Abstract
Riboswitches are RNA-based regulators of gene expression composed of a ligand-sensing aptamer domain followed by an overlapping expression platform. The regulation occurs at either the level of transcription (by formation of terminator or antiterminator structures) or translation (by presentation or sequestering of the ribosomal binding site). Due to a modular composition, these elements can be manipulated by combining different aptamers and expression platforms and therefore represent useful tools to regulate gene expression in synthetic biology. Using computationally designed theophylline-dependent riboswitches we show that 2 parameters, terminator hairpin stability and folding traps, have a major impact on the functionality of the designed constructs. These have to be considered very carefully during design phase. Furthermore, a combination of several copies of individual riboswitches leads to a much improved activation ratio between induced and uninduced gene activity and to a linear dose-dependent increase in reporter gene expression. Such serial arrangements of synthetic riboswitches closely resemble their natural counterparts and may form the basis for simple quantitative read out systems for the detection of specific target molecules in the cell.
Keywords: riboswitch design, synthetic biology, synthetic riboswitch, tandem riboswitch, theophylline aptamer, transcription regulation
Abbreviations
- bgaB
B. stearothermophilus-derived beta-galactosidase
- GFP
green fluorescent protein
- MU
Miller units
- PCR
polymerase chain reaction
- RS
riboswitch
Introduction
Riboswitches are versatile devices for synthetic biology applications. They consist of an aptamer domain that binds defined ligands with high affinity and an expression platform translating aptamer binding into an effect on gene expression.1 In synthetic biology, they are often used to design complex gene regulation circuits.2-5 In recent years there has been great effort in designing synthetic riboswitches that can in principle bind any ligand and regulate any step in gene expression of interest.6 Regulation of transcription is of special interest, as it allows for a tight regulation in an early step of gene expression.7 Furthermore, transcriptional control can be employed to regulate both protein expression as well as noncoding RNAs. In bacteria, riboswitches regulating transcription usually contain an intrinsic rho-independent terminator in the expression platform. Such an element is characterized by a stable, often GC-rich stem-loop-structure followed by a U-rich tail.8 Naturally occurring transcriptional riboswitches usually represent OFF-switches, where the binding of a ligand represses gene expression. This often leads to a feedback inhibition, where a certain metabolite produced by the riboswitch-controlled gene products shuts down its own synthesis.9 In synthetic biology, however, ON-switches that activate gene expression upon binding of a small inducer molecule are highly preferred.10 Two distinct strategies have been applied for the design of artificial transcriptional riboswitches. In the first approach, an in vitro selected theophylline-binding aptamer (see ref. 11) was fused via a short spacer region to a sequence complementary to the 3′-part of the aptamer. Together with this aptamer part, the spacer region and the complementary sequence lead to the formation of a stable intrinsic terminator in the absence of the aptamer ligand. In the presence of the ligand, the aptamer structure was stabilized and precluded terminator formation, leading to an increase in gene expression.12 In a similar setup based on natural elements, aptamer and terminator were connected via a P1-helix that was part of both regions.13,10 In an approach for designing ON-switches, aptamer and terminator from natural riboswitches required decoupling before an exchange of the aptamer domain was possible.10 This strategy can be challenging as decoupling requires mutations at positions that might be essential for proper function of the terminator. However, when designing purely synthetic riboswitches as in our preceding study (see ref. 12), it is hard to predict if these constructs will be functional at all. Natural transcriptional terminators tend to be shorter than those required for aptamer disruption in riboswitch design. As hairpin stability alone shows no direct correlation with riboswitch functionality, the efficiency of a designed terminator is hard to predict, although great efforts were made to solve this problem.14,15
Here, we analyzed which features of the first synthetic transcriptional riboswitches (see ref. 12) influence their functionality in Escherichia coli. As the synthetic terminator domain, in contrast to the aptamer, can vary in sequence and structure, the functionality of the isolated terminators was determined. Our data represent the first direct experimental proof for the hypothesis that riboswitch functionality is strongly affected by the stability of the terminator hairpin. Furthermore, we show that the folding pathway and the existence of possible folding traps in the terminator element are an equally important parameter. Therefore, both features have to be considered when designing functional riboswitches acting on transcription. In addition, we show that a fast and reliable way to enhance the activation ratio of transcriptional riboswitches is the design of constructs with several riboswitch copies. Such serial arrangements might be useful tools for the measurement of intracellular ligand concentration. The combination of optimized hairpin stability, absence of terminator folding traps and serial riboswitch arrangement should allow a more reliable and efficient design strategy for these synthetic regulatory RNA elements.
Results
Terminator composition is critical for riboswitch functionality
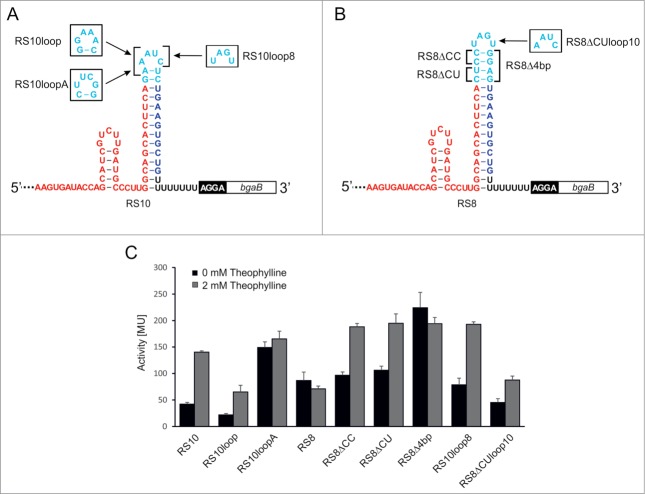
In our initial study for designing transcriptional ON-riboswitches that activate gene expression upon theophylline binding, 3 of 6 tested constructs from the in silico selection were able to regulate gene expression in a theophylline-dependent manner in E. coli. Here, we mainly focus on riboswitch 10 (RS10), as it was the best performer in the initial test set, showing a 3-fold activation.12 This riboswitch possesses a 14 base pair long terminator stem without mismatches and an AAUC tetraloop sequence in the resulting hairpin (Fig. 1A). While most naturally occurring terminators hold such tetraloops, 2 loop sequences, UUCG and GAAA, are found at high frequency.8,16 These loops are often closed by a C-G base pair. Replacing the AAUC loop sequence (closed by an A-U pair) by the sequence GAAA and a closing G-C pair reduced the background by 2-fold of the reporter gene activity in the absence of theophylline (RS10loop, Fig. 1A and C, see ref. 12), indicating that this loop enhances terminator performance and efficiently blocks expression of the downstream reporter gene. Encouraged by this result, we tested the most common terminator tetraloop UUCG with a closing C-G base pair for its ability to minimize read-through. The corresponding construct RS10loopA (Fig. 1A) did not show a reduced background activity, but a high overall activity, leading to a loss of theophylline-dependent gene regulation (Fig. 1C). Hence, this tetraloop seems to be not suitable for the terminator environment of RS10, indicating that specific features of an individual terminator might not be generally applicable for any structural context.
