Abstract
Gut microbiota regulated imbalances in the host's immune profile seem to be an important factor in the etiology of type 1 diabetes (T1D), and identifying bacterial markers for T1D may therefore be useful in diagnosis and prevention of T1D. The aim of the present study was to investigate the link between the early gut microbiota and immune parameters of non-obese diabetic (NOD) mice in order to select alleged bacterial markers of T1D. Gut microbial composition in feces was analyzed with 454/FLX Titanium (Roche) pyro-sequencing and correlated with diabetes onset age and immune cell populations measured in diabetic and non-diabetic mice at 30 weeks of age. The early gut microbiota composition was found to be different between NOD mice that later in life were classified as diabetic or non-diabetic. Those differences were further associated with changes in FoxP3+ regulatory T cells, CD11b+ dendritic cells, and IFN-γ production. The model proposed in this work suggests that operational taxonomic units classified to S24–7, Prevotella, and an unknown Bacteriodales (all Bacteroidetes) act in favor of diabetes protection whereas members of Lachnospiraceae, Ruminococcus, and Oscillospira (all Firmicutes) promote pathogenesis.
Keywords: gut microbiota, IFN-γ, NOD mice, regulatory immunity, Type 1 diabetes
Abbreviations
- CD
cluster of differentiation
- DC
dendritic cell
- FoxP3
forkhead box
- IFN
interferon
- MLN
mesenteric lymph node
- NKT
natural killer T cell
- PCA
principal component analysis
- PCoA
principal coordinate analysis
- PLN
pancreatic lymph node
- siLP
small intestinal lamina propria
- Treg
regulatory T cell
Introduction
The etiology of type 1 diabetes (T1D) is complex. The susceptibility to develop insulitis is polygenetic, particularly alleles encoding antigen-presenting cell functions (NOD MHC class II), immunoregulatory molecules (IL-2, IL21, and CTLA-4), and B-, T-, and Natural killer T (NKT) cell activation and homeostasis are found in strongly contributing loci linked to traits of clinical T1D.1,2 However, genetics cannot fully account for the development of diabetes. Differences in gut microbiota composition have received a great deal of attention, especially since it was published that disease incidence vary between non-obese diabetic (NOD) mice maintained in different facilities.3 This is supported by studies in germ-free NOD mice demonstrating the importance of gut microbiota as a disease regulator. The studies by Wen et al. and Suzuki et al. are frequently quoted as they reported a near 100% diabetes incidence in germ-free NOD mice.4,5
Experimental evidence from other studies in diabetic mouse models has confirmed that differences in gut microbiota partly explain the incidence of diabetes.3,6 For example, a recent study highlighted the diabetes protective association with presence of segmented filamentous bacteria (SFB) in NOD mice from different facilities.7 In a recent study, SFB was however only found to protect NOD males in contrast to females when mono-colonized, and the mice therefore regained the diabetes gender bias which is lost in germ-free conditions.8 Moreover, Lactobacillus and Bifidobacterium were found in higher abundance in feces from bio-breeding diabetes-resistant (BB-DR) rats before clinical onset of T1D compared with bio-breeding diabetes-prone (BB-DP) rats, which contained more Bacteroides, Eubacterium, and Ruminococcus ssp.9 Furthermore, diabetes incidence in BB-DP rats and NOD mice decrease after selective depletion of gut microbes with antibiotics in the postnatal period.10,11 This indicates that the early life gut microbiota is involved in the development of T1D. Few recent studies have therefore addressed the gut microbiota composition in children developing T1D, and found less abundance of Clostridium, Prevotella, Bifidobacterium, Roseburia, and butyrate producers in cases than controls while Bacteroides and the phylum Proteobacteria were relatively more abundant.12-14
The exact trigger of T1D and the mechanisms behind the protective or predisposing role of gut microbiota are still unknown. Studies in germ-free and gnotobiotic mice have provided evidence that stimulation afforded by the immune recognition of microorganisms is required for both pro- and anti-inflammatory mechanisms to evolve.15 The presence of Bacteroides fragiles,16 probiotic Bifidobacterium infantis,17 Lactobacillus reuteri,18,19 and several Clostridium spp. in the gut20 have for example been shown to increase the number and suppressive activity of regulatory T (Treg) cells; often associated with a decrease in pro-inflammatory cell numbers, pointing toward a profound effect of microbiota on immune homeostasis. Furthermore, Treg cell deficiency seems important in the pathogenesis of T1D.21-24 Consequently, altered immune regulation in T1D might very well be caused by intestinal dysbiosis, which is most intriguing as stimulation of the mucosal immune system by the gut microbiota may influence the way the immune system discriminates between self- and non-self antigens.
Current evidence on an autoimmune microbiome in human and animal models is still limited. Hence, it is important to identify bacterial markers as possible candidates for early diagnosis and as useful targets in prevention of T1D development. Due to an immense level of complexity that the mammalian gut microbiota vs. immune system cross talk is characterized by, the eventual bacterial markers will most likely not be consistent between various hosts, environments, diets etc. However, an effort should be taken in order to identify those markers even if their predictive value would be solely limited to defined conditions. Emerging evidence indicates that colonization pattern of the gut and early gut microbiota composition have a profound effect on the host's health status later in life. In this study we sought to investigate how the early gut microbiota is associated with changes in immune regulation that could directly link dysbiosis to pathogenesis of autoimmune diabetes.
