Abstract
Myoblast migration is essential for muscle development and repair; however, the factors that contribute to the polarity of migrating myoblasts are relatively unknown. We find that randomly migrating C2C12 myoblasts orient their centrosomes in the direction of migration. Using wounded monolayers, we further show that centrosome orientation is stimulated by the serum factor lysophosphatidic acid (LPA) and involves the rearward movement of the nucleus while the centrosome is maintained at the cell centroid. The rate of nuclear movement correlated with that of actin retrograde flow and both cytochalasin D and blebbistatin prevented nuclear movement and centrosome orientation. Actin-dependent rearward nuclear movement in fibroblasts is mediated by assembly of nuclear membrane nesprin-2G and SUN2 LINC complexes into transmembrane actin-associated nuclear (TAN) lines anchored by A-type lamins and emerin. In C2C12 myoblasts, depletion of nesprin-2G, SUN2 or lamin A/C prevented nuclear movement and endogenous nesprin-2G and a chimeric GFP-mini-nesprin-2G formed TAN lines during nuclear movement. Depleting nesprin-2G strongly interfered with directed cell migration and reduced the efficiency of myoblast fusion into multinucleated myotubes. Our results show that nuclear movement contributes to centrosome orientation and polarity for efficient migration and fusion of myoblasts. Given that mutations in the genes encoding A-type lamins, nesprin-2 and SUN2 cause Emery-Dreifuss muscular dystrophy and related myopathies, our results have implications for understanding the mechanism of disease pathogenesis.
Keywords: LINC complex; nuclear positioning; centrosome orientation; cell migration; muscle differentiation, TAN lines
Abbreviations
- CH
calponin homology
- EDMD
Emery-Dreifuss muscular dystrophy
- GFP
green fluorescent protein
- GFP-mN2G
GFP-mini-nesprin-2G
- LINC
linker of nucleoskeleton and cytoskeleton
- LPA
lysophosphatidic acid
- TAN lines
transmembrane actin-associated nuclear lines
Introduction
Rearward positioning of the nucleus is typical of migrating cells.1 In wounded monolayers of fibroblasts, this rearward positioning of the nucleus occurs by an active, actin-dependent process that precedes active cellular migration and results in the orientation of the centrosome toward the leading edge.2 Serum or the serum-derived factor lysophosphatidic acid (LPA) stimulates the formation and rearward movement of dorsal actin cables powered by myosin II to provide force to move the nucleus.2,3 Dorsal actin cables are coupled to the nuclear envelope by the linker of nucleoskeleton and cytoskeleton (LINC) complex proteins nesprin-2G (the giant isoform of nesprin-2) and SUN2, which accumulate along the cables to form linear assemblies called transmembrane actin-associated nuclear (TAN) lines that span the nuclear envelope.4,5 Nesprin-2G binds to actin cables through its paired calponin homology (CH) domains,4 but TAN line formation requires a second actin binding protein, the formin FHOD1, which interacts with spectrin repeats in nesprin-2G near its CH domains.6 TAN lines must be anchored to the nucleus to move it and this is achieved by interactions of nesprin-2G with SUN2 and SUN2 with A-type lamins (lamin A/C); depleting either SUN2 or lamin A/C causes nesprin-2G TAN lines to slip over an immobile nucleus.7 Anchorage of TAN lines to lamin A/C is also stabilized by the inner nuclear membrane proteins Samp1 and emerin.3,8 Emerin also interacts with cytoplasmic myosin IIB to regulate the direction of actin cable movement and resultant nuclear movement.3 Consistent with the importance of proper nuclear positioning for migration, TAN line components, including lamin A/C, nesprin-2G, Samp1 and emerin, are required for efficient migration of fibroblasts.3,4,8
Proteins involved in TAN line-dependent nuclear movement in fibroblasts are critical for skeletal muscle function. Skeletal muscle is a dynamic tissue and undergoes active remodeling and repair. During muscle regeneration, myosatellite cells are activated, proliferate, migrate, differentiate and fuse to form multinucleated myotubes. Defects in myogenesis lead to muscle disorders. Both lamin A/C and emerin are required for myoblast differentiation9 and mutations in the EMD and LMNA genes encoding emerin and lamin A/C, respectively, cause X-linked and autosomal dominant Emery-Dreifuss muscular dystrophy (EDMD) and related myopathies.10-13 Lamin A variants causing autosomal dominant EDMD or loss of emerin as occurs in X-linked EDMD both inhibit TAN line-dependent nuclear movement in fibroblasts.3,7 The roles of LINC complex proteins in myoblast differentiation are less clear, as overexpression of dominate negative SUN or SUN-interacting KASH domain of nesprins have no effect on differentiation under relaxed culture conditions and bypass inhibition of differentiation caused by cyclic strain.14 Nevertheless, sequence variants in SYNE1 and SYNE2 genes, the latter of which encodes the TAN line component nesprin-2, are associated with an EDMD-like disease and defective differentiation of myoblasts into myotubes,15 and knockout of the related nesprin-1 in mice causes an EDMD-like phenotype.16 Similarly, sequence alterations in SUN1 and SUN2 genes are associated with EDMD-like phenotypes and the variant proteins cause defective nuclear positioning when expressed in mouse fibroblasts.17 Lastly, knockout mouse studies have shown that nesprin-1 and SUN1/SUN2 play critical roles in anchoring nuclei in skeletal muscle.17-19
These results suggest a correlation between proteins involved in nuclear positioning and muscle functions and disease. Indeed, nuclei in mature muscle fibers exhibit specific locations: a small cluster of nuclei is localized under the neuromuscular junction while the remaining nuclei are positioned on the periphery of the muscle fiber spaced as far apart from each other as possible.20 In contrast, nuclei are positioned in the center of muscle fibers during muscle regeneration and this central position is a characteristic of myopathies and is used as a pathological marker for muscle diseases.21 Mechanisms of nuclear positioning during muscle differentiation are beginning to be explored and have implicated microtubules and microtubule associated proteins and motors.22-25
Centrosome orientation and nuclear positioning may also be important for muscle cell migration. Myoblast migration plays a critical role in skeletal muscle development26,27 and proper myoblast migration and is thought to contribute to muscle repair and regeneration.28 It is also essential for efficacy of myoblast transplantation therapies.29,30 However, it is unknown if myoblasts position their centrosomes during migration. Similarly, whether nuclei adopt specific positions in migrating myoblasts and how these positions are determined has not been explored. Here we examine the role of TAN line components in centrosome orientation and nuclear positioning in myoblasts and the consequence of disrupting nuclear positioning for myoblast migration and fusion to form myotubes.
