Abstract
Synaptic plasticity, learning, and memory require high temporal and spatial control of gene expression. These processes are thought to rely mainly on asymmetric mRNA transport to synapses. Already in the early days of studying mRNA transport, Wilhelm and Vale proposed a multi-step process in 1993. Since then, we have gained important novel insights into how these individual steps are controlled by research performed in various cell types and organisms. Here, we present the latest view on how dendritic mRNA localization is achieved and how local translation at the synapse is regulated. In particular, we propose that the recently observed heterogeneity of RNA-protein particle assembly in neurons might be the key for how precise gene expression in the brain is achieved. In addition, we focus on latest data dealing with translational activation of translationally repressed mRNPs at a synapse that experiences learning-induced changes in its morphology and function. Together, these new findings shed new light on how precise regulatory mechanisms can lead to synaptic plasticity and memory formation.
Keywords: mRNA transport, RBPs, Translational repression, mRNA unmasking, synaptic plasticity
Spatial accumulation of mRNAs in the cytoplasm has been generally recognized as a way to regulate gene expression. It represents—in addition to nuclear events—an important control mechanism with special relevance in polarized cells. Especially in neurons, with axons and dendrites reaching long distances from the cell body, localized mRNAs contribute significantly to synaptic plasticity and memory consolidation (reviewed in ref. 1). In this context, local protein synthesis allows to alter individual synapses both functionally as well as structurally in an experience-dependent manner. Research has employed various model systems (i.e., from whole organisms to a variety of cell types) in order to understand the mechanism(s) by which locally restricted mRNA accumulation and subsequent local protein synthesis is achieved. It is now generally accepted that selective mRNA transport together with regulated local translation at the synapse are central to this pathway. However, we are just starting to unravel the molecular mechanisms that enable this system to respond dynamically in an experience-shaped manner.
Wilhelm and Vale (1993)1 were the first to propose that mRNA transport consists of multiple, consecutive steps. Their model was based on the findings of Carson and colleagues, who described the formation and unidirectional transport of RNA particles after microinjection of MBP mRNA into oligodendrocytes.2 The following four-step model had been suggested at the time:
formation of ribonucleoproteins (mRNPs) by interaction of mRNA with RNA-binding proteins (RBPs);
transport of mRNPs to their destination site;
anchoring of mRNPs;
local translation.
Since then, this model has been modified accordingly.3-6 With slight amendments, these reviews still follow the same basic model while research has continued to add further mechanistic insight into individual steps. A more recent update has been presented in the concept of the “RNA signature,” i.e., intrinsic mRNA localization signals, combined with the sushi-belt model.7 According to this concept, specific mRNAs assemble into mRNPs based on their “RNA signature” and then patrol individual synapses instead of being anchored at the synapse.
In this review, we highlight several recent findings that have significantly extended the knowledge of individual steps of RNA localization in dendrites. In particular, we focus on the heterogeneity of neuronal mRNP particles, and discuss how the local availability of mRNAs for translation at their target site might be regulated.