Results
Intestinal and systemic Treg, CD11b+ dendritic cells (DCs), and IFNγ producing T cells are more abundant in non-diabetic NOD mice
The cumulative diabetes incidence in the 46 female NOD mice was 52%. Data collected from some of these mice were used for correlation analyses and results of the correlations between diabetes onset age and host immune markers are presented in Table 1. The significant correlations are furthermore shown in Figure 1. Forkhead box (FoxP3+) T cells correlated positively with delayed diabetes onset age in mesenteric lymph node (MLN) and spleen but not in the intestinal lamina propria. Cluster of differentiation (CD) 11b+ DCs and activated CD40+ DCs in MLN and spleen were also significantly associated with a late onset age but tolerogenic CD103+ DCs known to induce Treg were not. Interestingly, small intestinal lamina propria (siLP) interferon (IFN) γ and splenic IFNγ but not IL-10 producing T cells correlated positively to a delayed diabetes onset age. All the significant immune parameters were also significant when diabetic mice were statistically compared with non-diabetic mice except for CD40+ DCs (Table 1).
Table 1.
Correlation of immune parameters and diabetes onset age in NOD mice
| Correlation with diabetes onset age1 |
|||||
|---|---|---|---|---|---|
| Parameter | Organ | Group size | P value | R | T-test between diabetic vs. non-diabetic mice2 |
| Diabetes onset age | — | 46 | — | — | — |
| FoxP3/CD4 Treg | pancreatic lymph nodes | 13 | 0.090 | 0.489 | *<0.05 |
| FoxP3/CD4 Treg | small intestine lamina propria | 18 | NS | −0.197 | NS |
| FoxP3/CD4 Treg | colon lamina plopria | 12 | NS | −0.130 | NS |
| FoxP3/CD4 Treg | mesenteric lymph nodes | 27 | **<0.01 | 0.663 | ***<0.001 |
| FoxP3/CD4 Treg | spleen | 28 | ***<0.001 | 0.732 | ***<0.001 |
| CD11b/CD11c DC | spleen | 21 | ***<0.001 | 0.742 | ***<0.001 |
| CD11b/CD11c DC | mesenteric lymph nodes | 32 | **<0.01 | 0.463 | **<0.01 |
| CD40/CD11c DC | spleen | 21 | *<0.05 | 0.480 | NS |
| CD40/CD11c DC | mesenteric lymph nodes | 32 | *<0.05 | 0.374 | NS |
| CD103/11c DC | pancreatic lymph nodes | 13 | NS | −0.061 | NS |
| CD103/11c DC | mesenteric lymph nodes | 28 | NS | 0.197 | NS |
| INF-γ/CD4 Th1 | small intestine lamina propria | 25 | **<0.01 | 0.432 | ***<0.001 |
| INF-γ/CD4 Th1 | spleen | 23 | ***<0.001 | 0.780 | **<0.01 |
| IL-10/CD4 | small intestine lamina propria | 9 | NS | 0.244 | NS |
| IL-10/CD4 | spleen | 12 | NS | −0.209 | NS |
Results of Pearson correlation of the given parameter with diabetes onset age [days]. 2Results of T test between diabetic and non-diabetic mice for the given parameter.
Figure 1.
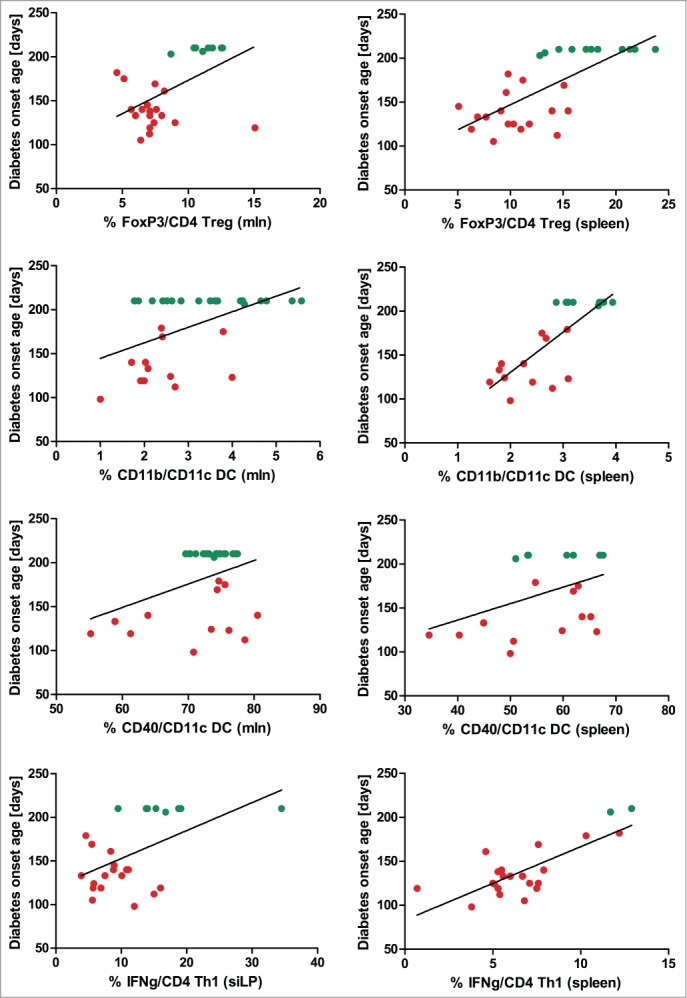
Diabetes onset age correlates with Treg, tolerogenic and activated DCs, and IFNγ producing (T)cells. Correlation analyses plots between diabetes onset age and immune cell subsets analyzed by flow cytometry in non-diabetic (green) and diabetic (red) NOD mice illustrate significant results presented in Table 1. DC = dendritic cells; MLN = mesenteric lymph node; siLP= small intestinal lamina propria; Th = T helper; Treg = regulatory T cells.