Results
The centrosome is oriented to a position between the nucleus and leading edge in migrating C2C12 myoblasts
Many migrating cells, including fibroblasts, neurons, astrocytes, endothelial cells and macrophages, orient their centrosomes to a position between the nucleus and the leading edge, thus defining the polarity of the cell.31,32 However, whether myoblasts orient their centrosomes during cell migration is unknown. To test this, we examined cell migration of murine satellite cell-derived C2C12 myoblasts, a cell line that has been widely used to study muscle cell differentiation. We expressed green fluorescent protein (GFP)-tagged centrin-2 to label centrosomes in C2C12 myoblasts and monitored their dynamics during single cell migration on fibronectin-coated surfaces. 33 As in other cell types, GFP-centrin-2 concentrated at the centrosome and was excluded from the nucleus in C2C12 myoblasts. It also showed dim diffuse cytoplasmic localization, allowing us to image centrosomes, nuclei and cell bodies without additional labeling. In directionally migrating C2C12 myoblasts, centrosomes were maintained in a position between the nuclei and the leading edges of the cells (Fig. 1A and Movie S1). We measured the angle between the nucleus-centrosome axis and the direction of cell migration and found that it was maintained within a narrow range near zero degrees for several hours when the cell was moving unidirectionally (Fig. 1B). Analysis of this angle in many cells revealed that in over 85% of directionally migrating cells it was less than 30 degrees (Fig. 1C). These results showed that the centrosome is highly oriented between the nucleus and leading edge in migrating myoblasts.
Figure 1.
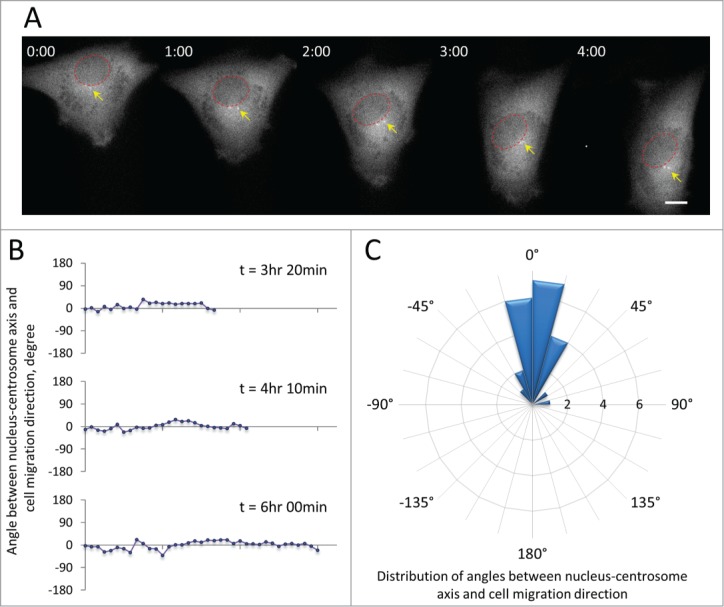
Centrosome orientation in C2C12 myoblasts expressing GFP-centrin-2 to label the centrosome. A, Frames from a time lapse movie showing the position of the centrosome (yellow arrows) between the nucleus (red circles) and the leading edge of a migrating C2C12 cell. Time is in hr:min. Bar,10 μm. B, Representative traces of the angle between the nucleus-centrosome axis and the direction of cell migration (defined as 0°) for 3 cells from movies such as in A. C, Rose plot showing the distribution of angles between the nucleus-centrosome axis and the direction of migration (defined as 0°) for C2C12 myoblasts (n = 22). The expanding rings indicate the frequency of cells exhibiting a given angle. Note that 19/22 (86%) of the cells maintain the centrosome within 30 degrees of the direction of migration.
Centrosome orientation in wounded monolayers of C2C12 myoblasts is stimulated by LPA and results from rearward movement of the nucleus
Wounded monolayers of serum-starved cells have been useful to identify external factors stimulating centrosome orientation and the cellular activities mediating centrosome orientation. In wounded monolayers of serum-starved NIH3T3 fibroblasts, the serum factor LPA stimulates centrosome orientation through an actomyosin pathway that moves the nucleus to the rear and a microtubule and dynein pathway that maintains the centrosome at the cell centroid.2,34 LPA also stimulates migration of activated mouse myoblasts,35 so we tested whether LPA induced centrosome orientation in serum-starved wounded monolayers of C2C12 cells. The centrosome was oriented in 44% of unstimulated cells (near the level of 33% expected from random orientation36) and increased to 65% in LPA-treated cells (Fig. 2, A and B). Measurement of the positions of the nucleus and the centrosome showed that LPA stimulated the rearward positioning of the nucleus while the centrosome remained near the cell centroid (Fig. 2C). Thus, centrosome orientation in C2C12 myoblasts resembled that previously described in fibroblasts.2 That LPA induced the movement of nuclei was confirmed by direct phase contrast imaging, which showed that the nucleus moved rearward while the leading and lagging edges of the cell remained stationary (Fig. 2D). The velocity of nuclear movement in C2C12 myoblasts measured from these movies was 0.48 ± 0.18 μm/min (n = 30), substantially faster than that previously reported for NIH3T3 fibroblasts (0.28 ± 0.09 μm/min).2
Figure 2.
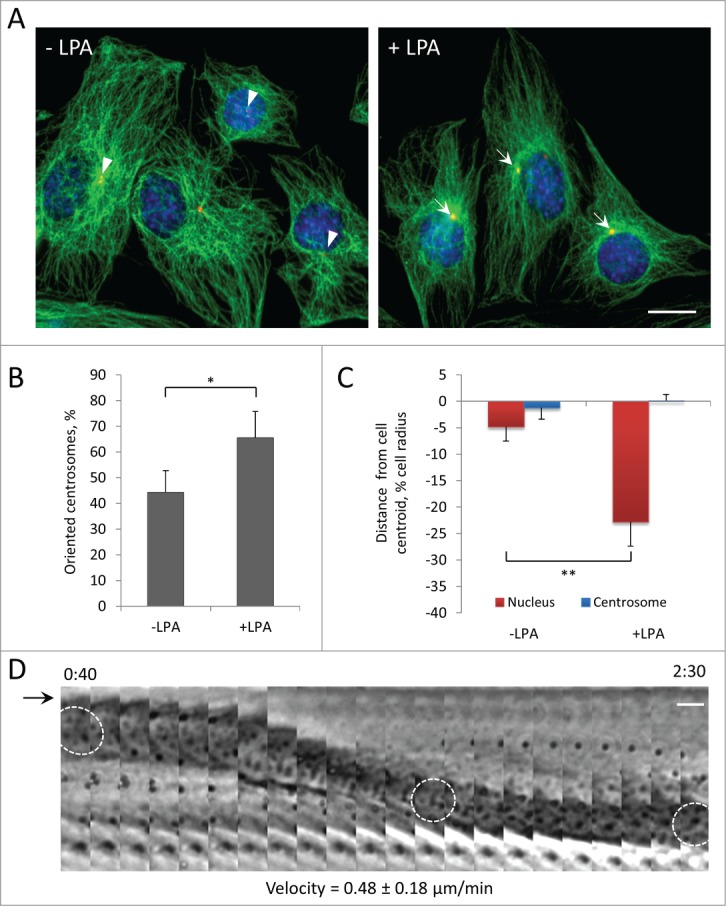
Centrosome orientation in wounded monolayers of serum-starved C2C12 myoblasts is stimulated by LPA and occurs by rearward movement of the nucleus. A, Immunofluorescence images of microtubules (green), pericentrin (red) and nuclei (blue) in C2C12 myoblasts at the edge of a wounded monolayer before and after 10 μM LPA stimulation for 2 hr. Arrows indicated oriented centrosomes; arrowheads indicate unoriented centrosomes. B, Quantification of centrosome orientation in C2C12 myoblasts before and after LPA stimulation. Centrosomes were scored “oriented” if they were within the pie-shaped region defined by the nucleus and the 2 sides of the leading edge; random orientation is 33% by this measure.36 C, Positions of the nucleus and the centrosome along the front-back cell axis for the cells treated as B. The cell centroid is defined as “0;" + values, toward the leading edge; - values, toward the cell rear. D, Kymograph showing the rearward movement of the nucleus (outlined in 3 panels) from a phase contrast movie of a wound edge C2C12 myoblast stimulated with LPA. The arrow denotes the leading edge of the cell, which does not extend with LPA treatment.2 Time is in hr:min after LPA treatment. The velocity of nuclear movement from 20 cells is indicated. Bars in A and D, 10 μm. Error bars in B and C are SD from 3 experiments (n > 30 cells per experiment). *, P < 0 .05; **, P < 0 .01 by t-test.