Figure 1.
Riboswitch variants and activity tests. Sequence and secondary structure of the functional riboswitch 10 (RS10), the non-functional riboswitch 8 (RS8) and tested mutants are shown in (A) and (B), respectively. Shine-Dalgarno sequence (black box) and bgaB reporter gene (white box) follow immediately downstream of the riboswitches. Aptamer sequence is shown in red, the spacer region in cyan, and the sequence complementary to the aptamer is indicated in blue, followed by a black U-stretch. These elements make up the intrinsic terminator element. Mutated positions are indicated. (C) Activity test of β-galactosidase reporter constructs with RS10 and RS8 mutants. Activities are indicated in Miller Units. The reduction in activity observed for RS8 and RS8Δ4bp is statistically not significant according to Student's t-test (p = 0.17).
When comparing functional and non-functional riboswitches in our previous study (see ref. 12), we hypothesized that the stability of the terminator stem seemed to be limiting for riboswitch function. Riboswitch 8 (RS8) carried the most stable terminator and showed a constitutive theophylline-independent repression of the reporter gene. Yet, the isolated terminator revealed a comparably low termination efficiency. This was not expected, as this riboswitch showed many structural similarities with the functional riboswitch RS10 (Fig. 1B). Both elements differ only in length and sequence of the spacer element in the terminator hairpin, but share the same complementarity to the aptamer sequence and a 4-base hairpin loop representing a common feature of natural E. coli terminators.16 As RS8 contains a longer spacer sequence (12 versus 8 nts) resulting in a much more stable terminator stem, we assumed that this construct formed a terminator structure unaffected by theophylline. If this hypothesis were true, the destabilization of the terminator stem should allow a ligand-dependent switch from terminator to aptamer structure as it was observed for RS10. We removed 2 base pairs in the terminator stem that were part of the spacer, resulting in constructs having deleted either 2 C-G base pairs (RS8ΔCC) or a C-G and a U-A base pair (RS8ΔCU) (Fig. 1B). When testing these constructs for regulating gene expression in a theophylline-dependent way, they showed a 2-fold activation of the β-galactosidase gene, while the original RS8 construct did not respond to theophylline (Fig. 1C). Accordingly, a massive destabilization of the terminator in RS8 should lead to a constitutive ON state. We therefore combined the 2 deletions in the terminator stem, leading to RS8Δ4bp (Fig. 1B). As expected, this construct showed a theophylline-independent high reporter gene activity (Fig. 1C). The slight reduction in the presence of theophylline that was also observed for the original RS8 turned out to be statistically not significant (p = 0.17).
Although the constructs RS8ΔCC and RS8ΔCU showed a 2-fold activation ratio, RS10 had a slightly better performance. As both switches differ also in their tetraloop sequences, the influence of these loop regions was analyzed by reciprocal exchange between RS10 and RS8ΔCU, resulting in RS10loop8 and RS8ΔCUloop10 (Fig. 1). While both constructs retained a 2-fold activation of the reporter gene, RS8ΔCUloop10 showed a general strong reduction in β-galactosidase activity, indicating that the loop of RS10 increases the transcription termination efficiency of this construct.
The activation of the RS8 construct by weakening the terminator hairpin strongly supports the hypothesis that the stability of the individual terminators has a dramatic impact on the functionality of the riboswitches. While the functional constructs have terminator hairpins with ΔG values between −21.9 and −24.8 kcal/mol, the constitutively active RS8Δ4bp has an unstable terminator with −18.1 kcal/mol (Table 1). On the other hand, the original RS8 carries the most stable terminator (−29.0 kcal/mol) that inhibits riboswitch refolding upon ligand binding. While the different hairpin stabilities of RS8Δ4bp and RS8 explain the observed gene expression levels, it remained unclear why RS10loopA with an intermediate hairpin stability of −24.3 kcal/mol is not functional. We therefore investigated the efficiencies of the individual terminator elements in more detail.
Table 1.
Energy values for terminator hairpin formation and folding traps. Functional riboswitches require that the values of both parameters lie within a relatively narrow range. For the theophylline-binding aptamer, the terminator stability should be higher than −18.1 kcal/mol and lower than −29.0 kcal/mol. Furthermore, the formation barrier of the functional terminator hairpin should not exceed 6.7 kcal/mol to prevent potential kinetic folding traps. The functional constructs are marked with “+”.
| Riboswitch | Terminator free energy (kcal/mol) | Terminator formation barrier (kcal/mol) | Riboswitch functionality |
|---|---|---|---|
| RS10 | −21.9 | 2.4 | + |
| RS10loop | −24.8 | 1.6 | + |
| RS10loopA | −24.3 | 10.0 | − |
| RS8 | −29.0 | 12.2 | − |
| RS8ΔCC | −22.1 | 2.9 | + |
| RS8ΔCU | −24.5 | 6.7 | + |
| RS8Δ4bp | −18.1 | 2.9 | − |
| RS10loop8 | −22.6 | 2.4 | + |
| RS8ΔCUloop10 | −23.9 | 2.9 | + |
Terminator function corresponds to riboswitch function
To study individual isolated terminator elements, the 5′-region of the aptamer that was not part of the terminator hairpin was removed. The resulting constructs were tested for β-galactosidase activity (Fig. 2A) as well as mRNA expression (Fig. 2B) to confirm a regulation at the level of transcription. To illustrate the maximal activity of the reporter, a construct lacking a terminator hairpin but still holding the U8-track was used (bgaBU8). As published before, constructs comprising either the isolated aptamer or the U8-track alone result in a constitutively active reporter gene.12 Terminators from functional riboswitches showed good (T10loop) or intermediate termination efficiencies (T10, T8ΔCC, T8ΔCU, T10loop8, T8ΔCUloop10), leading to lower reporter gene activities than those of the non-functional riboswitches RS10loopA, RS8 and RS8Δ4bp. In agreement with this, the Northern blot analyses revealed the most intense mRNA hybridization signals for these weaker terminator elements. Accordingly, the corresponding riboswitch constructs RS10loopA and RS8Δ4bp showed the highest reporter gene activity in the absence of theophylline (Fig. 1C). The non-functional RS8, however, which is also independent of theophylline, showed only an intermediate expression rate, although it carries the most stable hairpin (Fig. 1C; Table 1). This result indicates that hairpin stability is not directly correlated with transcription termination and cannot be taken as the sole criterion for riboswitch construction.
Figure 2.