Early microbial profiles clustered according to diabetes development
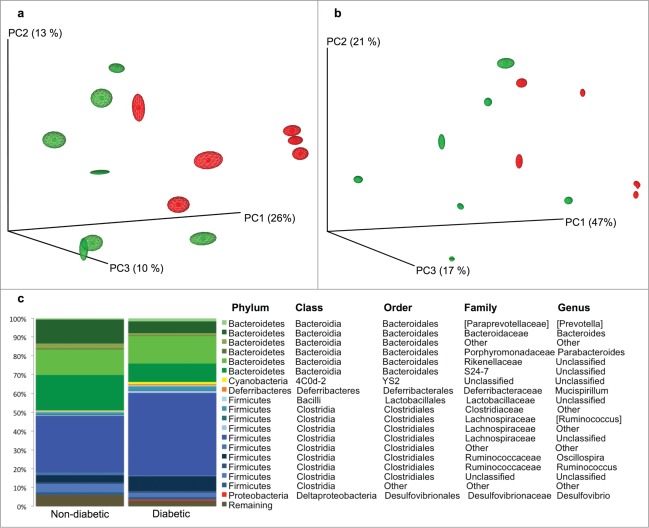
Fecal gut microbiota collected at weaning was used from 13 mice to compare the diabetic with the non-diabetic group. PCo analysis showed significant differences in the early bacterial community composition between NOD mice later classified as diabetic or non-diabetic at 30 weeks of age. The differences were significant for both weighted and unweighted UniFrac distance metrics (Fig. 2; ANOSIM: *p < 0.05, R = 0.341 and *p < 0.05, R = 0.296 for unweighted and weighted analysis respectively). The abundance of 3 bacterial taxa: unclassified genus from S24–7 family, unidentified taxon from order Bacteroidales, and genus Prevotella were significantly increased in the non-diabetic group. The abundance of an unclassified genus from the Lachnospiraceae family were in contrast significantly increased in the diabetic group together with a tendency for more Oscillospira and Ruminococcus (p = 0.05 and p = 0 .07 respectively) compared to the non-diabetic mice (Fig. 2 and Table S1). According to the qualitative analysis (g-test), no genera were categorically different between the 2 categories meaning that no genera were actually lacking in between the groups but it was only the abundance of genera that differed.
Figure 2.
Early gut microbiota in diabetic and non-diabetic NOD mice differ. PCoA plot based on unweighted (a) and weighted (b) distance matrices calculated from 10 rarefied (1400 reads per sample) OTU tables showing the separation between the fecal gut microbiota at weaning of NOD mice that later in life were diagnosed as diabetic (red, n = 6) or non-diabetic (green, n = 7). The degree of variation between 10 jackknifed replicates of PCoA is displayed with confidence ellipsoids around each sample. (c) The relative distribution of the major genera (20) determined for diabetic and non-diabetic NOD mice. Taxa denoted as "Unclassified" means that the reference database does not have an official taxonomy for this cluster. Taxa denoted as "Other" (unidentified), indicates ambiguity in the assignment meaning that more than one bacterial taxa could be assigned to this cluster.
The α diversity measure expressed by the observed species index was significantly increased in the non-diabetic group compared to the diabetic group (146 ± 23 vs. 109 ± 32 [OTU]; *p < 0.05; data not shown), what is in concordance with studies on humans.13,14
Early life gut microbiota correlates with diabetes onset age and immune parameters later in life
Significant results for correlations between all bacterial taxa (genus level) and diabetes onset age are presented in Table 2 and Figure 3. Significant correlations between immune parameters and taxa are also included in Table 2, but those that were not significantly associated with diabetes onset age or did not differ between diabetic and non-diabetic mice are not included in the table. Interestingly, the unclassified genus from Lachnospiraceae family, Oscillospira, and Ruminococcus, that were all more abundant in diabetic mice, correlated negatively with delayed diabetes onset age which suggests a role of these bacteria in diabetes development. Furthermore, these taxa correlated negatively with the proportion of splenic FoxP3+CD4+ Treg cells and CD11b+CD11c+ DCs, and while Lachnospiraceae correlated further negatively to CD40+CD11c+ DCs in the spleen, Oscillospira and Ruminococcus correlated negatively to CD40+CD11c+ DCs in MLN. This strongly indicates a diabetes protecting function of these immune cell subsets. In support of this, the unclassified genus belonging to the S24–7 family, the unidentified taxon from order Bacteroidales, and genus Prevotella that were all found more abundant in non-diabetic mice also correlated positively with delayed diabetes onset age, splenic FoxP3+CD4+ Treg cells, CD11b+CD11c+ DCs, and CD40+CD11c+ DCs. There was furthermore a tendency for S24–7 and Bacteroidales taxon to positively correlate with splenic IFN-γ+CD4+ Th1 cells, while a tendency was evident for Lachnospiraceae and Oscillospira to correlate negatively with splenic IFN-γ+CD4+ Th1 cells.
Table 2.