Nuclear movement in C2C12 myoblasts occurs at the same rate as actin retrograde flow and is dependent on actin and myosin
To test if actin retrograde flow might drive rearward nuclear movement, we first determined whether LPA stimulated actin retrograde flow in C2C12 myoblasts by monitoring actin cables by expressing the F-actin probe, LifeAct-GFP. LPA stimulated the formation of actin cables in the leading lamella as well as their retrograde movement (Fig. 3A). Measurement of the rate of retrograde flow of actin cables in the lamella of myoblasts revealed a rate (0.45 ± 0.28 μm/min) that was closely similar to that of nuclear movement (Fig. 3A). We next tested if actomyosin was required for nuclear movement in myoblasts by inhibiting actin polymerization with cytochalasin D and myosin II motor function with blebbistatin. Treatment with either cytochalasin D or blebbistatin prevented LPA-stimulated centrosome orientation in wounded monolayers of C2C12 myoblasts (Fig. 3, B and C). Both drugs blocked the rearward positioning of the nucleus compared to control cells without affecting the position of the centrosome, indicating an inhibition of nuclear movement (Fig. 3D). These results show that nuclear movement and centrosome orientation in myoblasts is actin and myosin dependent.
Figure 3.
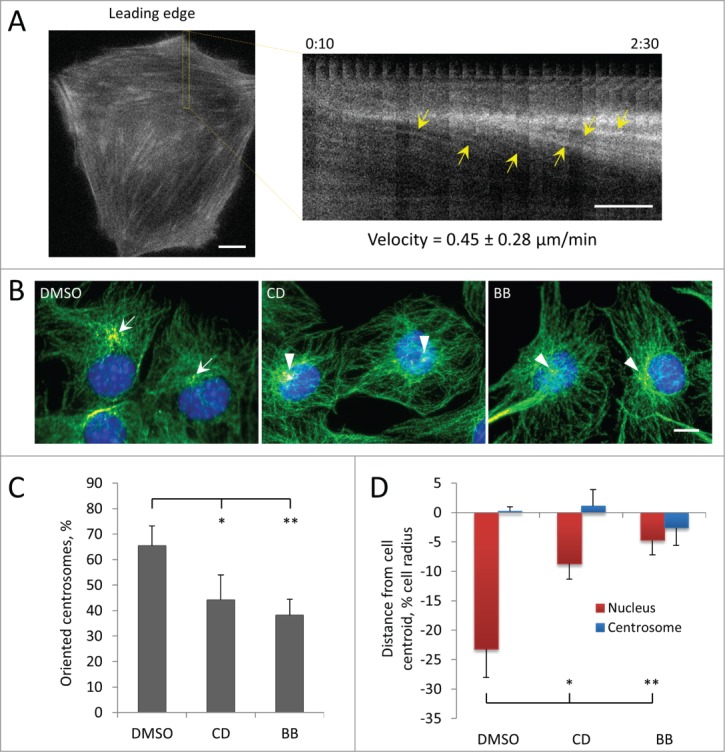
Centrosome orientation and nuclear movement in wounded monolayers of serum-starved C2C12 myoblasts depends on actin and myosin II. A, Initial panel (left) and kymograph (right) from the indicated region showing the rearward movement of actin cables (yellow arrows) in the lamella from a fluorescence movie of a LPA-stimulated, wound-edge C2C12 myoblast expressing LifeAct-mCherry. Time is in hr:min after LPA treatment. The velocity of actin cable movement from 12 cells is indicated. B, Immunofluorescence images of microtubules (green), pericentrin (red) and nuclei (blue) in LPA-stimulated C2C12 myoblasts at the edge of a wounded monolayer after treatment with vehicle (DMSO), 0.5 μM cytochalasin D (CD) or 25 μM blebbistatin (BB). Arrows indicate oriented centrosomes; arrowheads indicated unoriented centrosomes. C, Quantification of centrosome orientation in LPA-stimulated C2C12 myoblasts treated with actin and myosin II inhibitors as in B. D, Positions of the nucleus and the centrosome in LPA-stimulated C2C12 myoblasts treated with actin and myosin II inhibitors as in B. Bars in A and B, 10 μm. Error bars in C and D are SD from 3 experiments (n > 30 cells per experiment). *, P < 0 .05; **, P < 0 .01 by t-test.
Centrosome orientation and nuclear movement in C2C12 myoblasts require lamin A/C
Actin-dependent nuclear movement in fibroblasts depends on lamin A/C.7 These proteins act as the anchoring site for TAN lines by binding to SUN2, thus mechanically coupling moving actin to the nucleus. In the absence of lamin A/C, TAN lines slip over the nucleus and fail to move it.7 To examine the role of lamin A/C in myoblast nuclear movement, we depleted these proteins using siRNA. Specific siRNA oligonucleotides efficiently reduced the levels of lamin A/C (Fig. S1A) and caused significant deformation of the nuclei (Fig. S1C), indicative of a loss of lamin A/C.37 Reduction of lamin A/C prevented LPA-stimulated centrosome orientation in C2C12 myoblasts (Fig. 4A, B) and also blocked the rearward positioning of nuclei, indicating defective nuclear movement (Fig. 4C). Similar results were obtained in C2C12 cells stably expressing two distinct lamin A/C shRNAs (Fig. S1B,D-E). Taking together, these results show that lamin A/C is required for nuclear movement in myoblasts.
Figure 4.