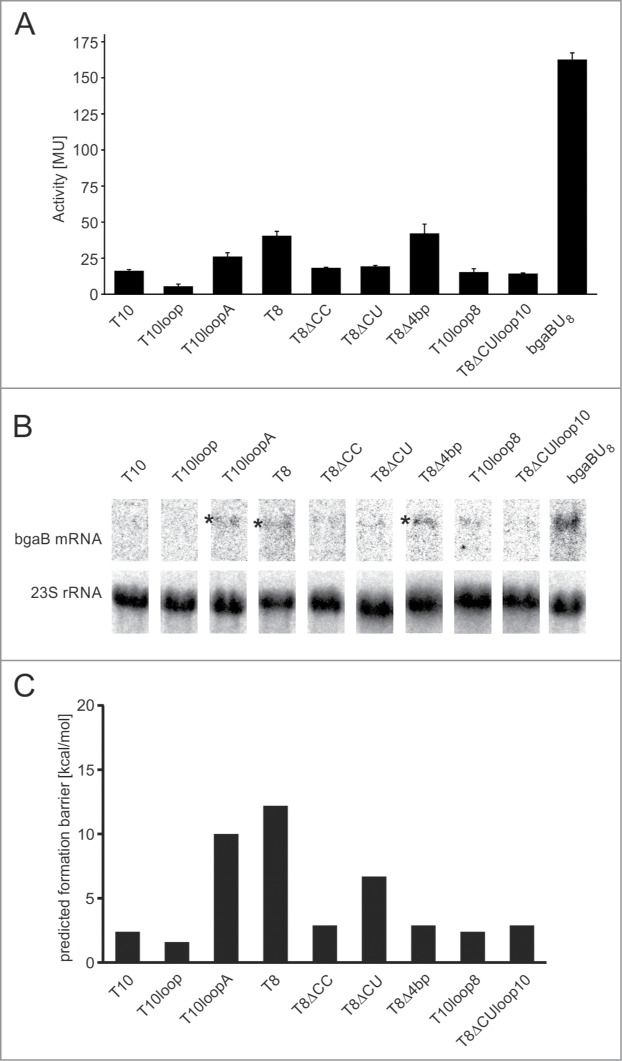
Analysis of isolated terminator sequences. (A) Activity test of the β-galactosidase reporter gene, indicating transcriptional read-through for isolated terminator constructs that leads to gene expression in the absence of theophylline. The positive control bgaBU8 is a construct lacking the terminator hairpin but still carries the U8 stretch. Activities are given in Miller Units. (B) Northern blot analysis of full-length β-galactosidase mRNA from terminator constructs. As internal control, the hybridization signal for 23S rRNA is shown. Asterisks indicate the position of mRNA signals where a sufficient amount of transcriptional read-though occurred. (C) Predicted energy barriers for terminator formation in the individual constructs in kcal/mol. Barriers were computed using the Pathfinder algorithm.21 Constructs with high formation barriers (T10loopA, T8, T8ΔCU) are predicted to get trapped in a meta-stable state.
In silico parameterization of transcriptional riboswitches
To find an answer why the individual riboswitch constructs performed so differently in the experiment, we conducted a bioinformatical analysis of their RNA transcripts. Similar to the total free energy values described above (Table 1), other terminator hairpin features, such as GC content or stem length did not show a consistent correlation with the background activity signal of the individual riboswitches or isolated terminators. As an additional parameter, the seed stability during hairpin formation was calculated for each terminator sequence. The nucleation of hairpins strongly correlates with the stability of the innermost base pairs, since these close the loop region.17 The stability of these seeds can be approximated by the folding energy of the loop and its adjacent base pairs forming the helix of the terminator hairpin.17–19 Our calculations suggest that the loops of most of the terminator hairpins need up to 4 base pairs to shift the equilibrium structure toward a closed loop conformation, i.e. to exhibit a negative free energy of nucleation. In this approximation, only the hairpin of T10loop seems to have a slightly better stabilization already after closing the first 2 base pairs due to the stabilizing effect of the tetraloop motif (Fig. S2). With the exception of T8Δ4bp, the seed stabilities of the remaining terminators show no significant differences and therefore do not allow to distinguish functional from non-functional riboswitch constructs.
As a next step, we took the sequence context of the terminator elements into account to check for possible kinetic effects during the transcription process. For each riboswitch transcript, a co-transcriptional folding trajectory was calculated with the program kinwalker (see ref. 20), setting the transcription at a uniform speed of 200 nt/s. In this simulation, kinwalker predicts RNA structure intermediates formed during the proceeding transcription. This allows a time-resolved identification of possible folding traps in the nascent RNA molecule. To determine the stability of putative traps, we calculated the energy barrier that separates them from the active terminator state, using the Pathfinder algorithm (see ref. 21) to compute an optimal possible unrestricted refolding path between the 2 structural states. For terminators T10loopA, T8 and T8ΔCU, considerable energy barriers of 10.0 kcal/mol, 12.2 kcal/mol and 6.7 kcal/mol were predicted. The occurrence of these rather stable folding barriers in the transcription simulation suggests the presence of kinetic folding traps. All other tested terminator hairpins do not show trapped states in this model (Figs. 2C, 3).
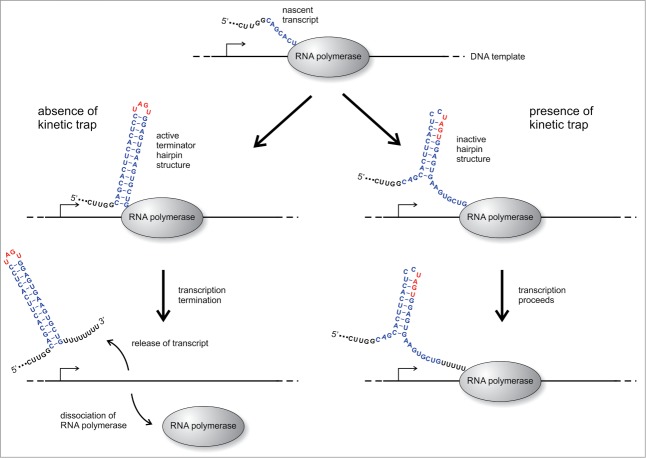
Figure 3.
Model for a kinetic folding trap. Some of the designed transcripts do not show terminator activity even in the absence of the aptamer platform, as it is shown here for terminator T8. Although having a nucleotide composition to form a stable terminator hairpin (helix part shown in blue, tetra loop depicted in red), their sequence also encodes for a predicted kinetically trapped structure state. In the absence of such a kinetic trap, the active terminator hairpin is expected to immediately form during transcription (left). This results in transcription termination along with release of premature RNA and dissociation of the polymerase from the template. If a predicted kinetic trap is present (right), the RNA co-transcriptionally folds into an inactive structure which interferes with terminator hairpin formation. The time needed to refold into the active terminator conformation is then dictated by the energy barrier separating both structure states. In cases of high energy barriers, transcription is not interrupted, since the terminator hairpin is formed only after the polymerase moved on.