Correlations between given host parameters and fecal gut microbiota taxa at weaning that were significantly associated with diabetes onset
| Taxa |
||||||
|---|---|---|---|---|---|---|
| Order | Family | Genus | Parameter | p value | R | Group size |
| Clostridiales | Lachnospiraceae | Unclassified | Diabetes onset age | *<0,05 | −0,672 | 13 |
| FoxP3/CD4 Treg | *<0,05 | −0,619 | 12 | |||
| CD11b/CD11c DC | *<0,05 | −0,665 | 13 | |||
| CD40/CD11c DC | **<0,01 | −0,731 | 13 | |||
| Clostridiales | Ruminococcaceae | Oscillospira | Diabetes onset age | *<0,05 | −0,583 | 13 |
| FoxP3/CD4 Treg | **<0,01 | −0,744 | 12 | |||
| CD11b/CD11c DC | *<0,05 | −0,615 | 13 | |||
| CD40/CD11c DC (MLN) | *<0,05 | −0,661 | 7 | |||
| INF-γ/CD4 Th1 | 0.066 | −0.724 | 7 | |||
| Clostridiales | Ruminococcaceae | Ruminococcus | Diabetes onset age | *<0,05 | −0,586 | 13 |
| FoxP3/CD4 Treg | *<0,05 | −0,720 | 12 | |||
| CD11b/CD11c DC | *<0,05 | −0,685 | 13 | |||
| CD40/CD11c DC (MLN) | *<0,05 | −0,614 | 7 | |||
| Bacteroidales | S24–7 | Unclassified | Diabetes onset age | **<0,01 | 0,769 | 13 |
| FoxP3/CD4 Treg | **<0,01 | 0,781 | 12 | |||
| INF-γ/CD4 Th1 | 0.081 | 0.670 | 7 | |||
| CD11b/CD11c DC | *<0.05 | 0.684 | 13 | |||
| CD40/CD11c DC | *<0,05 | 0,604 | 13 | |||
| Bacteroidales | Prevotellaceae | Prevotella | Diabetes onset age | **<0,01 | 0,718 | 13 |
| FoxP3/CD4 Treg | *<0,05 | 0,665 | 12 | |||
| CD11b/CD11c DC | *<0,05 | 0,704 | 13 | |||
| CD40/CD11c DC | 0,097 | 0,502 | 13 | |||
| Bacteroidales | Other | Other | Diabetes onset age | **<0,01 | 0,709 | 13 |
| FoxP3/CD4 Treg | **<0,01 | 0,732 | 12 | |||
| CD11b/CD11c DC | **<0,01 | 0,735 | 13 | |||
| CD40/CD11c DC | *<0,05 | 0,651 | 13 | |||
Correlations between splenic or mesenteric lymph nodes (MLN) host parameters at diabetes onset age and taxa relative abundance at genus level (94% sequence similarity OTUs) in samples collected at weaning verified with the Pearson's product-moment correlation coefficient. Results with p < 0.1 are included in the table.
Figure 3.
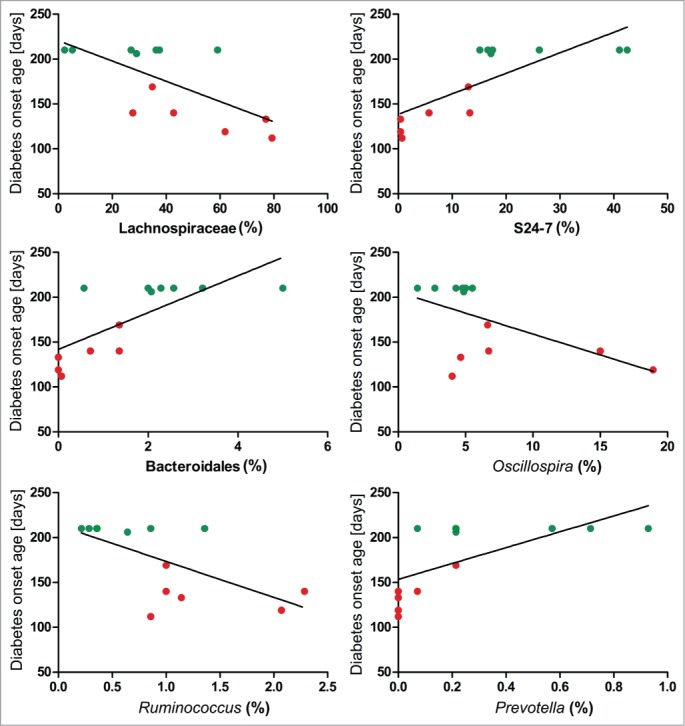
Gut microbial taxa correlates significantly to diabetes onset age. Correlation analyses plots between diabetes onset age and gut microbial taxa analyzed by 454 pyro-sequencing in non-diabetic (green, n = 7) and diabetic (red, n = 6) NOD mice illustrate significant results presented in Table 2. Testing was performed based on 1000 rarefied OTU tables unified to an equal number of reads per sample (1400).
Diabetic and non-diabetic NOD mice cluster separately according to early gut microbiota and correlating immune parameters in a PC analysis
In order to illustrate a combined effect of fecal microbes and the immune system on the development of diabetes a combined Principal component analysis (PCA) analysis was used solely with bacteria suspected to be alleged diabetes markers in early life and significantly correlating immune cell subsets. This illustrated a significant separation between diabetic and non-diabetic mice (Fig. 4, **p < 0.01 for PC1). The total variance explained with the first 3 components was 86.1%.
Figure 4.
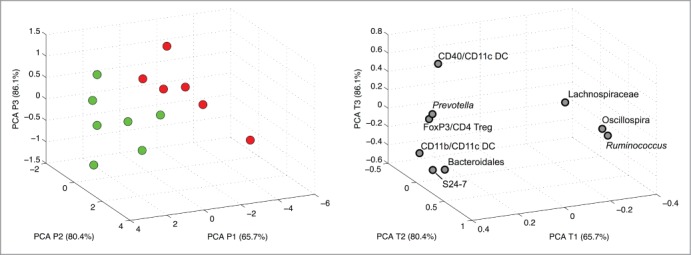
PCA using selected alleged bacterial and host immune markers. PCA scores (a) of non-diabetic (green, n = 7) and diabetic (red, n = 6) NOD mice and the corresponding PCA loadings (b) based on taxa and immune parameters that significantly correlated to diabetes onset age.