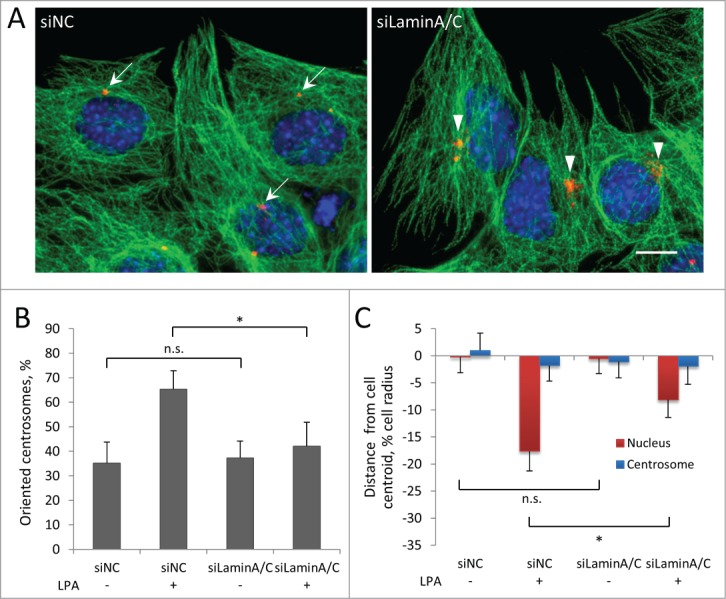
Centrosome orientation and nuclear movement in wounded monolayers of serum-starved C2C12 myoblasts require A-type lamins. A, Immunofluorescence images showing microtubules (green), pericentrin (red) and nuclei (blue) in LPA-stimulated C2C12 myoblasts at the edge of a wounded monolayer after treatment with the indicated siRNAs (siNC refers to control non-coding siRNA). Arrows indicate oriented centrosomes; arrowheads indicated unoriented centrosomes. Bar, 10 μm. B, Quantification of centrosome orientation in LPA-stimulated C2C12 myoblasts treated with the indicated siRNAs as in A. C, Positions of the nucleus and the centrosome in LPA-stimulated C2C12 myoblasts treated with the indicated siRNAs as in A. Error bars in B and C are SD from 3 experiments (n > 30 cells per experiment). ns, not significant; *, P < 0 .05 by t-test.
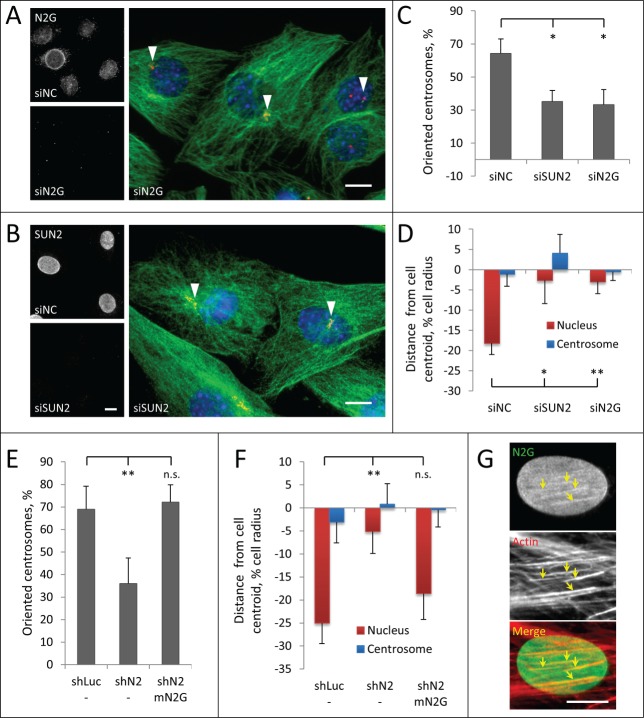
Centrosome orientation and nuclear movement in C2C12 myoblasts requires the LINC complex components nesprin-2G and SUN2
The LINC complex proteins nesprin-2G and SUN2 form TAN lines that couple the nucleus to actin filaments for movement in polarizing fibroblasts.4,5 To test whether TAN lines mediate movement of nuclei in myoblasts, we reduced the level of nesprin-2G and SUN2 using previously validated siRNAs.4,7 Nesprin-2G is essential for TAN line formation and it recruits SUN2 to TAN lines.4 SUN2 is required to anchor TAN lines to nuclear lamina.7 We confirmed by immunofluorescence microscopy that nesprin-2G and SUN2 were undetectable in nuclei of cells treated with the respective siRNAs (Fig. 5A, B). Depletion of either nesprin-2G or SUN2 blocked centrosome orientation and prevented rearward positioning of nuclei, indicating defective nuclear movement (Fig. 5A–D). Similar defects in centrosome orientation and nuclear movement were obtained with retrovirally expressed shRNAs targeting nesprin-2 and SUN2 to generate stable knockdown C2C12 cells (Fig. 5E, F; Fig. S2A–D). Depletion of either protein did not affect the overall organization of the actin cytoskeleton (Fig. S2E), indicating that, similar to NIH3T3 fibroblasts, defects in nuclear movement are unlikely caused by altered actin dynamics.4,7 The shRNA to nesprin-2 targeted the 3′-UTR and depleted nesprin-2G (Fig. S2A) but could also have depleted other nesprin-2 isoforms. To confirm the specific requirement of nesprin-2G, we re-expressed a chimeric GFP-mini-nesprin-2G (GFP-mN2G) retroviral construct that lacks most spectrin repeats but contains the actin-binding CH and KASH domains and rescues nuclear movement in nesprin-2G depleted fibroblasts.4 Re-expression of GFP-mN2G in nesprin-2 shRNA treated C2C12 cells rescued both centrosome orientation and nuclear movement (Fig. 5E, F). Localization of expressed GFP-mN2G (Fig. 5G) and endogenous nesprin-2G (Fig. S3) in LPA-stimulated cells revealed that it formed linear arrays on the nuclear surface that colocalized with dorsal actin cables, resembling the TAN lines on moving nuclei in mouse fibroblasts. These results show that nuclear movement in LPA-stimulated C2C12 myoblasts utilizes a nesprin-2G/SUN2/lamin A/C TAN line pathway similar to that in fibroblasts.
Figure 5.
Centrosome orientation and nuclear movement in wounded monolayers of serum-starved C2C12 myoblasts require nesprin-2G and SUN2. A, B (left panels), Immunofluoresence images of nesprin-2G and SUN2 in C2C12 myoblasts treated with the indicated siRNAs (siNC refers to control non-coding siRNA); (right panels), Immunofluorescence images of microtubules (green), pericentrin (red) and nuclei (blue) in LPA-stimulated C2C12 myoblasts at the edge of a wounded monolayer after treatment with the indicated siRNAs. Arrowheads indicate unoriented centrosomes. C, Quantification of centrosome orientation in LPA-stimulated C2C12 myoblasts treated with the indicated siRNAs. D, Positions of the nucleus and the centrosome in LPA-stimulated C2C12 myoblasts treated with the indicated siRNAs. E, Quantification of centrosome orientation in LPA-stimulated C2C12 myoblasts treated with retrovirally expressed shRNAs to luciferase (shLuc) or nesprin-2 (shN2) and re-expressing GFP-mini-nesprin-2G (mN2G). F, Quantification of centrosome and nuclear position in LPA-stimulated C2C12 myoblasts treated with the indicated shRNAs and re-expressing GFP-mini-nesprin-2G (mN2G). G, Immunofluorescence images of GFP-mN2G (GFP stained) and dorsal actin cables (phalloidin stained) on a nucleus of a C2C12 cell (leading edge of the cell is toward the top). Note the linear arrays (arrows) of GFP-mN2G that colocalize with dorsal actin cables. Bars in A, B and G, 10 μm. Error bars in C-F are SD from at least 3 experiments (n > 30 cells per experiment). ns, not significant; *, P < 0 .05, **, P < 0 .01 by t-test.