Neuronal RNA Particles are Heterogeneous
Intrinsic localization elements (LEs) within a given mRNA, commonly referred to as cis-acting signals, are recognized by specific RBPs (i.e., trans-acting factors), resulting in the formation of an mRNP. In neurons, cis-acting signals are usually located in the 3′-UTR of the transcript and can either simply be represented in the primary RNA sequence, e.g., in the hnRNP A2 response element (A2RE) or ZBP1 sequence (e.g., β-actin zipcode).8,9 Alternatively, they comprise more complicated secondary (and tertiary) structures in form of RNA stem loops, such as the Staufen recognition sequence (SRS) or possibly a combination of both primary sequences and higher order structures (Fig. 1). In this context, it is currently unknown whether a distinct LE might be recognized by one or more RBPs in a synergistic manner as recently proposed.10,11 Similarly, there is a long-lasting discussion about how complex and distinct the composition of individual mRNP particles might be, e.g., with respect to the associated RBPs or the number and (unique or diverse) identity of associated cargo mRNAs. Recent research has revealed that the composition of mRNP granules is far more heterogeneous than originally assumed. By biochemical analysis, characterizing the proteome of Staufen2 (Stau2) and Barentsz (Btz) containing RNA particles—two key RBPs involved in dendritic RNA transport—the majority of the associated proteins was found to be specific for the respective RBP.12 Interestingly, the individual composition of RNA granules is also seen on the level of associated RNAs as these individual RBP-containing granules have been shown to be associated with distinct sets of mRNAs. For instance, Arc mRNA was found to be preferentially associated with Btz. CaMKIIα mRNA, on the other hand, was primarily found in Staufen2-containing particles,12 even though it shares the same cis-acting signal for transport as Arc mRNA.8 Similarly, Farris et al. have detected CaMKIIα and Arc mRNA in distinct particles in the dentate gyrus using FISH.13 These data confirm earlier findings demonstrating that individual mRNPs preferentially contain only one RNA species.14,15 Applying sophisticated fluorescence microscopy, these two studies could also demonstrate that the copy number of an individual mRNA per dendritic particle is rather low (∼1–2 RNAs per particle).14,15 Whether this might indicate that certain mRNAs could possibly be transported as dimers, as previously reported for oskar mRNA in the Drosophila oocyte16 remains to be seen. Interestingly, the number of RNA molecules in an mRNP appears to be subject to dynamic changes. It depends on both the granule's relative position in the dendrite (with the soma and proximal region displaying higher numbers of RNA per particle than distally localized RNA granules), and on synaptic activity.17,18 This raises the intriguing possibility that local translation of distinct RNA species in the course of synaptic plasticity can be controlled by elaborate fine-tuning mechanisms in response to extrinsic signals. This is particularly interesting considering the fact that Arc mRNA, in contrast to other neuronal localized RNA species that are constitutively expressed (such as CaMKIIα or MAP2 mRNA), belongs to the family of immediately early genes (IEG), i.e., genes that are expressed quickly and only for a short time upon a specific stimulus. This implies that gene expression including RNA translation at the synapse can be “turned off” in order to keep the system responsive for further stimulation (see also below).
Figure 1.
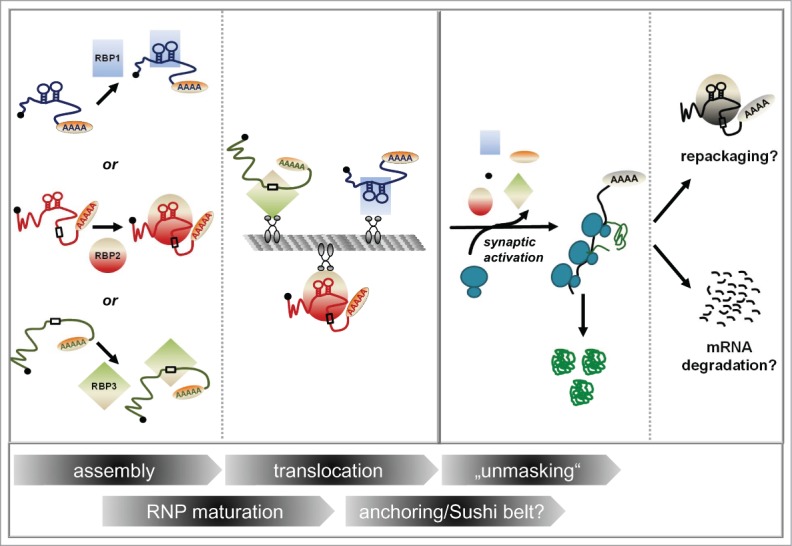
Distinct classes of mRNAs bearing distinct localization elements (LEs) are assembled with the help of specific RBPs (RBP1, RBP2, RBP3) into heterogeneous mRNPs. Those LEs are represented either as a linear primary sequence element (indicated by boxes) or a stem-loop for higher-order structures, respectively. mRNPs may change in their composition (“RNP maturation”) before and/or during their translocation process along microtubules (“translocation”) before they are either locally anchored or when they patrol synapses (anchoring/”sushi-belt?”). Translational repression is relieved upon synaptic activation (“unmasking”). Here, mRNA bound proteins (RBPs and probably also cap- and polyA-binding proteins, indicated by the little black and orange circle, respectively) dissociate from the mRNP, resulting in the mRNA being translated by ribosomes. Eventually, translation will terminate and the mRNA locally degraded or alternatively repacked into an mRNP.