Discussion
Bacterial markers for T1D may likely be useful in diagnosis and prevention of T1D. The most striking result presented here was the high abundance of S24–7 in feces from non-diabetic NOD mice, whereas the Lachnospiraceae in the Clostriales XIVa cluster was found primarily in the early diabetic mice at weaning. The Lachnospiraceae family was also in pre-diabetic BB-DP rats one of the most prominent differences in the gut microbiota compared with BB-DR rats,9 but in humans, this taxa decreased over time in cases and was therefore more abundant in controls at the time of diabetes onset.14 It is however also consistent with a previous study in which non-diabetic NOD mice had significantly fewer microaerophilic bacteria than diabetic NOD mice, primarily due to a difference in the Gram-positive microbiota.
The relevance of these 2 bacterial markers together with the minor but significant differences in Rumincoccus, Oscillospira, Bacteriodales, and Prevotella is further augmented by their simultaneous correlation to systemic FoxP3+ Treg cells. This is important because a high level of splenic Treg cells was clearly associated with diabetes protection in the NOD mice; an association that was also found in other rodent studies.25,26 In addition, jejunal biopsies from T1D patients showed reduced frequency of FoxP3+ Treg cells and CD103+ tolerogenic DCs defective of inducing FoxP3+ Treg cell differentiation.27 We therefore propose that a microbiota signature characterized by a decrease in S24–7, Bacteriodales, and Prevotella and an increase in Lachnospiraceae, Ruminococcus, and Oscillospira may contribute to the pathogenesis of T1D by decreasing FoxP3+ Treg cells that protects against diabetes. OTUs classified to S24–7 seem to be largely unexplored in mice models for the time being. De-novo clustering disclosed 243 species level OTUs falling into this taxon, what constituted to nearly 10% of all registered OTUs. Although, correlation performed using species level OTUs have confirmed several OTUs from this family correlating in a similar way, due to relatively shallow sequencing the results could not be cross-validated with a proper subsampling (data not shown). It is however not unlikely that within this broad family only a fraction of OTUs is influencing the result. However, in order to ensure more accurate identification reaching genus or species level within S24–7 family, not only deeper sequencing, but also updates in the databases along with the use of alternative molecular techniques are required. Certain Bacteriodetes species are already known to promote accumulation of Treg cells and has beside its anti-inflammatory properties also been found relatively more abundant in controls than human T1D children.14,16
Significantly reduced abundance of Prevotella in diabetic patients was previously observed in prediabetic Finnish children29 and recently confirmed in a cohort of Mexican children suffering from T1D30 which supports its possible anti-inflammatory properties. In contrast, Oscillospira was negatively associated with diabetes development and low level of Treg cells. This is in accordance with a recent study by Everard et al. showing that treatment with probiotic yeast resulted in (inter alia) reduction of Oscillospira abundance, what was further linked with reduced hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic mice. Moreover, this clade was positively correlated with the adiposity index and epididymal adipose tissue which consolidates its rather negative role in the development of an inflammatory disease state.31
All taxa in the early fecal gut microbiota that correlated to diabetes onset age and FoxP3+ Treg also correlated significantly to CD11b+ DCs in the spleen and there was a tendency (p = 0.07–0.08) for the major bacterial markers S24–7, Lachnospiraceae, and Bacteriodales to also correlate to splenic IFN-γ producing T cells. This suggests a combined protective role of these immune cell subsets. This seems in contrast to the fact that IFN-γ producing Th1 cells are involved in T1D pathogenesis. However, it is not known whether these IFN-γ producing T cells are pathogenic. In parallel, the sexual dimorphism in incidence is lost in IFN-γ knock out NOD mice and strikingly, the gene expression pattern in IFN-γ signaling pathway was shown to be unique in male specific pathogen free (SPF) NOD mice compared with SPF females and germ-free males and females in which the gender bias in incidence no longer exist.8 Male PLN and MLN also contained more IFN-γ producing T cells, and the authors suggest that IFN-γ negatively regulates Th cells required to activate B cells involved in the pathogenesis of T1D. Based on these studies together with our own data, it is likely that the bacterial taxa that strongly correlated with IFN-γ producing T cells mediate protection against autoimmune diabetes through induction of excessive IFN-γ production as this was characteristic for the non-diabetic NOD mice compared to the diabetic mice; this was significant in both siLP lymphocytes and splenocytes. This hypothesis is further supported by the failure to induce protection with Freund's Complete Adjuvant (CFA) in IFN-γ signaling-deficient NOD mice.32 That study indicated that IFN-γ signaling control the susceptibility of pathogenic T cells to the inhibitory activity of Treg cells, which in parallel to our results seem probable as Treg and IFN-γ producing T cells simultaneously increased with delayed diabetes onset age. This might also explain why attenuated diabetes incidence in vancomycin-treated NOD mice is accompanied by excessive IFN-γ production in siLP lymphocytes.11
A positive correlation between delayed diabetes onset age and the proportion of CD11b+ DCs in MLN and spleen at diabetes onset time indicates a more tolerogen phenotype of this subset compared with their pathogenic CD11b− counterparts that are responsible for breaking peripheral tolerance.33 An immunosuppressive milieu created by these tolerogenic DCs was previously documented to inhibit autoreactive T cells and suppress autoimmune diabetes in an in vivo transfer model.34 It is, thus, possible that the microbes characteristic for the signature profile of non-diabetic NOD mice mediate a protective effect through induction of immunosuppressive CD11b+ DCs in the gut, which migrate to islets to exert its inhibitory effect. However, Kriegel et al. found that the tolerogenic DCs had reduced levels of activation markers CD80 and CD86 which is in contrast to the high level of CD40+ DCs that in this study correlated positively to delayed diabetes onset age. These cells were however not significantly different between diabetic and non-diabetic NOD mice. Also, another study found an increased proportion of the CD11b+CD11c+ DC subset within spleen in NOD, but not in congenic non-obese diabetes-resistant (NOR) mice, compared with BALB/c and C57BL/6 mice.35 This was further reinforced by data showing increased percentage of CD11b+ DCs during insulitis. Thus, the phenotype of the CD11b+ DCs associated with late diabetes onset needs further investigation.