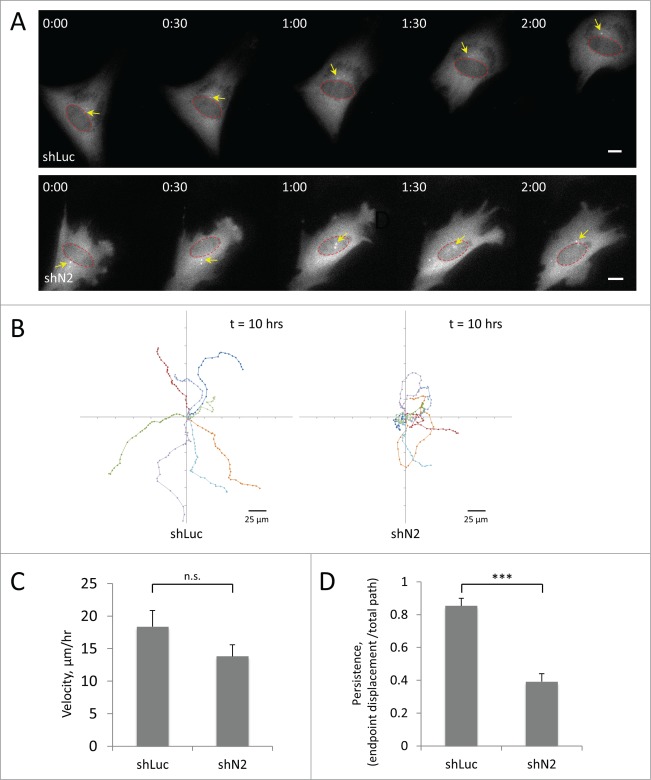
Depletion of the TAN line component nesprin-2G in C2C12 myoblasts interferes with directed cell migration
To test the effect of proper nuclear positioning in cell migration, we depleted the TAN line component nesprin-2G with shRNA and examined the ability of single myoblasts to migrate on a fibronectin-coated surface. As described above, we expressed GFP-centrin-2 to visualize the centrosome, the nucleus and the cell body. We found that treatment with nesprin-2 shRNA strongly inhibited cell migration (Fig. 6A, Movie S2, S3). By analyzing the position of the cell centroid of individual cells over time, we found that while control cells moved persistently over long periods, those treated with shRNA against nesprin-2 failed to undergo directional migration and frequently changed directions (Fig. 6B). Compared to control cells, the migration velocity of C2C12 depleted of nesprin-2G was reduced by approximately 20% but the difference was not statistically significant (Fig. 6C). Persistence of migration (endpoint displacement/total path length) was highly significantly reduced by >50 % (Fig. 6D). While the centrosome was oriented in cells treated with control shRNA (Fig. 6A, Movie S2; also see Fig. 1), in cells treated with nesprin-2 shRNA the centrosome failed to adopt a specific position and frequently changed position relative to the nucleus (Fig. 6A, Movie S3). These results indicate that disrupting normal centrosome orientation and nuclear positioning interferes with directional migration in C2C12 myoblasts.
Figure 6.
Knockdown of the TAN line component nesprin-2G affects directional persistence of C2C12 cell migration. A, Panels from movies of C2C12 myoblasts treated with retroviral shRNAs to luciferase (shLuc) or nesprin-2 (shN2) and expressing GFP-centrin-2 (to label the centrosome, arrows). Red circles indicate nuclei. Time is in hr:min. Bars, 10 μm. B, Eight representative traces of migration paths of C2C12 myoblasts treated with the indicated shRNAs. Positions were plotted every 20 min. C, Quantification of the velocity of C2C12 myoblasts treated with the indicated shRNAs. D, Quantification of the persistence of C2C12 myoblasts treated with the indicated shRNAs. A value of “1” would indicate a cell moving in a straight line. Error bars in C and D are SD from 3 experiments in which >20 cells were analyzed per experiment. ns, not significant; ***, P < 0 .001 by t-test.
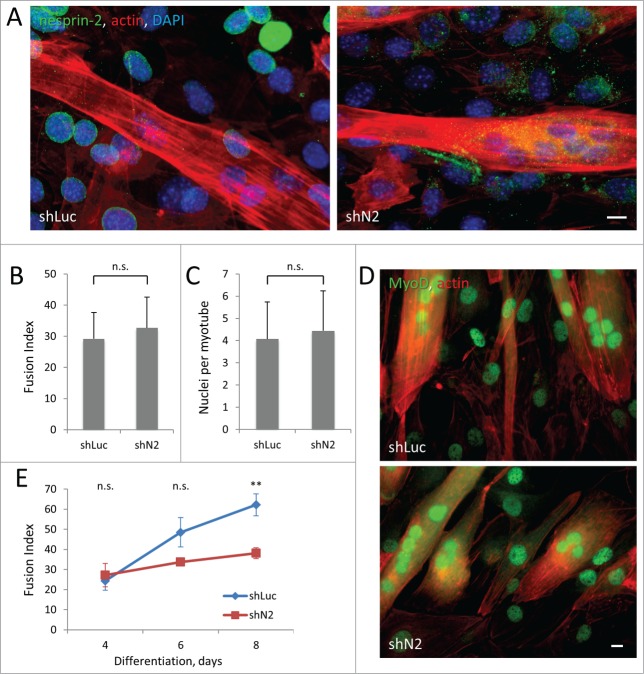
Depletion of nesprin-2G in C2C12 myoblasts reduces the efficiency of myotube formation
Nesprin-2 knockout mice appear to develop normally, suggesting that nesprin-2 is not required for normal muscle development.15 However, mutations in the SYNE2 gene encoding nesprin-2 cause EDMD-like disease,15 suggesting a role of nesprin-2 in the maintenance of adult muscle. Activated satellite cells need to migrate considerable distances to the site of tissue damage so directed cell migration is essential for proper skeletal muscle repair.38 Our finding that nesprin-2G is required for persistent myoblast cell migration raised the possibility that nesprin-2G may be required for myotube formation. We tested this by inducing differentiation of C2C12 cells stably expressing nesprin-2 or control shRNAs (Fig. 7A). We found that the initial differentiation and fusion of myoblasts was unaffected by depletion of nesprin-2 assayed 4 d after induction, based on quantification of fusion index (percentage of number of nuclei inside myotubes to the number of total nuclei) and number of nuclei per myotube (Fig. 7B, C). The positioning of nuclei in nesprin-2-depleted myotubes appeared normal compared to control myotubes; nuclei clustered close to the center of myotubes with spherical shape and aligned along the length of elongated myotubes (Fig. 7D). In myotubes carrying a SUN2 R620C mutation that associates with EDMD, centrosome components failed to redistribute to the nuclear envelope during differentiation.17 However, in nesprin-2 depleted-myoblasts pericentrin redistributed to the nuclear envelop normally during differentiation (Fig. S4A), suggesting early differentiation is normal in these cells. Consistently, immunofluorescence labeling of MyoD, a key regulator of muscle differentiation and an early marker for myogenic commitment, showed that depletion of nesprin-2 did not affect myogenic commitment of the cells (Fig. 7D and S4B-D). However, when differentiation was allowed to proceed for longer intervals, we observed an increasing difference in the fusion index between controls and cells depleted of nesprin-2, which became significant at 8 d of differentiation (Fig. 7E). The decreased fusion index was caused by an increased number of myotubes (Fig. S5A), as the average number of nuclei in each myotubes was not significantly changed by loss of nesprin-2 (Fig. S5B). These results suggest that although nesprin-2 is not required for myoblast differentiation or fusion per se, it contributes to the efficiency of myotube formation consistent with its effect on directional migration.