In summary, research over the last few years has highlighted how heterogeneous and diverse individual mRNPs can be. Recently, the list of potential candidates of localized mRNAs has significantly grown. A large-scale sequencing approach revealed 2550, mostly previously undetected, local mRNAs in axons and dendrites of the hippocampal neuropil, with most of these mRNAs coding for proteins involved in synaptic function.19 It can be expected that (many of) these mRNAs are regulated both spatially and temporally (possibly using different mechanisms) in order to achieve a precise control of gene expression at the synapse.
Dendritic mRNPs are Translationally Repressed
mRNA transport to the synapse has been suggested to be tightly linked with translational repression thereby ensuring local protein synthesis (ref. 20; see also accompanying News and Views, ref. 21). Consequently, this model implies the need for translational repression to be relieved upon synaptic activity (see below). However, we are just beginning to understand how the translation of the multitude of different neuronal localized mRNAs is orchestrated. Several different mechanisms how mRNA translation is controlled in neurons have been described (for extensive reviews, see refs. 22 and 23). One well-accepted mechanism is the association of RBPs as translational regulators with the mRNA (see ref. 24). For example, the RBP Fragile X Mental Retardation Protein (FMRP) has been recently shown to bind directly to the 80S ribosome in vitro in a manner that blocks tRNA binding and therefore interferes with translation.25 In addition, miRNAs have been suggested to be involved in the translational control of mRNA.26-28 Importantly, RBPs and miRNAs may also act together to modulate miRNA-mediated repression. For example, degradation of high-affinity mRNA targets of HuD in response to mTORC1 signaling has recently been shown to result in increased concentrations of freely available HuD that can now sequester miR-129. This relieves miR-129 mediated repression of Kv1.1 translation.29
It is still experimentally unresolved whether transported mRNAs in dendrites are already associated with ribosomes. On one hand, it was recently suggested that neuronal mRNPs are associated with polyribosomes. These are thought to be stalled during the translation elongation phase and to become re-activated upon synaptic activity,30 arguing for at least partial translation before translational repression. On the other hand, many localized transcripts are reported to have several predominantly nuclear proteins associated.12,31,32 As the ribosome is likely to remove those nuclear proteins, this supports the hypothesis that mRNPs are kept in a translationally repressed state.
An almost puzzling phenomenon has been inferred by recent findings that some mRNAs localizing to dendrites contain introns in their 3′-UTR suggesting additional levels of regulation of mRNA localization and stability.33,34 Current reports even refer to up to 16% of all human 3′-UTRs (reviewed in ref. 35) as being annotated to contain an intron. The most prominent example so far is the Arc mRNA containing 2 intronic sequences in its 3′-UTR.34 Recently it has been suggested that localization and stability of the Arc mRNA is regulated by eIF4AIII, a member of the exon junction complex (EJC).34 The EJC, consisting of Btz, eIF4AIII, Magoh and Y14, is deposited onto mRNA upon pre-mRNA splicing in the nucleus and is also supposed to be removed upon translation.36-38 eIF4AIII and Magoh, two other core members of the EJC, have also been identified as part of Btz-containing particles indicating translational repression of the associated mRNAs.