Taken together, the 2 parameters hairpin stability and folding trap can explain the observed activation ratios in the tested riboswitches, while the seed stability appears to play a less prominent role. The combination of the 2 indicative parameters could help to improve the design of transcription regulating riboswitches (see Table 1 and discussion).
Tandem arrangement allows fast and easy enhancement of activation ratio
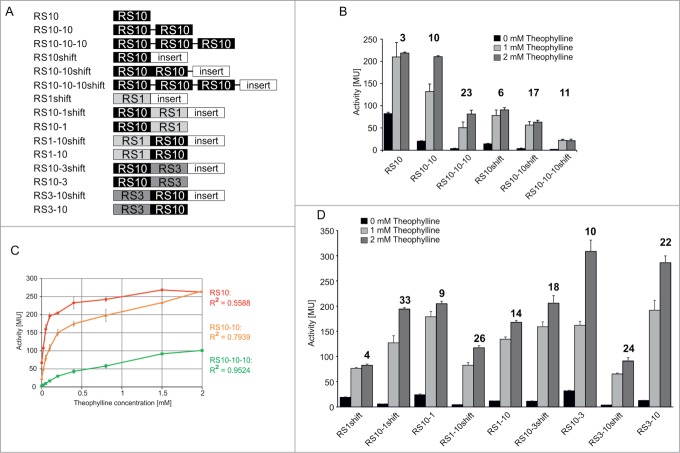
As demonstrated above, base replacements and/or deletions in the terminator stem allowed the conversion of an inactive construct into a functional riboswitch. However, for most of the functional constructs, the activation ratios at high theophylline concentrations remained moderate and could not be further improved by these rational optimization steps. To target this problem, we applied an alternative approach. For several gene regulating RNA elements, tandem construction has been shown to be an easy strategy for increasing activation ratio in splicing (see ref. 22), RNA cleavage (see refs. 23–24), ribosome shunting (see ref. 25) or trans-regulation of transcription.26 One advantage of riboswitches acting at the transcriptional level is that each expression platform represents an independent functional unit. Hence, we tested whether a serial arrangement of riboswitches leads to an increased activation ratio in our constructs. We generated constructs consisting of one, 2 or 3 copies of RS10 in a row (Fig. 4A). To test the functionality of these arrangements, coding sequences for β-galactosidase (Fig. 4) as well as GFP (Fig. S3) were used as reporter genes. Furthermore, as the close proximity of the terminator U stretch and the downstream Shine-Dalgarno sequence leads to a strong theophylline-independent activity of the reporter gene due to an enhanced translation rate (see ref. 27), we additionally designed serial RS constructs based on a modified riboswitch RS10shift.12 In these constructs, a 19 nt insertion increases the distance between riboswitches and Shine-Dalgarno sequence, leading to a dramatic reduction of activity without theophylline (Fig. 4A, B). For all constructs, reporter gene activity was measured in the presence and absence of 1 or 2 mM theophylline (Figs. 4B and S3). With increasing number of riboswitches, the activity of the downstream reporter gene in the absence of theophylline was reduced. Furthermore, in most cases, an increase of activation ratio could be observed. While the single riboswitch construct showed a 3- (RS10) or 6-fold activation (RS10shift) of reporter activity in the presence of theophylline, the tandem constructs allowed a 10- (RS10–10) to 17-fold activation (RS10–10shift). For the constructs with 3 serial riboswitches (tridems), only the construct RS10–10–10 with β-galactosidase reporter showed a further increased activation ratio (23-fold), in contrast to the RS10–10–10shift tridem (11-fold). Similarly, the activation ratio in the tandem riboswitch with GFP reporter showed an increase from 3- towards 10-fold. In the tridem version, however, the activation ratio dropped down to 7-fold (Fig. S3).
Figure 4.
Serial riboswitch constructs. (A) Schematic representation of tandem and tridem constructs upstream of the reporter gene (β-galactosidase or GFP), in 5′ to 3′ orientation. All transcripts start with a 3 nt-leader upstream of the first aptamer domain. For cloning reasons, several constructs carry a 6 nt-gap between the repeats, indicated by a black line. If no line is present, riboswitches directly follow each other. When indicated, an inert 19 nt region of the pBAD multiple cloning site (see ref.12) is inserted between riboswitch and Shine-Dalgarno sequence (RSshift constructs). The same arrangement was chosen for constructs with GFP reporter. (B) β-galactosidase activity of constructs with up to 3 serial riboswitch copies. While in the RS10 constructs the response ratio is increasing with additional riboswitch copies, the RS10–10–10shift construct shows a drop in activation compared to the corresponding tandem. (C) β-galactosidase activity as a function of theophylline concentration for the constructs RS10 (red), RS10–10 (orange) and RS10–10–10 (green). The linear regression coefficient R2 (colored accordingly) indicates that the linearity increases with the number of riboswitches. (D) β-galactosidase activity of tandem combinations of RS10 with RS1 or RS3, respectively. The approximate maximal activation ratio for all constructs is given as a number above each column set.
As shown in Figure 4B, the impact of the different theophylline concentrations was higher for tandem or tridem constructs than for the single riboswitch. Particularly RS10 repeats showed a theophylline dose-dependent increase in reporter gene activity. Hence, we tested for RS10, RS10–10 and RS10–10–10 whether such a linear correlation can be found over a broader range of theophylline concentrations (Fig. 4C). While the single riboswitch RS10 showed a steep increase and fast saturation of β-galactosidase activity at low concentrations of theophylline, the increase of activity became more linear in the tandem and tridem riboswitches. Correspondingly, the linear regression coefficient R2 increased from 0.56 for the single riboswitch to 0.95 for the tridem riboswitch RS10–10–10.
Besides tandem constructs based on identical RS10 repeats, we also fused one copy of RS10 to the equally functional theophylline riboswitches RS1 and RS3 (Fig. 4; Fig. S1). While RS1 has a terminator hairpin of identical length with only a slight difference in GC content (7 vs. 6 GC pairs in RS10), RS3 has a hairpin with 18 bp, but carries one single nucleotide bulge (Fig. S1). However, in terms of stability, these terminators have ΔG values comparable to the RS10 terminator (T1: −21 kcal/mol; T3: −25.8 kcal/mol; T10: −21.9 kcal/mol) in the hairpin region.12 These riboswitches were combined with one copy of RS10 in different orders, leading to the constructs RS10-1, RS10-1shift, RS1-10, RS1-10shift, RS10-3, RS10-3shift, RS3-10 and RS3-10shift (Fig. 4A). Again, all combinations showed a strongly increased activation ratio of the reporter gene for β-galactosidase, resulting in 9- to 33-fold activation compared to the 4- to 6-fold activation of the single riboswitches RS1shift (Fig. 4D), RS10shift (Fig. 4B) or RS3shift.12 The effect of the 19 nt insertion in the shifted constructs is again evident. Compared to the non-shifted tandems, these constructs consistently show a reduced activity in the absence of theophylline. Taken together, the activation enhancement observed for the homogeneous repeats consisting of identical RS10 copies is also visible for heterogeneous tandem repeats. Nevertheless, the shifted combinations of RS1 or RS3 with RS10 show surprising differences. While RS10–3shift and RS3–10shift have an activation ratio of 18 and 24, respectively, the RS10–1shift and RS1–10shift constructs show even better ratios of 33 and 26, respectively. The absolute values in Miller units, however, are rather similar for these constructs (Fig. 4D). Yet, a dose-dependent increase of β-galactosidase activity similar to that of the homogeneous RS10–10 tandem is visible for all 4 heterogeneous riboswitch combinations.