Figure 7.
Stable knockdown of nesprin-2 in C2C12 cells reduces efficiency of myoblast fusion without affecting MyoD expression. A. Immunofluorescence of nesprin-2G (green) and staining of actin (red) and nuclei (blue) in C2C12 expressing control shLuciferase (shLuc) and nesprin-2 shRNA (shN2) 4 d after induction of differentiation. Note reduction of nesprin-2 levels and formation of multinucleated myotubes. B, C. Fusion index (ratio of number of nuclei in myotubes to total number of nuclei multiplied by 100) and number of nuclei per myotube in C2C12 cells differentiated for 4 d. At least 2000 total nuclei were count for each condition. D) MyoD immunofluorescence (green) and actin (red) staining of C2C12 cells differentiated for 4 d. E. Fusion index of C2C12 myoblasts expressing control Luciferase (shLuc) and nesprin-2 (shN2) shRNAs induced to differentiate for different times. Statistical analysis was performed to compare fusion index of shN2 to shLuc controls. Bars in A and D, 10 μm. Error bars in B,C,E are SD from 3 experiments. ns, not significant; **, P < 0 .01 by t-test.
Discussion
Factors contributing to the polarity of migrating cells have been studied in several different cell types. In many cases, a major factor contributing to the front-rear polarity of migrating cells is the orientation of the centrosome to a specific position relative to the leading edge and nucleus and disrupting this positioning leads to defects in directed cell migration.31,32, 39 However, previous studies did not address whether migrating cells of the striated muscle lineage also exhibit specific positioning of the centrosome. We found that C2C12 myoblasts orient their centrosome during either single cell migration or when migrating as a group in wounded monolayers. Disrupting nuclear movement, which prevents centrosome orientation, by depleting nesprin-2 dramatically affected the directional persistence of migrating myoblasts without affecting their velocity, consistent with the idea that proper centrosome orientation contributes to the establishment of polarity necessary for directional migration.
By analyzing the relative contributions of centrosome and nuclear positioning using the wounded monolayer system, we found that myoblasts establish the anterior positioning of their centrosomes by actively moving their nucleus rearward. The mechanism for this nuclear movement strongly resembles that described earlier in fibroblasts2,4, 7: it depends on actomyosin and LINC complex components nesprin-2G, SUN2 and A-type lamins and involves the formation of TAN lines on the dorsal surface of the nucleus. As in fibroblasts, an artificial chimera of nesprin-2G, GFP-mN2G, was capable of restoring nuclear movement in nesprin-2 depleted C2C12 cells, suggesting that the minimal requirements for nesprin are the actin-binding CH domains and the SUN-binding KASH domain. Additionally, because the shRNA to nesprin-2 targets all nesprin-2 isoforms, this result suggests that other isoforms of nesprin-2 are not required. One significant difference between nuclear movement in myoblasts and fibroblasts is the faster rate of nuclear movement in myoblasts, reflecting the faster rate of actin flow in these cells.
Our results further show that interfering with myoblast polarity and migration by disrupting the LINC complex does not directly affect the transcriptional program of muscle differentiation or the initial fusion of myoblasts into multinucleated myotubes. However, the efficiency of fusion was reduced at long time intervals presumably because migration becomes more important as the density of myoblasts is reduced as they fuse to form myotubes. Previous studies have established a correlation between myoblast cell migration and myotube formation.40,41 In general, decreasing migration reduces myotube formation, although there is at least one example in which reducing migration enhanced fusion.42 During development, there is dramatic migration of muscle precursors during the initial formation of muscle, and incorporation of satellite cells into damaged muscle can occur at sites distant from the injury.38,43 Surprisingly, knockout of nesprin-2 in mice does not lead to developmental defects, suggesting either that the migration of muscle precursors during development differs from that of myoblasts derived from satellite cells during muscle repair or that there are redundant factors that come into play during development.
Mutations in the genes encoding many of the components contributing to nuclear movement through TAN line formation (nesprin-2G and SUN2) and anchorage (A-type lamins and emerin) are known to cause EDMD and related myopathies. Previous studies on TAN lines were restricted to fibroblasts raising the question whether this correlation was relevant for muscle disease. While additional studies are necessary, our new findings that myoblasts also use TAN lines to establish nuclear position and centrosome orientation increases the likelihood that this altered nuclear positioning through LINC complex components may be a contributing factor to pathogenesis.
Materials and Methods
Reagents
LPA was obtained from Avanti Polar Lipids (Alabaster, AL) and rhodamine phalloidin was from (Life Technologies, Grand Island, NY). All other chemicals, unless noted, were from Sigma-Aldrich (St. Louis, MI). Nesprin-2G antibody (specific for the CH domains) was previously described.4 Tyrosinated α-tubulin rat mAb (YL1/2) was from the European Collection of Animal Cell Cultures (Salisbury, UK). Pericentrin mouse antibody and MyoD rabbit antibodies were from BD Transduction Laboratories (San Jose, CA). GFP chicken antibody was from Millipore (Billerica, MA). Lamin A/C mouse mAb (MANLAC1) was from the MDA Monoclonal Antibody Resource at the Wolfson Center for Inherited Neuromuscular Disease (Oswestry, UK). SUN2 antibodies were from Abcam (Cambridge, MA). GAPDH rabbit antibody was from Santa Cruz (Santa Cruz, CA).
