center (OARDC) of The Ohio State University.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Sjogren YM, Tomicic S, Lundberg A, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E, Jenmalm MC. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy: J Brit Soc Allergy Clin Immunol 2009; 39:1842-51; PMID:19735274; http://dx.doi.org/ 10.1111/j.1365-2222.2009.03326.x [DOI] [PubMed] [Google Scholar]
- 2. Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009; 31:677-89; PMID:19833089; http://dx.doi.org/ 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 3. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008; 4:337-49; PMID:18854238; http://dx.doi.org/ 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol 2002; 168:171-8; http://dx.doi.org/ 10.4049/jimmunol.168.1.171 [DOI] [PubMed] [Google Scholar]
- 5. Ibnou-Zekri N, Blum S, Schiffrin EJ, von der Weid T. Divergent patterns of colonization and immune response elicited from two intestinal Lactobacillus strains that display similar properties in vitro. Infect Immun 2003; 71:428-36; PMID:12496193; http://dx.doi.org/ 10.1128/IAI.71.1.428-436.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rudin A, Lundell AC. Infant B cell memory and gut bacterial colonization. Gut Microbes 2012; 3:474-5; PMID:22892691; http://dx.doi.org/ 10.4161/gmic.21419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lundell AC, Bjornsson V, Ljung A, Ceder M, Johansen S, Lindhagen G, Tornhage CJ, Adlerberth I, Wold AE, Rudin A. Infant B cell memory differentiation and early gut bacterial colonization. J Immunol 2012; 188:4315-22; PMID:22490441; http://dx.doi.org/ 10.4049/jimmunol.1103223 [DOI] [PubMed] [Google Scholar]
- 8. Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004; 303:1662-5; PMID:15016999; http://dx.doi.org/ 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- 9. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010; 328:1705-9; PMID:20576892; http://dx.doi.org/ 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emer Infect Dis 2006; 12:304-6; PMID:16494759; http://dx.doi.org/ 10.3201/eid1202.050006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, Network WH-cGRS . 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 2012; 12:136-41; PMID:22030330; http://dx.doi.org/ 10.1016/S1473-3099(11)70253-5 [DOI] [PubMed] [Google Scholar]
- 12. Vesikari T, Prymula R, Schuster V, Tejedor JC, Cohen R, Bouckenooghe A, Damaso S, Han HH. Efficacy and immunogenicity of live-attenuated human rotavirus vaccine in breast-fed and formula-fed European infants. Pediatr Infect DisJ 2012; 31:509-13; PMID:22228235; http://dx.doi.org/ 10.1097/INF.0b013e3182489cac [DOI] [PubMed] [Google Scholar]
- 13. Esposito DH, Tate JE, Kang G, Parashar UD. Projected impact and cost-effectiveness of a rotavirus vaccination program in India, 2008. Clin Infect Dis: An Off Pub Infect Dis Soc Am 2011; 52:171-7; PMID:21288839; http://dx.doi.org/ 10.1093/cid/ciq094 [DOI] [PubMed] [Google Scholar]
- 14. Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. New Eng J Med 2010; 362:289-98; PMID:20107214; http://dx.doi.org/ 10.1056/NEJMoa0904797 [DOI] [PubMed] [Google Scholar]
- 15. Franco MA, Greenberg HB. Role of B cells and cytotoxic T lymphocytes in clearance of and immunity to rotavirus infection in mice. J Virol 1995; 69:7800-6; PMID:7494291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lagos R, Fasano A, Wasserman SS, Prado V, San Martin O, Abrego P, Losonsky GA, Alegria S, Levine MM. Effect of small bowel bacterial overgrowth on the immunogenicity of single-dose live oral cholera vaccine CVD 103-HgR. J Infect Dis 1999; 180:1709-12; PMID:10515838; http://dx.doi.org/ 10.1086/315051 [DOI] [PubMed] [Google Scholar]