Discussion
Design criteria for synthetic terminators in the riboswitch design context
Using a computational algorithm, we have been able to design functional ON-riboswitches acting at the level of transcription.12 These synthetic regulatory elements are composed of an in vitro selected aptamer for theophylline and an overlapping synthetic intrinsic terminator that includes the 3′-part of the aptamer. While the terminator structure is formed in the ligand-free state, binding of theophylline stabilizes the aptamer conformation and thus precludes the formation of the terminator. According to this hypothesis, the stability of the terminator hairpin should have a strong impact on the competition between these 2 mutually exclusive states. Among the originally tested constructs, RS10 showed the best theophylline-dependent activation ratio (3-fold expression difference between induced and uninduced state). As this riboswitch still shows a considerable gene activation in the absence of theophylline, a stabilization of the terminator hairpin should lead to a reduction of occasional read-through during transcription, increasing the activation ratio. Hence, an adjustment of the terminator structure to the consensus of naturally occurring terminators should enhance its functionality. Studies on such in vivo terminator elements identified several features critical for high termination efficiency.14,15 These are, among others, the genetic environment of the terminator as well as length and composition of the downstream-located U-stretch. As these features are identical in our design, differences in termination efficiency must reside within the terminator hairpins. In natural terminators, GAAA tetraloops closed by a C-G base pair are found rather frequently and are associated with high termination efficiencies.14,15 Accordingly, a corresponding construct T10loop, carrying the GAAA sequence, shows an improved termination efficiency compared to the original T10 element. Interestingly, the most frequent tetraloop sequence, UUCG (see refs. 8,16) did not lead to such an effect. However, a direct correlation between frequency of a certain loop sequence and termination efficiency in natural terminators was not shown yet. As these elements are optimized for the specific needs in the expression of individual genes and strongly depend on the genetic environment, natural terminators show a considerable variation in their effects on transcription. Hence, statistical parameters of terminators are not a reliable tool for the design of highly efficient synthetic terminators that have to function in a riboswitch environment. On the other hand, design principles for terminators with stems of up to 8 base pairs (see ref. 15) are also valid for the terminators in the synthetic riboswitches. While the tested terminators show only slight differences in the calculated seed stabilities (Fig. S2), their predicted folding behavior and the general stability of the terminator hairpin seem to be a more important parameter (Fig. 2C; Table 1).
Interestingly, the combined evaluation of hairpin stability and the prediction of folding traps allows a rather robust prediction of riboswitch functionality (Table 1). Predicted folding traps are structures that interfere with terminator formation (Figs. 2C, 3) and seem to be quite common for synthetic riboswitches, as they were also described for artificial elements regulating translation.28 The most efficient terminator element is found in RS10loop. No folding trap is predicted and the hairpin stability allows formation of the aptamer structure upon theophylline binding. Hence, this construct represents a functional and efficient riboswitch. Most of the other riboswitches carry terminators with intermediate efficiencies (Fig. 2A). Concerning the first parameter hairpin stability, these terminators support a theophylline-induced formation of the aptamer fold. The folding trap parameter, however, shows for 2 of these constructs, RS10loopA and RS8ΔCU, that the presence of a trapping structure is suggested, with folding barriers of 10.0 and 6.7 kcal/mol, respectively (Fig. 2C; Table 1). The possible trap of RS8ΔCU is less robust and can be overcome, leading to the formation of a functional terminator structure. The corresponding riboswitch is functional as it allows both the formation of a terminator in the absence of theophylline as well as its disruption upon binding of the ligand. The predicted folding trap that prevents the formation of the terminator of RS10loopA, however, yields a higher formation barrier that probably inhibits efficient refolding into the terminator structure during transcription. Consequently, this riboswitch does not respond to the presence of theophylline and is locked in a constitutive ON state.
The most stable folding trap is calculated for the terminator used in the RS8 transcript. Here, the trap structure consists of a stable upstream located hairpin stem (−25.4 kcal/mol; Fig. 3, right panel) that is separated from the equally stable terminator folding intermediate (−25.5 kcal/mol) by an energy barrier as high as 12.2 kcal/mol. This barrier presumably prevents refolding from the intermediate conformation into the functional terminator. Furthermore, the high stability of the terminator hairpin structure (−29.0 kcal/mol) may outcompete the aptamer structure even with bound ligand so that no response of the riboswitch to theophylline is observed. Yet, the overall activity of this riboswitch is lower than that of the other 2 inactive constructs RS10loopA and RS8Δ4bp (Fig. 1C). As secondary structure elements in the region upstream of the ribosomal binding site can interfere with the efficiency of translation (see refs. 29–30), it is very likely that a similar effect due to the highly stable structure of the T8 terminator is the reason for the observed reduced reporter gene expression in this construct. The last non-functional riboswitch RS8Δ4bp is also not fulfilling both criteria. While a formation barrier of 2.9 kcal/mol does not result in a folding trap, the terminator hairpin shows a strongly reduced stability (−18.1 kcal/mol), due to the deletion of 4 base pairs in the stem region. Thus, terminator formation is rather inefficient, and the constitutively high gene expression indicates an inefficient termination of transcription (Fig. 1C, Fig. 2A, B).
Taken together, if the hairpin is too stable, the formation of the ligand-bound aptamer structure is not possible. If the stability of the hairpin is too low, then the terminator is not formed, even in the absence of the ligand, and the construct is in a permanent ON state. Hence, the stabilities of the terminator hairpin and the aptamer have to be balanced and adjusted to the stability of the chosen aptamer domain in the constructs. Consequently, a terminator stability that turned out to be functional in combination with the theophylline aptamer is not necessarily suited for a different aptamer. While these parameters make it possible to distinguish functional from non-functional theophylline-dependent riboswitch constructs, it is important to note that the translation of the regulated downstream ORF may also be affected by these structured elements in the 5′-UTR. This complicates the quantitative interpretation of riboswitch-mediated gene regulation. Nevertheless, the parameters of terminator stability and putative formation barriers leading to folding traps have to be considered for the construction of a functional riboswitch acting on transcription and can even be combined with serial arrangements of riboswitches to further improve the functionality (Fig. 5, see discussion below).