Cell culture, plasmids, microinjection and transfection
C2C12 myoblasts and HEK293T cells (from ATCC, Manassas, VA) were cultured in Dulbecco's Modified Eagle's Medium (Life Technologies) with 10% fetal bovine serum (Gemini, West Sacramento, CA), 20 mM HEPES (pH 7.4), penicillin and streptomycin (Thermo Scientific, Waltham, MA). C2C12 cells were serum-starved at 40–60% confluence for 3–4 d to obtain confluent monolayers of cells. Starving the cells at low density was critical to avoid myoblast differentiation and densely packed monolayers, and the long starvation period was required to fully starve the cells and make their nuclear movement dependent on added LPA. Serum-starved monolayers of cells were wounded and stimulated with 10 μM LPA for 1 hr to analyze TAN line formation4 or for 2 hrs to analyze nuclear movement and centrosome orientation.2,44 For live cell imaging, 20 μM LPA was added to initiate nuclear movement. For live cell imaging of single cell migration, GFP-centrin-2 transfected C2C12 cells were plated on fibronectin-coated (4 μg/ml) imaging dishes for 4 hr before imaging. Myoblast differentiation was induced by changing the medium of confluent C2C12 monolayers to differentiation medium (Dulbecco's Modified Eagle's Medium with 2% horse serum (Gemini), 20 mM HEPES, pH 7.4, penicillin and streptomycin). Differentiation medium was replaced every 24 hrs.
Centrin-2 cDNA was obtained from NIH3T3 cells and cloned into a pEGFP vector with 5′-BamHI and 3′-NotI sites. Other plasmids have been previously described.4 GFP-centrin-2 plasmid was transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. LifeAct-mCherry and GFP-mini-nesprin-2G were microinjected into nuclei of cells and were allowed to express for 1–3 hrs. siRNAs (20 μM) were transfected with using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer's instructions. All siRNA oligonucleotides were from Shanghai GenePharma Co., Ltd (Shanghai, China) and have been previously described.4,7 We noticed significantly increased cell death in myoblasts treated with nesprin-2 siRNAs (targeting either nesprin-2G or all nesprin-2 isoforms) used at concentration previously optimized for fibroblasts; however, by reducing the amount of siRNA to 4 μM and increasing the number of seeded cells, we obtained confluent monolayers of cells with efficient depletion of nesprin-2G. shRNAs were cloned in to pSUPER.retro.puro vector (Oligoengine, Seattle, WA) with 5’-BglII and 3’-HindIII sites and GFP-mini-N2G was cloned into pMSCV vector (Clontech Laboratories, Mountain View, CA) with NotI sites. Viruses were produced in HEK293T cells by calcium phosphate transfection. Harvested retroviruses were frozen and stored at −80°C. Viruses were added to subconfluent cultures of C2C12 cells in the presence of 2 μg/ml polybrene (Millipore, Billerica, MA) and re-plated on the next day. See Supplemental Table 1 for shRNA sequences.
Microscopy
Immunofluorescence microscopy and live cell imaging were done as described previously.3 Briefly, cells on coverslips were fixed in 4 % paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), permeabilized with 0.3 % Triton X-100 and blocked with 5% normal goat serum (MP Biomedicals, Santa Ana, CA). Coverslips were mounted with Fluoromount-G (Southern Biotech, Birmingham, AL) after staining with primary and secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI). All secondary antibodies for immunofluorescence were from Jackson ImmunoResearch (West Grove, PA) and used at 1:200 dilution. Phase contrast live cell movies were acquired at 5 min per frame with a temperature controller (37°C) on a Nikon TE300 inverted microscope with a CoolSNAP HQ CCD camera. Fluorescent live cell movies were acquired at 4 min per frame on a Nikon Eclipse Ti microscope with an iXon X3 CCD camera and CO2 (5%) and temperature (37°C) controls. Images were processed using ImageJ (NIH, Bethesda, MD), Nikon NIS Element and custom software.3
Western blot analysis
SDS cell lysates were separated using 10% or 4–12% gradient NuPAGE Bis-Tris gels (Life Technologies) and transferred to nitrocellulose membrane. After staining with primary and IRDye-conjugated secondary antibodies (Li-Cor, Lincoln, NE), membranes were scanned and recorded with an Odyssey Infrared Imager (Li-Cor).
Data analysis
Centrosome orientation and nuclear and centrosomal positions were measured with custom software as previously described.3 The angel between the nucleus-centrosome axis and the direction of cell migration in live cell movies were manually tracked and calculated with custom software (available under request). Cell traces and velocity of cell migration were calculated by tracking the center positions of the nuclei with the same software. Results were exported to Microsoft Excel for statistical analysis and plotting. The velocity of TAN lines and nuclear movement were calculated from the slopes of lines in kymographs generated using NIS-Elements and ImageJ.3
For differentiation assays, myotubes were defined as cells that had 2 or more nuclei. Total number of nuclei and number of nuclei in myotubes were counted for each image. At least 2000 nuclei were counted for each experiment. Levels of myoD expression were assessed by determining the percentage of nuclei exhibiting myoD staining (n > 250 cells per experiment) and the fluorescence intensity of myoD staining per nuclei using rectangle ROI tool in ImageJ (n > 25 cells per experiment).
Statistical analysis was performed in Microsoft Excel 2010. Unpaired two-tailed Student's t-tests were used to calculate p values. In all figures p values were labeled as: n.s., P ≥ 0 .05; *, P < 0 .05; **, P < 0 .01; ***, P < 0 .001. All plots were created using Microsoft Excel.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Glenn Morris (Wolfson Center for Inherited Neuromuscular Disease, Wrexham, UK) for MANLAC1 antibody.