- 17. Levine MM. Immunogenicity and efficacy of oral vaccines in developing countries: lessons from a live cholera vaccine. BMC Biol 2010; 8:129; PMID:20920375; http://dx.doi.org/ 10.1186/1741-7007-8-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hallander HO, Paniagua M, Espinoza F, Askelof P, Corrales E, Ringman M, Storsaeter J. Calibrated serological techniques demonstrate significant different serum response rates to an oral killed cholera vaccine between Swedish and Nicaraguan children. Vaccine 2002; 21:138-45; PMID:12443672; http://dx.doi.org/ 10.1016/S0264-410X(02)00348-1 [DOI] [PubMed] [Google Scholar]
- 19. Ferreira RB, Antunes LC, Finlay BB. Should the human microbiome be considered when developing vaccines? PLoS Pathogens 2010; 6:e1001190; PMID:21124987; http://dx.doi.org/ 10.1371/journal.ppat.1001190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Serazin AC, Shackelton LA, Wilson C, Bhan MK. Improving the performance of enteric vaccines in the developing world. Nat Immunol 2010; 11:769-73; PMID:20720580; http://dx.doi.org/ 10.1038/ni0910-769 [DOI] [PubMed] [Google Scholar]
- 21. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PloS One 2012; 7:e36957; PMID:22606315; http://dx.doi.org/ 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rubaltelli FF, Biadaioli R, Pecile P, Nicoletti P. Intestinal flora in breast- and bottle-fed infants. J Perinatal Med 1998; 26:186-91; PMID:9773376; http://dx.doi.org/ 10.1515/jpme.1998.26.3.186 [DOI] [PubMed] [Google Scholar]
- 23. Alvarez-Olmos MI, Oberhelman RA. Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin Infect Dis: An Off Pub Infect Dis Soc Am 2001; 32:1567-76; PMID:11340528; http://dx.doi.org/ 10.1086/320518 [DOI] [PubMed] [Google Scholar]
- 24. Isolauri E, Joensuu J, Suomalainen H, Luomala M, Vesikari T. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine 1995; 13:310-2; PMID:7631519; http://dx.doi.org/ 10.1016/0264-410X(95)93319-5 [DOI] [PubMed] [Google Scholar]
- 25. Holscher HD, Czerkies LA, Cekola P, Litov R, Benbow M, Santema S, Alexander DD, Perez V, Sun S, Saavedra JM, et al. Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: a randomized, double-blind, controlled trial. JPEN J Parenter Enter Nutr 2012; 36:106S-17S; PMID:22237870; http://dx.doi.org/ 10.1177/014860-7111430817 [DOI] [PubMed] [Google Scholar]
- 26. Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int J food Microbiol 1998; 42:39-44; PMID:9706796; http://dx.doi.org/ 10.1016/S0168-1605(98)00056-7 [DOI] [PubMed] [Google Scholar]
- 27. Gueimonde M, Sakata S, Kalliomaki M, Isolauri E, Benno Y, Salminen S. Effect of maternal consumption of lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J Pediatr Gastroenterol Nutr 2006; 42:166-70; PMID:16456409; http://dx.doi.org/ 10.1097/01.mpg.0000189346.25172.fd [DOI] [PubMed] [Google Scholar]
- 28. Saif LJ, Ward LA, Yuan L, Rosen BI, To TL. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch Virol Supplementum 1996; 12:153-61; PMID:9015112 [DOI] [PubMed] [Google Scholar]