Our proposed model is that bacterial members belonging to S24–7, Bacteriodales, and Prevotella act in favor of diabetes protection while Lachnospiraceae, Ruminococcus, and Oscillospira promote pathogenesis. Their regulatory effect may be mediated through or together with changes in FoxP3+ Treg cells, CD11b+ DCs, and IFN-γ production. This model is illustrated in the PCA plots that demonstrate how these parameters direct diabetes onset what further indicates the importance of the early life events in the intestinal environment in the development of diabetes.
Methods
The experiments were carried out in accordance with the Council of Europe Convention European Treaty Series (ETS) 123 on the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes, and the Danish Animal Experimentation Act (LBK 1306 from 11/23/2007). The study was approved by the Animal Experiments Inspectorate, Ministry of Justice, Denmark.
Animals and diet
NOD/BomTac mice (Taconic, NY, USA) were purchased and mated in a 2 y time interval, and an overall number of 46 female offspring were group-housed at our barrier animal facility (Faculty of Health and Medical Sciences, University of Copenhagen, Frederiksberg, Denmark) under standard conditions and used for the analyses. Mice had free access to Altomin 1324 diet (Brogaarden, Lynge, Denmark) and water. From ten weeks of age, measurements of tail blood glucose commenced twice a week. A mouse was considered diabetic and killed when blood glucose levels exceeded 12 mmol/l on 2 consecutive days. At the age of 30 weeks (210 days), all remaining mice were killed by cervical dislocation. Two mice were diabetic at 30 weeks of age (203 and 206 days) but were included in the non-diabetic group throughout the analyses as these 2 mice were very late diabetic and thus resembled the non-diabetic group in both microbiota and immune parameters. The diabetes incidence for male mice was very low and data on diabetic males were therefore too insufficient to include.
Cell Isolation and Flow Cytometry
Immediately after the mice were killed when diagnosed as diabetic or at 30 weeks of age, organs were placed in cold PBS. Single cell suspensions from spleen, MLN, PLN, and siLP were prepared, PMA/ionomycin stimulated, and analyzed by flow cytometry as described previously.11 The following immune cell subsets were analyzed as follows: Tregs: % FoxP3+ cells in CD4+ cell gate, tolerogenic DCs: % CD11b+ cells or % CD103+ cells in CD11c+ cell gate, activated DCs: % CD40+ cells in CD11c+ cell gate, Th1 cells: % IFN-γ+ producing cells in CD4+ cell gate, and suppressive T cells: % IL-10+ producing cells in CD4+ cell gate. All dead cells were first gated out using 7-AAD viability staining kit (eBioscience, San Diego, CA) except on intracellular stained cells. All antibodies were purchased from eBiosciences. The immunological analyses were performed using an Accuri C6 flow cytometer and software (Accuri Cytometers Inc., Ann Arbor, MI). Same template with fixed gates was used in each analysis to ensure reproducibility between test days. The number of samples collected and used in the correlation analyses for each immune cell subset is shown in Table 1. The analyses were performed with Pearson correlation coefficient.
Tag-encoded pyrosequencing
Feces samples from 16 NOD mice were collected at 4 weeks of age at weaning. The composition of the bacterial community of feces samples was determined using tag-encoded 454/FLX Titanium (Roche) pyro-sequencing of the V3 and V4 region of the 16S rRNA (rRNA) gene by the National High Throughput DNA Sequencing Center, University of Copenhagen, Denmark as described in Hansen et al.11
Pyrosequencing data analysis
Quantitative Insight Into Microbial Ecology (QIIME version 1.7.0) was used to analyze the pyrosequencing data (NCBI accession numbers: no. SRS471978, no. SRS270214, no. SRS2704(25)-SRS2704(35), and no. SRS2704(38)-SRS2704(40)).
Initial analysis steps such as quality control, de-noising, chimera filtering and operational taxonomic unit (OTU) picking were conducted as previously described.36 High quality sequences purged from chimeric reads were clustered at 97% relatedness using UCLAST.37 The representative sequences from each cluster were aligned with pyNAST38 and subjected to the Greengenes reference database (version 12.10). For each category of data sets, a set of 1000 subsampled OTU tables was generated in order to unify the number of reads per sample.