Figure 5.
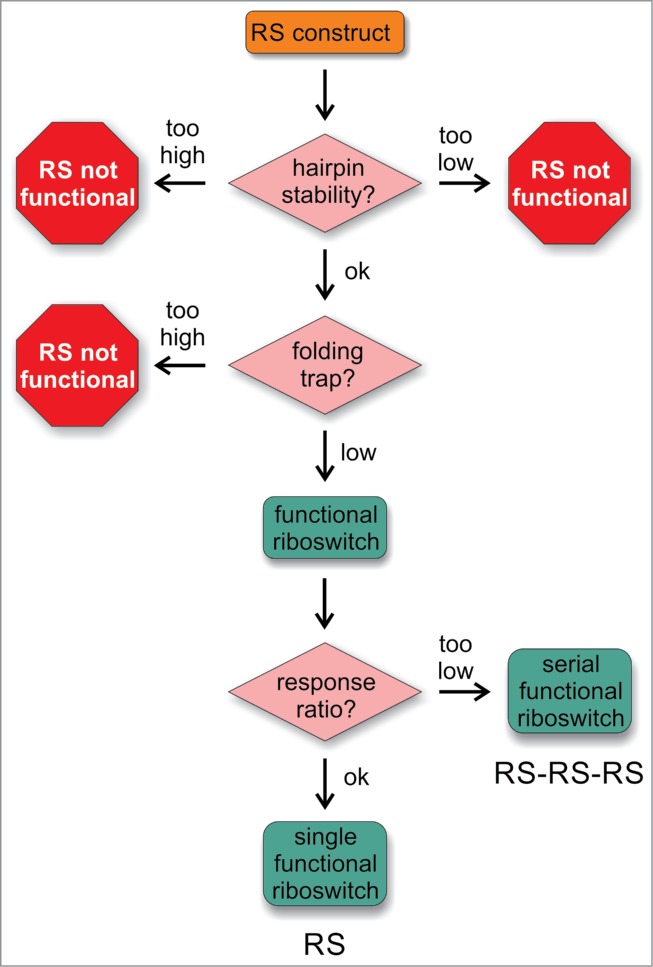
Flow chart for the construction of transcriptional riboswitches. In order to obtain functional riboswitches that act in a ligand-dependent manner, 2 parameters have to be considered. The stability of the terminator hairpin has to be adjusted to the stability of the competing structure of the ligand-bound aptamer. If the terminator is too stable, the riboswitch is in a permanent OFF state and not functional. If the hairpin stability is too low, the terminator will not form, even in the absence of the ligand, resulting again in a non-functional construct. Having a compatible stability of the terminator, possible folding traps have to be considered. If the energy barrier for a refolding into a functional terminator is too high, the riboswitch will be not functional. Only the absence of such traps or a low folding barrier will lead to the formation of a functional construct that regulates a ligand-dependent gene expression. Finally, the response ratio is considered. If it is not sufficient in the single riboswitch construct (RS), a serial arrangement of riboswitch copies (RS-RS-RS) can be used to increase the ON/OFF ratio.
Increasing the activation ratio by tandem and tridem constructs
A tandem arrangement of regulatory elements for gene expression is a frequent tool in nature as well as biotechnology. Many recombinant gene expression systems have to deal with problems arising due to the use of highly active promoters. In such systems, tandem combinations of strong natural intrinsic terminators are a convenient way to achieve an efficient stop of transcription downstream of the desired open reading frame.31,32,15 In naturally occurring riboswitches, tandem arrangements are observed as well. One type of architecture identified in the metE mRNA of Bacillus clausii consists of 2 distinct transcriptional OFF-switches responding to different ligands. The 5′-located element binds S-adenosylmethionine, while the second riboswitch recognizes coenzyme B12.33 The independent gene regulation by these 2 elements represents a Boolean NOR gate, inhibiting the synthesis of metE in the presence of one of the ligands.34 The second type of tandem arrangement is found in glycine riboswitches, where only the aptamer domain is present in 2 copies, followed by a single regulatory domain (containing a terminator or a ribosomal binding site).35-37 A third naturally occurring tandem arrangement is represented by a TPP riboswitch repeat.38,33 Surprisingly, while the tandems consisting of complete riboswitch repeats show an independent function of each unit, the glycine riboswitches with 2 neighboring aptamer domains exhibit a cooperative binding behavior.35,39 Our synthetic serial riboswitches show a rather linear increase of activity with increasing amounts of theophylline, especially in the tridem arrangement (Fig. 4C). Hence, these elements obviously do not show cooperative binding effects, similar to their natural tandem counterparts consisting of complete riboswitch repeats.33,36
The simple repeat of identical or nearly identical riboswitch elements leads to a considerable improvement of the functionality of our synthetic regulators. The serial arrangement of individual terminators causes a drastically reduced read-through during transcription, and, consequently, a very low reporter gene activity in the absence of theophylline. Furthermore, these constructs carry 2 to 3 aptamer domains that require ligand binding to prevent terminator formation. Due to this increase in ligand binding sites, a higher amount of theophylline is necessary to activate gene expression. Consequently, both background as well as absolute activity of the reporter gene in the ON state are decreasing in these constructs. Due to an increased ratio between these OFF and ON states, the overall sensitivity of the serial riboswitches is elevated. As the enhanced activation ratio of the serial riboswitches is primarily the result of a reduced gene expression in the OFF state when no theophylline is present, a background close to zero inevitably limits any further increase in activation and can even lead to a reduction as observed in the case of RS10–10–10 compared to RS10–10–10shift (Fig. 4B). Both constructs differ in their overall activity, because in RS10–10–10, the ribosomal binding site is located immediately downstream of the riboswitch U-stretch that has a strong positive impact on translation efficiency.12,27 Accordingly, gene expression is reduced in the shifted counterpart. In this tridem, a further increase of the activation ratio compared to the tandem is not possible, as the theophylline-independent activity is already close to zero in RS10–10shift. RS10–10, on the other hand, still exhibits a considerable background signal that is further reduced in RS10–10–10.
In the constructs with the GFP reporter gene, a different effect of the serial arrangement of riboswitches was observed (Fig. S3). In RS10–10, the activation ratio as well as the absolute activity of the reporter gene was increased, while in RS10–10–10, both values dropped again. As described for the constructs with β-galactosidase reporter, the reduction in absolute activity might be the result of an increased number of theophylline binding sites that require a higher ligand concentration for activation of all riboswitch repeats. Furthermore, the reporter gene in use seems to have an impact on the result as well. Fowler et al.40 and Kötter et al.41 observed that the insertion of secondary structure elements in the 5′-UTR of a GFP gene can lead to a reduction in gene expression and, consequently, fluorescence intensity. Obviously, some of the riboswitch elements provoke a similar reduction.