- 29. Zhang W, Azevedo MS, Gonzalez AM, Saif LJ, Van Nguyen T, Wen K, Yousef AE, Yuan L. Influence of probiotic Lactobacilli colonization on neonatal B cell responses in a gnotobiotic pig model of human rotavirus infection and disease. Veter Immunol Immunopathol 2008; 122:175-81; PMID:18023882; http://dx.doi.org/ 10.1016/j.vetimm.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vlasova AN, Chattha KS, Kandasamy S, Liu Z, Esseili M, Shao L, Rajashekara G, Saif LJ. Lactobacilli and Bifidobacteria promote immune homeostasis by modulating innate immune responses to human rotavirus in neonatal gnotobiotic pigs. PloS One 2013; 8:e76962; PMID:24098572; http://dx.doi.org/ 10.1371/journal.pone.0076962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chattha KS, Vlasova AN, Kandasamy S, Rajashekara G, Saif LJ. Divergent immunomodulating effects of probiotics on T cell responses to oral attenuated human rotavirus vaccine and virulent human rotavirus infection in a neonatal gnotobiotic piglet disease model. J Immunol 2013; 191:2446-56; PMID:23918983; http://dx.doi.org/ 10.4049/jimmunol.1300678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sinkora M, Stepanova K, Butler JE, Francis D, Santiago-Mateo K, Potockova H, Karova K, Sinkorova J. Ileal Peyer's patches are not necessary for systemic B cell development and maintenance and do not contribute significantly to the overall B cell pool in swine. J Immunol 2011; 187:5150-61; PMID:22013120; http://dx.doi.org/ 10.4049/jimmunol.1101879 [DOI] [PubMed] [Google Scholar]
- 33. Narvaez CF, Franco MA, Angel J, Morton JM, Greenberg HB. Rotavirus differentially infects and polyclonally stimulates human B cells depending on their differentiation state and tissue of origin. J Virol 2010; 84:4543-55; PMID:20164228; http://dx.doi.org/ 10.1128/JVI.02550-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deal EM, Lahl K, Narvaez CF, Butcher EC, Greenberg HB. Plasmacytoid dendritic cells promote rotavirus-induced human and murine B cell responses. J Clin Invest 2013; 123:2464-74; PMID:23635775; http://dx.doi.org/ 10.1172/JCI60945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology 2005; 115:153-62; PMID:15885120; http://dx.doi.org/ 10.1111/j.1365-2567.2005.02159.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iliev ID, Kitazawa H, Shimosato T, Katoh S, Morita H, He F, Hosoda M, Saito T. Strong immunostimulation in murine immune cells by Lactobacillus rhamnosus GG DNA containing novel oligodeoxynucleotide pattern. Cell Microbiol 2005; 7:403-14; PMID:15679843; http://dx.doi.org/ 10.1111/j.1462-5822.2004.00470.x [DOI] [PubMed] [Google Scholar]
- 37. Menard O, Gafa V, Kapel N, Rodriguez B, Butel MJ, Waligora-Dupriet AJ. Characterization of immunostimulatory CpG-rich sequences from different Bifidobacterium species. App Environ Microbiol 2010; 76:2846-55; PMID:20208019; http://dx.doi.org/ 10.1128/AEM.01714-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimosato T, Kitazawa H, Katoh S, Tomioka Y, Karima R, Ueha S, Kawai Y, Hishinuma T, Matsushima K, Saito T. Swine Toll-like receptor 9(1) recognizes CpG motifs of human cell stimulant. Biochimica Et Biophysica Acta 2003; 1627:56-61; PMID:12759192; http://dx.doi.org/ 10.1016/S0167-4781(03)00048-4 [DOI] [PubMed] [Google Scholar]