Taxa differences
Determining differences in taxa composition between gut microbiota of diabetic and non-diabetic mice was performed using otu_category_significance.py script. The G test of independence (g_test) and ANOVA determining respectively: qualitative (presence/absence) and quantitative (relative abundance) association of OTUs in the given category, were calculated based on 1000 subsampled OTU tables rarefied to an equal number of reads (1400 per sample). Principal coordinate analysis (PCoA) plot was generated with Jackknifed Beta Diversity workflow based on 10 unweighted UniFrac distance metrics calculated using 10 subsampled OTU tables. The minimum number of sequences taken for each jackknifed subset was set to 1400. Two samples were withdrawn from the plot due to low reads number. Analysis of similarities (ANOSIM) was used to evaluate group differences using unweighted uniFrac distance metrics that were generated based on rarefied (1400 reads per sample) OTU tables. Alpha diversity measure expressed with an observed species (sequence similarity 97% OTUs) value was computed for rarefied OTU tables (1400 reads per sample) using α rarefaction workflow. Testing on the differences in α diversity was conducted using t-test employing the non-parametric (Monte Carlo) method (999 permutations) that is implemented in the compare α diversity workflow.
Microbiota correlation analysis
Correlations between host parameters at diabetes onset age and taxa relative abundance summarized to the genus level in samples collected at weaning were verified with the Pearson's product-moment correlation coefficient implemented in the otu_category_significance.py script (QIIME 1.7.0). Testing was performed based on 1000 rarefied OTU tables unified to an equal number of reads per sample that were constituting 85% of the most read-indigent sample within the subset. An in-house Matlab (Mathworks) script was used to examine all vectors from subsampled OTU-tables with a Jarque-Bera test for normal distribution. One sample was excluded from all microbiota analyses as an outlier because its PCoA values were below mean ± 2 x SD of all samples and changed significant results to/from non-significant results.
PCA based on selected variables
The singular value decomposition data matrix factorization was used to perform the PCA using an in-house MATLAB (Mathworks) script. The pre-processing step included an auto-scaling. The data matrix was composed of values expressing genus level relative distribution of bacteria that significantly correlated with diabetes onset age together with corresponding immune cell measures.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The CHemometric ANalysis Center (CHANCE), University of Copenhagen is kindly acknowledged for assistance in data treatment.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was funded by and carried out as part of CHANCE, CHemometric ANalysis Center at the University of Copenhagen. The project was also funded by Carlsberg Foundation. It was further supported by CALAR, the Center for Applied Laboratory Animal Research.
References
- 1. Todd JA. Etiology of Type 1 Diabetes. Immunity 2010; 32:457-67; PMID:20412756; http://dx.doi.org/ 10.1016/j.immuni.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 2. Wicker LS, Todd JA, Peterson LB. Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol 1995; 3:179-200; http://dx.doi.org/ 10.1146/annurev.iy.13.040195.001143 [DOI] [PubMed] [Google Scholar]
- 3. Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world-recent facts and figures. Immunol Today 1993; 14:193-6; PMID:8517916; http://dx.doi.org/ 10.1016/0167-5699(93)90160-M [DOI] [PubMed] [Google Scholar]
- 4. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008; 455:1109-13; PMID:18806780; http://dx.doi.org/ 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawaguchi-Miyashita M, Shimizu K, Nanno M, Shimada S, Watanabe T, Koga Y, Matsuoka Y, Ishikawa H, Hashimoto K, Ohwaki M. Development and cytolytic function of intestinal intraepithelial T lymphocytes in antigen-minimized mice. Immunology 1996; 89:268-73; PMID:8943725; http://dx.doi.org/ 10.1046/j.1365-2567.1996.d01-740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohsugi T, Kurosawa T. Increased incidence of diabetes mellitus in specific pathogen-eliminated offspring produced by embryo transfer in NOD mice with low incidence of the disease. Lab Anim Sci 1994; 44:386-8; PMID:7983856 [PubMed] [Google Scholar]
- 7. Kriegel MA, Sefik E, Hill JA, Wu H-J, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. PNAS 2011; 108:11548-53; PMID:21709219; http://dx.doi.org/ 10.1073/pnas.1108924108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender Bias in Autoimmunity Is Influenced by Microbiota. Immunity 2013; 39:400-12; PMID:23973225; http://dx.doi.org/ 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. 2009; 3:536-48; PMID:19225551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brugman S, Klatter FA, Visser JTJ, Wildeboer-Veloo ACM, Harmsen HJM, Rozing J, Bos NA. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 2006; 49:2105-8; PMID:16816951; http://dx.doi.org/ 10.1007/s00125-006-0334-0 [DOI] [PubMed] [Google Scholar]
- 11. Hansen CHF, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012; 55:2285-94; PMID:22572803; http://dx.doi.org/ 10.1007/s00125-012-2564-7 [DOI] [PubMed] [Google Scholar]