The most interesting effect of these composite riboswitch arrangements, however, is the fact that the increasing number of ligand-binding sites leads to linear and dose-dependent response in reporter gene expression (Fig. 4C). The same phenomenon is discussed for natural arrangements consisting of complete riboswitches responding to the same ligand, as in the case of tandem TPP riboswitches.33 Compared to single riboswitch elements, these combinations can sense various ligand concentrations with a correlating signal output.
Taken together, our data indicate that terminator hairpin stability and predicted folding traps have a profound impact on transcriptional riboswitch functionality, corroborating our previous hypothesis.12 The a priori determination of these structural parameters and the serial combination of individual riboswitch copies should facilitate the construction of functional synthetic transcriptional riboswitches. Furthermore, tandem and tridem arrangements of such riboswitch constructs greatly improve the response ratio upon ligand binding. While natural riboswitch tandems probably lead to a fine-tuning of gene expression in response to fluctuating metabolite concentrations, the synthetic counterparts seem to be a valuable way to achieve a dose-dependent activation rate. Such riboswitch repeats might represent useful building blocks for gene regulation in synthetic biology.
Material and Methods
Chemicals
Oligonucleotides were purchased from biomers.net, dNTPs from Jena Biosciences. LB medium was purchased from Applichem; theophylline was obtained from Sigma-Aldrich. Ampicillin and Arabinose were purchased from Carl Roth.
Quantitative analysis for reporter gene activity
Quantitative analysis (3 independent experiments) of β-galactosidase activity and GFP fluorescence was done as described using E. coli Top10 with starter cultures grown between 16 and 24 h to obtain identical OD600.12
Northern blot analysis
Northern blot analysis was performed according to Wachsmuth et al.12
Primer sequences used for hybridization were:
bgaB: 5′-GGAGCAATAACTACTTTGTATTTTG-3′
23S rRNA (control): 5′-ACGACGGACGTTAGCACCCG-3′
Prediction of hairpin stabilities and kinetic folding traps
Thermodynamic calculation of terminator hairpin structures was done with RNAfold and RNAeval from the ViennaRNA Package v2.1.6 (see ref.42) using default parameters. Prediction of cotranscriptional folding trajectories for the RNA transcripts with and without aptamer part were computed with kinwalker.20 Intermediate structures along the predicted folding trajectories with base pairs that conflict with the respective terminator hairpin were considered kinetic folding traps. A refolding of these trapped structure states toward a functional terminator hairpin requires the RNA to surmount an energy barrier, induced by opening of conflicting base pairs prior the formation of the actual terminator hairpin. Here, the energy barrier is determined by the free energy difference between the trapped structure and the saddle point along the refolding path, i.e., the structure that exhibits the highest free energy. The higher this energy barrier is, the more time the RNA requires to surpass the saddle point upon refolding into its functional state. While most of the constructed transcripts readily fold into their terminator hairpin structure, kinwalker predicts kinetic traps for T8, T10loopA, and, to a lower degree, for T8ΔCU. These traps were taken to compute formation barriers of functional terminator hairpins via the Pathfinder algorithm.21 In contrast to that, intermediate structures prior the formation of the terminator hairpin loop were taken as refolding start point in all other cases.
Assessment of terminator seed stability
The seed stability of the terminator hairpin strongly depends on the gain of free energy in the transition toward the complete secondary structure. Thus, we analyzed the individual energy contributions of successively closing the innermost base pairs for all terminator constructs. Starting with the first base pair that closes the loop, free energies of the partial terminator hairpins with up to 5 consecutive base pairs were computed. Here, we utilized the RNAeval program which implements the Turner 2004 energy model and parameters for RNA secondary structures.42
Statistical test for significance
To investigate whether expression differences are significant, a Student's t-test was performed. All shown changes of reporter gene activity were highly significant with p < 0.001 or p < 0.01, while those of RS8 and RS8Δ4bp were not significant (p > 0.1).
Plasmid construction
Riboswitch and terminator constructs were generated by QuickChange site directed mutagenesis (Stratagene). For tandem arrangement, DNA with riboswitch sequences was amplified by PCR and fused to bgaB reporter in pBAD2_bgaB (see ref. 43) by QuickChange mutagenesis. Serial RS10 constructs were fused to a GFP reporter gene using a restriction site for SpeI. Plasmid constructs were isolated using GeneJETTM Plasmid Miniprep Kit (Thermo Scientific). Correctness of all constructs was verified by sequencing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Franziska Kretzschmar for assistance in the cloning of tandem riboswitch constructs. We thank Manuela Mießler for experimental support.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (MO 634/9–1 to MM, STA 850/15–1 to PFS) and by the European Commission under the Environment Theme of the 7th Framework Program for Research and Technological Development (grant agreement no.: 323987 to SF).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell 2011; 43:867-79; PMID:21925376; http://dx.doi.org/ 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kang Z, Zhang C, Zhang J, Jin P, Zhang J, Du G, Chen J. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 2014; 98:3413-24; PMID:24519458; http://dx.doi.org/ 10.1007/s00253-014-5569-y [DOI] [PubMed] [Google Scholar]
- 3. Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell 2011; 43:915-26; PMID:21925380; http://dx.doi.org/ 10.1016/j.molcel.2011.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weigand JE, Wittmann A, Suess B. RNA-based networks: using RNA aptamers and ribozymes as synthetic genetic devices. Methods Mol Biol 2012; 813:157-68; PMID:22083741; http://dx.doi.org/ 10.1007/978-1-61779-412-4_9 [DOI] [PubMed] [Google Scholar]
- 5. Wittmann A, Suess B. Engineered riboswitches: Expanding researchers' toolbox with synthetic RNA regulators. FEBS Lett 2012; 586:2076-83; PMID:22710175; http://dx.doi.org/ 10.1016/j.febslet.2012.02.038 [DOI] [PubMed] [Google Scholar]
- 6. Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem Bio 2010; 5:139-48; http://dx.doi.org/ 10.1021/cb900278x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chappell J, Takahashi MK, Meyer S, Loughrey D, Watters KE, Lucks J. The centrality of RNA for engineering gene expression. Biotechnol J 2013; 8:1379-95; PMID:24124015; http://dx.doi.org/ 10.1002/biot.201300018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol 1990; 216:835-58; PMID:1702475; http://dx.doi.org/ 10.1016/S0022-2836(99)80005-9 [DOI] [PubMed] [Google Scholar]