- 39. Massacand JC, Kaiser P, Ernst B, Tardivel A, Burki K, Schneider P, Harris NL. Intestinal bacteria condition dendritic cells to promote IgA production. PloS One 2008; 3:e2588; PMID:18596964; http://dx.doi.org/ 10.1371/journal.pone.0002588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 2007; 26:812-26; PMID:17570691; http://dx.doi.org/ 10.1016/j.immuni.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 41. Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, Zimmermann VS, Davoust J, Ricciardi-Castagnoli P. Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J Exp Med 1997; 185:317-28; PMID:9016880; http://dx.doi.org/ 10.1084/jem.185.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tezuka H, Abe Y, Asano J, Sato T, Liu J, Iwata M, Ohteki T. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity 2011; 34:247-57; PMID:21333555; http://dx.doi.org/ 10.1016/j.immuni.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 43. Krieg AM, Wagner H. Causing a commotion in the blood: immunotherapy progresses from bacteria to bacterial DNA. Immunol Today 2000; 21:521-6; PMID:11071532; http://dx.doi.org/ 10.1016/S0167-5699(00)01719-9 [DOI] [PubMed] [Google Scholar]
- 44. Takahashi N, Kitazawa H, Shimosato T, Iwabuchi N, Xiao JZ, Iwatsuki K, Kokubo S, Saito T. An immunostimulatory DNA sequence from a probiotic strain of Bifidobacterium longum inhibits IgE production in vitro. FEMS Immunol Med Microbiol 2006; 46:461-9; PMID:16553822; http://dx.doi.org/ 10.1111/j.1574-695X.2006.00064.x [DOI] [PubMed] [Google Scholar]
- 45. Blaas SH, Stieber-Gunckel M, Falk W, Obermeier F, Rogler G. CpG-oligodeoxynucleotides stimulate immunoglobulin A secretion in intestinal mucosal B cells. Clin Exp Immunol 2009; 155:534-40; PMID:19220839; http://dx.doi.org/ 10.1111/j.1365-2249.2008.03855.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cognasse F, Acquart S, Beniguel L, Sabido O, Chavarin P, Genin C, Garraud O. Differential production of immunoglobulin classes and subclasses by mucosal-type human B-lymphocytes exposed in vitro to CpG oligodeoxynucleotides. Clin Chem Lab Med: CCLM/FESCC 2005; 43:22-31; PMID:15653438; http://dx.doi.org/ 10.1515/CCLM.2005.003 [DOI] [PubMed] [Google Scholar]
- 47. Defrance T, Vanbervliet B, Briere F, Durand I, Rousset F, Banchereau J. Interleukin 10 and transforming growth factor beta cooperate to induce anti-CD40-activated naive human B cells to secrete immunoglobulin A. J Exp Med 1992; 175:671-82; PMID:1371300; http://dx.doi.org/ 10.1084/jem.175.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Menard O, Butel MJ, Gaboriau-Routhiau V, Waligora-Dupriet AJ. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. App Environ Microbiol 2008; 74:660-6; PMID:18083875; http://dx.doi.org/ 10.1128/AEM.01261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Ginkel FW, Wahl SM, Kearney JF, Kweon MN, Fujihashi K, Burrows PD, Kiyono H, McGhee JR. Partial IgA-deficiency with increased Th2-type cytokines in TGF-beta 1 knockout mice. J Immunol 1999; 163:1951-7; PMID:10438931 [PubMed] [Google Scholar]
- 50. Sato A, Hashiguchi M, Toda E, Iwasaki A, Hachimura S, Kaminogawa S. CD11b +Peyer's patch dendritic cells secrete IL-6 and induce IgA secretion from naive B cells. J Immunol 2003; 171:3684-90; PMID:14500666; http://dx.doi.org/ 10.4049/jimmunol.171.7.3684 [DOI] [PubMed] [Google Scholar]
- 51. Franco MA, Greenberg HB. Immunity to rotavirus in T cell deficient mice. Virology 1997; 238:169-79; PMID:9400590; http://dx.doi.org/ 10.1006/viro.1997.8843 [DOI] [PubMed] [Google Scholar]
- 52. Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 2008; 8:411-20; PMID:18469830; http://dx.doi.org/ 10.1038/nri2316 [DOI] [PubMed] [Google Scholar]
- 53. Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Ann Rev Immunol 2010; 28:243-73; PMID:20192805; http://dx.doi.org/ 10.1146/annurev-immunol-030409-101314 [DOI] [PubMed] [Google Scholar]