These results clearly show that nuclear events are linked with cytoplasmic mRNA targeting and translational control.39,40 Interestingly, data obtained in the Drosophila oocyte indicate that these nuclear events can be even a pre-requisite for correct mRNA localization. Deposition of the EJC by pre-mRNA splicing provides a transport signal for the oskar mRNA in the oocyte.41,42 While it had been shown before that splicing of the first intron and presence of the EJC are required for the correct localization of the mRNA to the posterior pole,42 the Ephrussi lab recently identified a sequence flanking the first intron, termed SOLE (spliced oskar localization element). Upon pre-mRNA splicing, it generates a 28 nt stem-loop structure that is required for oskar mRNA motility. So far, no analogous localization element has been identified in neurons, though this represents a thought-provoking possibility to be considered.
mRNP Transport to its Destination
Currently, it is not clear how and to what extent RNPs undergo changes in composition once they have been exported to the cytoplasm. Theoretically, mRNPs can be subject to dynamic changes at any time point (Fig. 1). For sure, motor proteins together with potential adaptor proteins are subsequently recruited to those mRNPs in the cytoplasm. We have recently reviewed different possibilities how transport into the dendritic arbour and entry into dendritic spines at the activated synapse might occur.7 In brief, it is generally assumed that dendritic localization of mRNA is achieved by interaction of mRNPs with microtubules and their associated motor proteins. In order to enter the dendritic spines in the region of activated synapses, two alternative models have been proposed. In one model, the last step of translocation is performed by the actin cytoskeleton known to be present at the base of dendritic spines. Alternatively, actively growing microtubules that extend into the head of dendritic spines have been suggested to be responsible for entry into dendritic spines.43,44 Several groups have reported that movement of RNA granules depend on kinesins and myosins (reviewed in ref. 45). Importantly, while some data indicate that FMRP directly links the mRNA to kinesin,46 no adaptors or mediators linking other mRNPs to motor proteins have been identified so far. Here, we would like to refer the interested reader to detailed reviews covering this topic (e.g., refs. 45, 47, and 48).
In the past, several laboratories have tackled the task to study the dynamic behavior of mRNA granules as they move along dendrites by live cell imaging using (at least in parts) overexpression.49-51 Recently, the Singer lab has gone one step further to visualize the dynamic behavior of a single endogenous mRNA (β-actin) in the mouse employing the MS2 system.17 Their data indicate that a subset (∼10%) of RNA granules appear to traffic actively with an average speed of ∼1.3 μm/sec. These particles exhibit a slight net anterograde movement—a similar phenomenon had been described earlier by Bullock et al. in Drosophila for dynein-mediated transport along microtubules.52 It will be interesting to see whether similar “classes” or mRNAs might display a similar kinetic behavior and how this is integrated with synaptic activation.
Local Availability of mRNA for Translation at its Destination Site
As discussed above, mRNA “on the move” is translationally repressed. Hence, mechanisms are required to “unmask” the mRNA upon synaptic activity in order to make it available or accessible for local translation. Most recently, two back-to-back publications by the Singer laboratory provide first indications on how this activation might be achieved in response to synaptic stimulation. First, using single molecule FISH, they could elegantly demonstrate that β-actin mRNA appears “obscured” by a proteinaceous complex.18 Similar to protease treatment, synaptic activity resulted in increased accessibility of the mRNA. Their data also indicate that this masking of mRNA might indeed, at least in parts, be executed by associated repressive RBPs, such as FMRP or ZBP1.18 In their second paper, they employed live cell imaging using their endogenous MS2-system to illustrate that the process of synaptic activation possibly includes rearrangements of mRNPs.17 First, they detect an increased rate of particle splitting over merging upon synaptic activation. Second, the number of particles carrying a single mRNA increased, suggesting at least partial disassembly of large mRNP particles upon activation.