- 9. Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 2008; 22:3383-90; PMID:19141470; http://dx.doi.org/ 10.1101/gad.1747308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ceres P, Trausch JJ, Batey RT. Engineering modular 'ON' RNA switches using biological components. Nucleic Acids Res 2013; 41:10449-61; PMID:23999097; http://dx.doi.org/ 10.1093/nar/gkt787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science 1994; 263:1425-9; PMID:7510417; http://dx.doi.org/ 10.1126/science.7510417 [DOI] [PubMed] [Google Scholar]
- 12. Wachsmuth M, Findeiß S, Weissheimer N, Stadler PF, Mörl M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res 2013; 41:2541-51; PMID:23275562; http://dx.doi.org/ 10.1093/nar/gks1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ceres P, Garst AD, Marcano-Velázquez JG, Batey RT. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol 2013; 2:463-72; PMID:23654267; http://dx.doi.org/ 10.1021/sb4000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cambray G, Guimaraes JC, Mutalik VK, Lam C, Mai Q, Thimmaiah T, Carothers JM, Arkin AP, Endy D. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res 2013; 41:5139-48; PMID:23511967; http://dx.doi.org/ 10.1093/nar/gkt163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y, Liu P, Nielsen AA, Brophy JA, Clancy K, Peterson T, Voigt CA. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 2013; 10:659-64; PMID:23727987; http://dx.doi.org/ 10.1038/nmeth.2515 [DOI] [PubMed] [Google Scholar]
- 16. Hoon MJ. de Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comp Biol 2005; 1:e25; http://dx.doi.org/ 10.1371/journal.pcbi.0010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pörschke D. Thermodynamic and kinetic parameters of an oligonucleotide hairpin helix. Biophys Chem 1974; 1:381-6; PMID:23260427; http://dx.doi.org/ 10.1016/0301-4622(74)85008-8 [DOI] [PubMed] [Google Scholar]
- 18. Zhang W, Chen S. Exploring the complex folding kinetics of RNA hairpins: II. Effect of sequence, length, and misfolded states. Biophys J 2006; 90:778-87; PMID:16272439; http://dx.doi.org/ 10.1529/biophysj.105.062950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang W, Chen S. Exploring the complex folding kinetics of RNA hairpins: I. General folding kinetics analysis. Biophys J 2006; 90:765-77; PMID:16272440; http://dx.doi.org/ 10.1529/biophysj.105.062935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geis M, Flamm C, Wolfinger MT, Tanzer A, Hofacker IL, Middendorf M, Mandl C, Stadler PF, Thurner C. Folding kinetics of large RNAs. J Mol Biol 2008; 379:160-73; PMID:18440024; http://dx.doi.org/ 10.1016/j.jmb.2008.02.064 [DOI] [PubMed] [Google Scholar]
- 21. Lorenz R, Flamm C, Hofacker IL. 2D Projections of RNA folding Landscapes. Grosse I, Neumann S, Posch S, Shreiber F, Stadler PF. (Eds.) German Conference on Bioinformatics 2009:11-20. Bonner Köllen Verlag. [Google Scholar]
- 22. Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res 2007; 35:4179-85; PMID:17567606; http://dx.doi.org/ 10.1093/nar/gkm425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A 2010; 107:8531-6; http://dx.doi.org/ 10.1073/pnas.1001721107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A 2009; 104:14283-8; http://dx.doi.org/ 10.1073/pnas.0703961104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogawa A. Ligand-dependent upregulation of ribosomal shunting. Chembiochem 2013; 14:1539-43, 1509; http://dx.doi.org/ 10.1002/cbic.201300362 [DOI] [PubMed] [Google Scholar]
- 26. Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A 2011; 108:8617-22; http://dx.doi.org/ 10.1073/pnas.1015741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J, Deutscher MP. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc Natl Acad Sci U S A 1992; 89:2605-9; PMID:1372983; http://dx.doi.org/ 10.1073/pnas.89.7.2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mishler DM, Gallivan JP. A family of synthetic riboswitches adopts a kinetic trapping mechanism. Nucleic Acids Res 2014; 42:6753-61; PMID:24782524; http://dx.doi.org/ 10.1093/nar/gku262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res 2014; 42:2646-59; PMID:24234441; http://dx.doi.org/ 10.1093/nar/gkt1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voges D, Watzele M, Nemetz C, Wizemann S, Buchberger B. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem Biophys Res Comm 2004; 318:601-14; PMID:15120642; http://dx.doi.org/ 10.1016/j.bbrc.2004.04.064 [DOI] [PubMed] [Google Scholar]
- 31. Brosius J, Ullrich A, Raker MA, Gray A, Dull TJ, Gutell RR, Noller HF. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 1981; 6:112-8; PMID:7025054; http://dx.doi.org/ 10.1016/0147-619X(81)90058-5 [DOI] [PubMed] [Google Scholar]
- 32. Schumann W, Ferreira LCS. Production of recombinant proteins in E. coli. Genet Mol Biol 2004; 27:442-53; http://dx.doi.org/ 10.1590/S1415-47572004000300022 [DOI] [Google Scholar]
- 33. Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science 2006; 314:300-4; PMID:17038623; http://dx.doi.org/ 10.1126/science.1130716 [DOI] [PubMed] [Google Scholar]
- 34. Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol 2007; 10:176-81; PMID:17383225; http://dx.doi.org/ 10.1016/j.mib.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 2004; 306:275-9; PMID:15472076; http://dx.doi.org/ 10.1126/science.1100829 [DOI] [PubMed] [Google Scholar]
- 36. Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 2007; 13:573-82; PMID:17307816; http://dx.doi.org/ 10.1261/rna.407707; PMID:17307816; http://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 2005; 15:342-8; PMID:15919195; http://dx.doi.org/ 10.1016/j.sbi.2005.05.003 [DOI] [PubMed] [Google Scholar]
- 38. Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. Reconstruction of regulatory and metabolic pathways in metal-reducing delta-proteobacteria. Genome Biol 2004; 5:R90; PMID:15535866; http://dx.doi.org/ 10.1186/gb-2004-5-11-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Breaker RR. Riboswitches and the RNA World. Cold Spring Harb PerspectBiol 2012; 4:pii: a003566; PMID:21106649; http://dx.doi.org/ 10.1101/cshperspect.a003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. ChemBioChem 2008; 9:1906-11; PMID:18613182; http://dx.doi.org/ 10.1002/cbic.200700713 [DOI] [PubMed] [Google Scholar]
- 41. Kötter P, Weigand JE, Meyer B, Entian K, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res 2009; 37:e120; PMID:19592423; http://dx.doi.org/ 10.1093/nar/gkp578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA package 2.0. Algorithms Mol Biol 2011; 6:26; PMID:22115189; http://dx.doi.org/ 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klinkert B, Cimdins A, Gaubig LC, Roßmanith J, Aschke-Sonnenborn U, Narberhaus F. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J Biotechnol 2012; 160:55-63; PMID:22285954; http://dx.doi.org/ 10.1016/j.jbiotec.2012.01.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.