Funding
This study was supported by NIH grants R56NS059352 and R01GM099481.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Gundersen GG, Worman HJ. Nuclear positioning. Cell 2013; 152:1376-89; PMID:23498944; http://dx.doi.org/ 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 2005; 121:451-63; PMID:15882626; http://dx.doi.org/ 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- 3. Chang W, Folker ES, Worman HJ, Gundersen GG. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol Biol Cell 2013; 24:3869-80; PMID:24152738; http://dx.doi.org/ 10.1091/mbc.E13-06-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 2010; 329:956-9; PMID:20724637; http://dx.doi.org/ 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2011; 2:173-81; PMID:21818410; http://dx.doi.org/ 10.4161/nucl.2.3.16243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kutscheidt S, Zhu R, Antoku S, Luxton GW, Stagljar I, Fackler OT, Gundersen GG. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol 2014; 16:708-15; PMID:24880667; http://dx.doi.org/ 10.1038/ncb2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci U S A 2011; 108:131-6; PMID:21173262; http://dx.doi.org/ 10.1073/pnas.1000824108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci 2012; 125:1099-105; PMID:22349700; http://dx.doi.org/ 10.1242/jcs.087049 [DOI] [PubMed] [Google Scholar]
- 9. Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev 2006; 20:486-500; PMID:16481476; http://dx.doi.org/ 10.1101/gad.1364906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 1999; 21:285-8; PMID:10080180; http://dx.doi.org/ 10.1038/6799 [DOI] [PubMed] [Google Scholar]
- 11. Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr., Spudich S, De Girolami U, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. New Engl J Med 1999; 341:1715-24; PMID:10580070; http://dx.doi.org/ 10.1056/NEJM199912023412302 [DOI] [PubMed] [Google Scholar]
- 12. Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Human Mol Genetics 2000; 9:1453-9; PMID:10814726; http://dx.doi.org/ 10.1093/hmg/9.9.1453 [DOI] [PubMed] [Google Scholar]
- 13. Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 1994; 8:323-7; PMID:7894480; http://dx.doi.org/ 10.1038/ng1294-323 [DOI] [PubMed] [Google Scholar]
- 14. Brosig M, Ferralli J, Gelman L, Chiquet M, Chiquet-Ehrismann R. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. Int J Biochem Cell Biol 2010; 42:1717-28; PMID:20621196; http://dx.doi.org/ 10.1016/j.biocel.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007; 16:2816-33; PMID:17761684; http://dx.doi.org/ 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- 16. Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 2009; 18:607-20; PMID:19008300; http://dx.doi.org/ 10.1093/hmg/ddn386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, et al. Muscular Dystrophy-Associated SUN1 and SUN2 Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization. PLoS Genet 2014; 10:e1004605; PMID:25210889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A 2009; 106:10207-12; PMID:19509342; http://dx.doi.org/ 10.1073/pnas.0812037106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development 2007; 134:901-8; PMID:17267447; http://dx.doi.org/ 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- 20. Bruusgaard JC, Liestol K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J Physiol 2003; 551:467-78; PMID:12813146; http://dx.doi.org/ 10.1113/jphysiol.2003.045328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Folker ES, Baylies MK. Nuclear positioning in muscle development and disease. Front Physiol 2013; 4:363; PMID:24376424; http://dx.doi.org/ 10.3389/fphys.2013.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cadot B, Gache V, Vasyutina E, Falcone S, Birchmeier C, Gomes ER. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep 2012; 13:741-9; PMID:22732842; http://dx.doi.org/ 10.1038/embor.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 2012; 484:120-4; PMID:22425998; http://dx.doi.org/ 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson MH, Holzbaur EL. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci 2012; 125:4158-69; PMID:22623723; http://dx.doi.org/ 10.1242/jcs.108688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Folker ES, Schulman VK, Baylies MK. Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development 2012; 139:3827-37; PMID:22951643; http://dx.doi.org/ 10.1242/dev.079178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Capers CR. Multinucleation of skeletal muscle in vitro. J Biophys Biochem Cytol 1960; 7:559-66; PMID:13807523; http://dx.doi.org/ 10.1083/jcb.7.3.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hughes SM, Blau HM. Migration of myoblasts across basal lamina during skeletal muscle development. Nature 1990; 345:350-3; PMID:2111464; http://dx.doi.org/ 10.1038/345350a0 [DOI] [PubMed] [Google Scholar]
- 28. Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development 2012; 139:641-56; PMID:22274696; http://dx.doi.org/ 10.1242/dev.068353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hill E, Boontheekul T, Mooney DJ. Designing scaffolds to enhance transplanted myoblast survival and migration. Tissue Eng 2006; 12:1295-304; PMID:16771642; http://dx.doi.org/ 10.1089/ten.2006.12.1295 [DOI] [PubMed] [Google Scholar]
- 30. Skuk D, Goulet M, Tremblay JP. Transplanted myoblasts can migrate several millimeters to fuse with damaged myofibers in nonhuman primate skeletal muscle. J Neuropathol Exp Neurol 2011; 70:770-8; PMID:21865885; http://dx.doi.org/ 10.1097/NEN.0b013e31822a6baa [DOI] [PubMed] [Google Scholar]
- 31. Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol 2008; 9:860-73; PMID:18946475; http://dx.doi.org/ 10.1038/nrm2522 [DOI] [PubMed] [Google Scholar]
- 32. Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol 2011; 23:579-88; PMID:21885270; http://dx.doi.org/ 10.1016/j.ceb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaz R, Martins GG, Thorsteinsdottir S, Rodrigues G. Fibronectin promotes migration, alignment and fusion in an in vitro myoblast cell model. Cell Tissue Res 2012; 348:569-78; PMID:22427060; http://dx.doi.org/ 10.1007/s00441-012-1364-1 [DOI] [PubMed] [Google Scholar]
- 34. Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol 2009; 19:1065-74; PMID:19540120; http://dx.doi.org/ 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cencetti F, Bruno G, Blescia S, Bernacchioni C, Bruni P, Donati C. Lysophosphatidic acid stimulates cell migration of satellite cells. A role for the sphingosine kinase/sphingosine 1-phosphate axis. FEBS J 2014; 281:4467-78; PMID:25131845; http://dx.doi.org/ 10.1111/febs.12955 [DOI] [PubMed] [Google Scholar]
- 36. Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, Gundersen GG. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol 2001; 11:1536-41; PMID:11591323; http://dx.doi.org/ 10.1016/S0960-9822(01)00475-4 [DOI] [PubMed] [Google Scholar]
- 37. Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 1999; 147:913-20; PMID:10579712; http://dx.doi.org/ 10.1083/jcb.147.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schultz E, Jaryszak DL, Valliere CR. Response of satellite cells to focal skeletal muscle injury. Muscle Nerve 1985; 8:217-22; PMID:4058466; http://dx.doi.org/ 10.1002/mus.880080307 [DOI] [PubMed] [Google Scholar]
- 39. Elric J, Etienne-Manneville S. Centrosome positioning in polarized cells: Common themes and variations. Exp Cell Res 2014; PMID:25218948 [DOI] [PubMed] [Google Scholar]
- 40. Jansen KM, Pavlath GK. Molecular control of mammalian myoblast fusion. Methods Mol Biol 2008; 475:115-33; PMID:18979241; http://dx.doi.org/ 10.1007/978-1-59745-250-2_7 [DOI] [PubMed] [Google Scholar]
- 41. Goetsch KP, Myburgh KH, Niesler CU. In vitro myoblast motility models: investigating migration dynamics for the study of skeletal muscle repair. J Muscle Res Cell Motil 2013; 34:333-47; PMID:24150600; http://dx.doi.org/ 10.1007/s10974-013-9364-7 [DOI] [PubMed] [Google Scholar]
- 42. Bondesen BA, Jones KA, Glasgow WC, Pavlath GK. Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. FASEB J 2007; 21:3338-45; PMID:17488951; http://dx.doi.org/ 10.1096/fj.06-7070com [DOI] [PubMed] [Google Scholar]
- 43. Bentzinger CF, von Maltzahn J, Dumont NA, Stark DA, Wang YX, Nhan K, Frenette J, Cornelison DD, Rudnicki MA. Wnt7a stimulates myogenic stem cell motility and engraftment resulting in improved muscle strength. J Cell Biol 2014; 205:97-111; PMID:24711502; http://dx.doi.org/ 10.1083/jcb.201310035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gomes ER, Gundersen GG. Real-time centrosome reorientation during fibroblast migration. Methods Enzymol 2006; 406:579-92; PMID:16472689; http://dx.doi.org/ 10.1016/S0076-6879(06)06045-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.