Currently, (at least) two models exist to explain how mRNA is retained at or recruited to synapses subsequent to transport. In the synaptic anchoring model, the local actin cytoskeleton and/or associated (motor) proteins have been suggested to act as an anchor to retain mRNAs at the synapse.4 In Drosophila, dynein was described to anchor transcripts after apical transport.53 However, the list of anchoring molecules might not be complete yet. In the alternative sushi-belt model, mRNAs continuously patrol individual synapses supported by bidirectional transport until they are recruited by an active synapse.7 Detailed insights into how mRNA is “unmasked” and made available for translation at the synapse upon synaptic stimulation will certainly improve our understanding of memory formation. Possible underlying mechanisms can include, for example, dissociation of repressive RBPs via posttranslational modifications and/or by termination of miRNA-mediated translational silencing. In future, it will be interesting to see, how synaptic activity is mechanistically linked to unpacking events and what exact molecular changes occur on the level of mRNPs.
Fate of mRNA after Translation
In order to achieve synaptic plasticity, it is not only important to “turn on” translation at the synapse by processes such as unmasking, but also to switch off protein synthesis at indicated times. What is the fate of mRNA once it has been translated? This aspect of mRNA metabolism in neurons is not yet well studied. In principle, (at least) two scenarios are possible: An mRNA can either face degradation after it has served its purpose, or it can be re-packaged into mRNPs in association with repressive RBPs in order to switch off translation (Fig. 1).
A mechanism implying translation and synaptic activity dependent mRNA degradation has recently been proposed by the group of Oswald Steward. They showed that newly synthesized mRNA accumulates at site of synaptic activation in the hippocampus in response to NMDA receptor activation. This is mediated by direct transport of newly synthesized mRNA to activated synapses in combination with preferential degradation of mRNA at synaptically inactive domains.13
So far, only indirect evidence for the hypothesis of mRNA repackaging exists. Based on recent studies by McKnight et al.,54,55 mRNP formation is suggested to be reversible.23 The McKnight lab showed that at least in a cell-free system, low complexity domains of RBPs are able to mediate the formation of hydrogel-like particles resembling RNA granules in a reversible manner. This is in agreement with data presented by Singer and colleagues,18 who interpret the initial burst and subsequent decrease of β-actin mRNA accessibility after chemical stimulation of long-term potentiation (cLTP) as “re-masking.” It would indeed be tempting to speculate that “masked” is the default state of localized mRNA (as suggested by 18) and “unmasking” needs to be actively induced. Future work will show whether this also applies to other dendritic mRNAs and whether mRNP assembly and disassembly is in general a dynamic, reversible process at the synapse. As it has been shown that phosphorylation of translational repressors can result in their dissociation and subsequent translation of their associated mRNA,20 one could speculate that phosphorylation or other reversible post-translational modifications play a role in dendritic RNP (re-)assembly.
Conclusions
For some, it might come as a surprise that the—at the time—visionary model for mRNA transport by Wilhelm and Vale1 after more than two decades is generally still in accordance with today's view. While our understanding of some of these steps have not significantly progressed in the last few years (e.g., transport along microtubules or anchoring/recruitment at the synapse), other steps have been elucidated in fascinating detail, not least due to advances in imaging or biochemical techniques. First, neuronal RNA granules are much more heterogeneous in composition and therefore consequently in assembly than previously thought. One could easily imagine that a modular composition of RNA granules, possibly mediated by distinct classes of intrinsic RNA sorting signals (“RNA signatures”) would allow for precise differential neuronal gene expression of operational groups of mRNAs both in space and time. The recruitment of distinct mRNA regulators (i.e., RBPs, miRNAs) to distinct transcripts based on their individual “RNA signature” could therefore allow control of local mRNA availability at the synapse upon synaptic stimulation. In future, it will be interesting to see these regulatory principles unravelled and to learn if other, so far less understood steps, are also differentially regulated.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank member of the Kiebler lab as well as D. Dormann and J. Medenbach for critical reading of the manuscript. We apologize to the colleagues whose work could not be cited due to space constraints.