- 54. Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol 2002; 3:822-9; PMID:12154359; http://dx.doi.org/ 10.1038/ni829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blutt SE, Warfield KL, Lewis DE, Conner ME. Early response to rotavirus infection involves massive B cell activation. J Immunol 2002; 168:5716-21; PMID:12023371; http://dx.doi.org/ 10.4049/jimmunol.168.11.5716 [DOI] [PubMed] [Google Scholar]
- 56. Zhang W, Azevedo MS, Wen K, Gonzalez A, Saif LJ, Li G, Yousef AE, Yuan L. Probiotic Lactobacillus acidophilus enhances the immunogenicity of an oral rotavirus vaccine in gnotobiotic pigs. Vaccine 2008; 26:3655-61; PMID:18524434; http://dx.doi.org/ 10.1016/j.vaccine.2008.04.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yasui H, Kiyoshima J, Ushijima H. Passive protection against rotavirus-induced diarrhea of mouse pups born to and nursed by dams fed Bifidobacterium breve YIT4064. J Infect Dis 1995; 172:403-9; PMID:7622883; http://dx.doi.org/ 10.1093/infdis/172.2.403 [DOI] [PubMed] [Google Scholar]
- 58. Chae CS, Kwon HK, Hwang JS, Kim JE, Im SH. Prophylactic effect of probiotics on the development of experimental autoimmune myasthenia gravis. PloS One 2012; 7:e52119; PMID:23284891; http://dx.doi.org/ 10.1371/journal.pone.0052119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Avery DT, Bryant VL, Ma CS, de Waal Malefyt R, Tangye SG. IL-21-induced isotype switching to IgG and IgA by human naive B cells is differentially regulated by IL-4. J Immunol 2008; 181:1767-79; PMID:18641314; http://dx.doi.org/ 10.4049/jimmu-nol.181.3.1767 [DOI] [PubMed] [Google Scholar]
- 60. Yuan L, Ward LA, Rosen BI, To TL, Saif LJ. Systematic and intestinal antibody-secreting cell responses and correlates of protective immunity to human rotavirus in a gnotobiotic pig model of disease. J Virol 1996; 70:3075-83; PMID:8627786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schultz M, Gottl C, Young RJ, Iwen P, Vanderhoof JA. Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J Pediat Gastroenterol Nutr 2004; 38:293-7; PMID:15076629; http://dx.doi.org/ 10.1097/00005176-200403000-00012 [DOI] [PubMed] [Google Scholar]
- 62. Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Robins-Browne RM, Tang ML. Prenatal administration of Lactobacillus rhamnosus has no effect on the diversity of the early infant gut microbiota. Pediat Allergy Immunol: Off Pub Eur Soc Pediat Allergy Immunol 2012; 23:255-8; PMID:22136660; http://dx.doi.org/ 10.1111/j.1399-3038.2011.01239.x [DOI] [PubMed] [Google Scholar]
- 63. Meyer RC, Bohl EH, Kohler EM. Procurement and maintenance of germ-free seine for microbiological investigations. Appl Microbiol 1964; 12:295-300; PMID:14199016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parreno V, Hodgins DC, de Arriba L, Kang SY, Yuan L, Ward LA, To TL, Saif LJ. Serum and intestinal isotype antibody responses to Wa human rotavirus in gnotobiotic pigs are modulated by maternal antibodies. J Gen Virol 1999; 80 (Pt 6):1417-28; PMID:10374959 [DOI] [PubMed] [Google Scholar]
- 65. Azevedo MS, Yuan L, Pouly S, Gonzales AM, Jeong KI, Nguyen TV, Saif LJ. Cytokine responses in gnotobiotic pigs after infection with virulent or attenuated human rotavirus. J Virol 2006; 80:372-82; PMID:16352562; http://dx.doi.org/ 10.1128/JVI.80.1.372-382.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.