- 12. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 2010; 6:e25792-2; http://dx.doi.org/ 10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2013; 62:1238-44; PMID:23274889; http://dx.doi.org/ 10.2337/db12-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, ty HHO, et al. Toward defining the autoimmune microbiome for type 1 diabetes. The ISME Journal 2010; 5:82-91; PMID:20613793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hill DA, Artis D. Intestinal Bacteria and the Regulation of Immune Cell Homeostasis. Annu Rev Immunol 2010; 28:623-67; PMID:20192812; http://dx.doi.org/ 10.1146/annurev-immunol-030409-101330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 2010; 107:12204-9; PMID:20566854; http://dx.doi.org/ 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Brien F, Lyons A, Sherlock G, MacSharry J, Kiely B. PLOS Pathogens: Commensal-Induced Regulatory T Cells Mediate Protection against Pathogen-Stimulated NF-κB Activation. PLoS Pathog 2008; 8:e1000112-e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri–induced Regulatory T cells Protect against an Allergic Airway Response in Mice. Am J Respir Crit Care Med 2009; 179:186-93; PMID:19029003; http://dx.doi.org/ 10.1164/rccm.200806-951OC [DOI] [PubMed] [Google Scholar]
- 19. Livingston M, Loach D, Wilson M, Tannock GW, Baird M. Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol Cell Biol 2009; 88:99-102; PMID:19786979; http://dx.doi.org/ 10.1038/icb.2009.71 [DOI] [PubMed] [Google Scholar]
- 20. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011; 331:337-41; PMID:21205640; http://dx.doi.org/ 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Billiard F, Litvinova E, Saadoun D, Djelti F, Klatzmann D, Cohen JL, Marodon G, Salomon BL. Regulatory and effector T cell activation levels are prime determinants of in vivo immune regulation. J Immunol 2006; 177:2167-74; http://dx.doi.org/ 10.4049/jimmunol.177.4.2167 [DOI] [PubMed] [Google Scholar]
- 22. Herbelin A, Gombert JM, Lepault F, Bach JF, Chatenoud L. Mature mainstream TCR α β+CD4+ thymocytes expressing L-selectin mediate "active tolerance" in the nonobese diabetic mouse. J Immunol 1998; 161:2620-8 [PubMed] [Google Scholar]
- 23. Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 Costimulation Is Essential for the Homeostasis of the CD4+CD25+Immunoregulatory T Cells that Control Autoimmune Diabetes. Immunity 1999; 12:431-40; http://dx.doi.org/ 10.1016/S1074-7613(00)80195-8 [DOI] [PubMed] [Google Scholar]
- 24. You S, Leforban B, Garcia C, Bach J-F, Bluestone JA, Chatenoud L. Adaptive TGF-β-dependent regulatory T cells control autoimmune diabetes and are a privileged target of anti-CD3 antibody treatment. Proc Natl Acad Sci U S A 2007; 104:6335-40; PMID:17389382; http://dx.doi.org/ 10.1073/pnas.0701171104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petzold C, Riewaldt J, Watts D, Sparwasser T, Schallenberg S, Kretschmer K. Foxp3(+) regulatory T cells in mouse models of type 1 diabetes. J Diabetes Res 2012; 2013:940710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tucker CF, Nebane-Ambe DL, Chhabra A, Parnell SA, Zhao Y, Alard P, Kosiewicz MM. Decreased frequencies of CD4 +CD25 +Foxp3 +cells and the potent CD103 +subset in peripheral lymph nodes correlate with autoimmune disease predisposition in some strains of mice. Autoimmunity 2011; 44:453-64; PMID:21604973; http://dx.doi.org/ 10.3109/08916934.2011.568553 [DOI] [PubMed] [Google Scholar]
- 27. Badami E, Sorini C, Coccia M, Usuelli V, Molteni L, Bolla AM, Scavini M, Mariani A, King C, Bosi E, et al. Defective Differentiation of Regulatory FoxP3+ T Cells by Small-Intestinal Dendritic Cells in Patients With Type 1 Diabetes. Diabetes 2011; 60:2120-4; PMID:21646390; http://dx.doi.org/ 10.2337/db10-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuño MI. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-controlstudy. BMC Medicine 2013; 11:46; PMID:23433344; http://dx.doi.org/ 10.1186/1741-7015-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, et al. Gut Microbiome Metagenomics Analysis Suggests a Functional Model for the Development of Autoimmunity for Type 1 Diabetes. PLoS ONE 2011; 6:e25792; PMID:22043294; http://dx.doi.org/ 10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Esther Mejia-Leon M, Petrosino JF, Ajami NJ, Dominguez-Bello MG, Calderon de la Barca AM. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep 2014; 4:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. Saccharomyces boulardii Administration Changes Gut Microbiota and Reduces Hepatic Steatosis, Low-Grade Inflammation, and Fat Mass in Obese and Type 2 Diabetic db/db Mice. mBio 2014; 5:e01011-14-e01011-14; PMID:24917595; http://dx.doi.org/ 10.1128/mBio.01011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mori Y, Kodaka T, Kato T, Kanagawa EM, Kanagawa O. Critical role of IFN- in CFA-mediated protection of NOD mice from diabetes development. Int Immunol 2009; 21:1291-9; PMID:19778991; http://dx.doi.org/ 10.1093/intimm/dxp097 [DOI] [PubMed] [Google Scholar]
- 33. Katz JD, Ondr JK, Opoka RJ, Garcia Z, Janssen EM. Cutting Edge: Merocytic Dendritic Cells Break T Cell Tolerance to Cell Antigens in Nonobese Diabetic Mouse Diabetes. J Immunol 2010; 185:1999-2003; PMID:20644171; http://dx.doi.org/ 10.4049/jimmunol.1001398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kriegel MA, Rathinam C, Flavell RA. Pancreatic islet expression of chemokine CCL2 suppresses autoimmune diabetes via tolerogenic CD11c+ CD11b+ dendritic cells. Proc Natl Acad Sci U S A 2012; 109:3457-62; PMID:22328150; http://dx.doi.org/ 10.1073/pnas.1115308109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steptoe RJ, Ritchie JM, Harrison LC. Increased generation of dendritic cells from myeloid progenitors in autoimmune-prone nonobese diabetic mice. J Immunol 2002; 168:5032-41; http://dx.doi.org/ 10.4049/jimmunol.168.10.5032 [DOI] [PubMed] [Google Scholar]
- 36. Krych L, Hansen CHF, Hansen AK, van den Berg FWJ, Nielsen DS. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS ONE 2012; 8:e62578-8; http://dx.doi.org/ 10.1371/journal.pone.0062578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460-1; PMID:20709691; http://dx.doi.org/ 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 38. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26:266-7; PMID:19914921; http://dx.doi.org/ 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.