Funding
The work highlighted in this article was supported by the Austrian Science Funds (I590-B09, SFB F43), the ESF Program RNAQuality (I 127-B12), and an HFSP network grant (RGP24/2008).
References
- 1. Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol 1993; 123:269-74; PMID:8408211; http://dx.doi.org/ 10.1083/jcb.123.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol 1993; 123:431-41; PMID:7691830; http://dx.doi.org/ 10.1083/jcb.123.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 2005; 6:363-75; PMID:15852043; http://dx.doi.org/ 10.1038/nrm1643 [DOI] [PubMed] [Google Scholar]
- 4. Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell 2009; 136:719-30; PMID:19239891; http://dx.doi.org/ 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science 2009; 326:1212-6; PMID:19965463; http://dx.doi.org/ 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 2007; 8:776-89; PMID:17848965; http://dx.doi.org/ 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- 7. Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J 2011; 30:3540-52; PMID:21878995; http://dx.doi.org/ 10.1038/emboj.2011.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell 2008; 19:2311-27; PMID:18305102; http://dx.doi.org/ 10.1091/mbc.E07-09-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci 2003; 23:8859-66; PMID:14523087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol 2011; 9:e1000611; PMID:21526221; http://dx.doi.org/ 10.1371/journal.pbio.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Löffler P, Längst G, Merkl R, Urlaub H, et al. . The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev 2014; 28:749-64; PMID:24696456; http://dx.doi.org/ 10.1101/gad.236513.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, et al. . Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5:1749-62; PMID:24360960; http://dx.doi.org/ 10.1016/j.celrep.2013.11.023 [DOI] [PubMed] [Google Scholar]
- 13. Farris S, Lewandowski G, Cox CD, Steward O. Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. J Neurosci 2014; 34:4481-93; PMID:24671994; http://dx.doi.org/ 10.1523/JNEUROSCI.4944-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIα and β-actin RNAs in low copy numbers. EMBO Rep 2011; 12:1077-84; PMID:21869818; http://dx.doi.org/ 10.1038/embor.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A 2012; 109:4645-50; PMID:22392993; http://dx.doi.org/ 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jambor H, Brunel C, Ephrussi A. Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 2011; 17:2049-57; PMID:22028360; http://dx.doi.org/ 10.1261/rna.2686411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 2014; 343:422-4; PMID:24458643; http://dx.doi.org/ 10.1126/science.1239200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 2014; 343:419-22; PMID:24458642; http://dx.doi.org/ 10.1126/science.1242939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 2012; 74:453-66; PMID:22578497; http://dx.doi.org/ 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512-5; PMID:16306994; http://dx.doi.org/ 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- 21. Dahm R, Kiebler M. Cell biology: silenced RNA on the move. Nature 2005; 438:432-5; PMID:16306974; http://dx.doi.org/ 10.1038/438432a [DOI] [PubMed] [Google Scholar]
- 22. Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron 2009; 61:10-26; PMID:19146809; http://dx.doi.org/ 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell 2014; 157:26-40; PMID:24679524; http://dx.doi.org/ 10.1016/j.cell.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci 2013; 36:243-70; PMID:23701460; http://dx.doi.org/ 10.1146/annurev-neuro-062912-114322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell 2014; 54:407-17; PMID:24746697; http://dx.doi.org/ 10.1016/j.molcel.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79:351-79; PMID:20533884; http://dx.doi.org/ 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- 27. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9:102-14; PMID:18197166; http://dx.doi.org/ 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 28. Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010; 65:373-84; PMID:20159450; http://dx.doi.org/ 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 2013; 202:53-69; PMID:23836929; http://dx.doi.org/ 10.1083/jcb.201212089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci U S A 2013; 110:16205-10; PMID:24043809; http://dx.doi.org/ 10.1073/pnas.1307747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen Tv, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics 2007; 6:798-811; PMID:17289661; http://dx.doi.org/ 10.1074/mcp.M600346-MCP200 [DOI] [PubMed] [Google Scholar]
- 32. di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, Mercanti D, Molinari M, Bagni C, Achsel T. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol 2009; 184:423-35; PMID:19188494; http://dx.doi.org/ 10.1083/jcb.200807033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron 2011; 69:877-84; PMID:21382548; http://dx.doi.org/ 10.1016/j.neuron.2011.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 2007; 130:179-91; PMID:17632064; http://dx.doi.org/ 10.1016/j.cell.2007.05.028 [DOI] [PubMed] [Google Scholar]
- 35. Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ. Introns in UTRs: why we should stop ignoring them. Bioessays 2012; 34:1025-34; PMID:23108796; http://dx.doi.org/ 10.1002/bies.201200073 [DOI] [PubMed] [Google Scholar]
- 36. Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell 2009; 137:536-48; PMID:19410547; http://dx.doi.org/ 10.1016/j.cell.2009.02.042 [DOI] [PubMed] [Google Scholar]
- 37. Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 2002; 12:1060-7; PMID:12121612; http://dx.doi.org/ 10.1016/S0960-9822(02)00902-8 [DOI] [PubMed] [Google Scholar]
- 38. Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 2002; 21:3536-45; PMID:12093754; http://dx.doi.org/ 10.1093/emboj/cdf345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol 2007; 18:186-93; PMID:17459736; http://dx.doi.org/ 10.1016/j.semcdb.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 40. Farina KL, Singer RH. The nuclear connection in RNA transport and localization. Trends Cell Biol 2002; 12:466-72; PMID:12441250; http://dx.doi.org/ 10.1016/S0962-8924(02)02357-7 [DOI] [PubMed] [Google Scholar]
- 41. Ghosh S, Marchand V, Gáspár I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol 2012; 19:441-9; PMID:22426546; http://dx.doi.org/ 10.1038/nsmb.2257 [DOI] [PubMed] [Google Scholar]
- 42. Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 2004; 428:959-63; PMID:15118729; http://dx.doi.org/ 10.1038/nature02521 [DOI] [PubMed] [Google Scholar]
- 43. Hu X, Viesselmann C, Nam S, Merriam E, Dent EW. Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci 2008; 28:13094-105; PMID:19052200; http://dx.doi.org/ 10.1523/JNEUROSCI.3074-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. . Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 2009; 61:85-100; PMID:19146815; http://dx.doi.org/ 10.1016/j.neuron.2008.11.013 [DOI] [PubMed] [Google Scholar]
- 45. Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci 2006; 26:7139-42; PMID:16822968; http://dx.doi.org/ 10.1523/JNEUROSCI.1821-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell 2008; 14:926-39; PMID:18539120; http://dx.doi.org/ 10.1016/j.devcel.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 2009; 10:682-96; PMID:19773780; http://dx.doi.org/ 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- 48. Xing L, Bassell GJ. mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic 2013; 14:2-14; PMID:22913533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol 2007; 500:433-47; PMID:17120280; http://dx.doi.org/ 10.1002/cne.21189 [DOI] [PubMed] [Google Scholar]
- 50. Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell 1999; 10:2945-53; PMID:10473638; http://dx.doi.org/ 10.1091/mbc.10.9.2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 2011; 8:165-70; PMID:21240280; http://dx.doi.org/ 10.1038/nmeth.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol 2012; 14:416-23; PMID:22366687; http://dx.doi.org/ 10.1038/ncb2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell 2005; 122:97-106; PMID:16009136; http://dx.doi.org/ 10.1016/j.cell.2005.04.033 [DOI] [PubMed] [Google Scholar]
- 54. Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. . Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012; 149:768-79; PMID:22579282; http://dx.doi.org/ 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 55. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. . Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; http://dx.doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]


