Abstract
The RNA-binding protein Lin28 regulates the expression of the let-7 family of microRNAs (miRNAs) during early embryonic development. Lin28 recruits the 3′ terminal uridylyl transferase (TUTase) Zcchc11 (TUT4) and/or Zcchc6 (TUT7) to precursor let-7 RNA (pre-let-7) to selectively block let-7 biogenesis. Uridylated pre-let-7 is targeted for decay by the downstream exonuclease Dis3l2 thereby preventing processing to mature let-7. Activation of this oncogenic pathway via up-regulation of Lin28 expression promotes cellular transformation, drives tumorigenesis in mouse models, and is frequently observed in a wide variety of cancer. Recent proof-of-principle experiments showed that Zcchc11 knockdown inhibits the tumorigenicity of Lin28-expressing human cancer cells and established this enzyme as a possible new therapeutic target for human malignancies. However, there are currently no known pharmacological agents capable of targeting this novel enzyme. In this study we developed and applied a sensitive biochemical assay that monitors Zcchc11 activity. Using this assay we performed an automated high-throughput screen of ∼15,000 chemicals to identify putative TUTase inhibitors. Several of these small molecules were validated as specific inhibitors of Zcchc11 activity. Our results demonstrate the feasibility of screening for TUTase inhibitors and present a relatively simple platform that can be exploited for future drug discovery efforts aimed at restoring let-7 expression in cancer.
Keywords: cancer, let-7, Lin28, microRNA, miRNA, TUT4, TUTase, uridylation, Zcchc11
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs that function as negative regulators of target gene expression by directing Argonaute-containing complexes to sites in the 3′ untranslated region (3′ UTR) of complementary mRNAs for translation suppression and/or mRNA decay. The links between miRNA and cancer is now well established with miRNAs being directly involved in cancer initiation, progression, and metastasis.1-3 In particular, the let-7 miRNA family members function as tumor suppressors in multiple different tumor types by inhibiting expression of oncogenes and key regulators of several mitogenic pathways including RAS, MYC, and HMGA2.4-6 The let-7 miRNAs are frequently down-regulated in many different types of cancers and low let-7 expression is associated with poor patient survival.7,8 Restoration of let-7 expression was shown to effectively inhibit cancer growth in mouse models of lung and breast cancers.9,10 These findings imply that let-7 may be used as a next-generation cancer therapeutic target.11
Recent studies have uncovered that let-7 miRNA expression is post-transcriptionally regulated. miRNAs are typically generated by a processing pathway that involves the cleavage of primary miRNA transcripts (pri-miRNAs) by the Microprocessor to generate pre-miRNAs that are subsequently processed by Dicer into mature ∼22 nucleotide (nt) miRNAs.12 We and other groups found that in mouse embryonic stem cells (ESCs) and several human cancer cell lines, the paralogous RNA-binding proteins Lin28A or Lin28B, selectively block the maturation of let-7 miRNA.13-15 Lin28A/B are frequently over-expressed in cancers and their elevated expression often correlates with poor prognosis.16-18 Indeed, Lin28A/B are classical oncogenes that can promote cellular transformation and tumorigenesis when ectopically expressed in vitro and in vivo.16,19-21 Moreover, depletion of Lin28A or Lin28B expression results in decreased cell proliferation, and invasion in cancer cells and reverts tumorigenesis in mouse model.16,21,22 Importantly, this oncogenic effect of Lin28A/B can be abrogated when let-7 is reintroduced into these cells, suggesting that Lin28A/B-mediated cellular transformation is directly dependent on let-7 levels.16 Therefore, the Lin28-let-7 oncogenic pathway represents an important novel therapeutic target for effective cancer treatment.
Two 3′ terminal uridylyl transferases (TUTases), Zcchc11 (TUT4) and Zcchc6 (TUT7), are key mediators in the Lin28 blockade of let-7 biogenesis.23-25 These enzymes are recruited by Lin28 to let-7 precursors (pre-let-7) where they catalyze the addition of an oligo(U)-tail. Uridylated pre-let-7 is resistant to Dicer processing and degraded by the 3′–5′ exonuclease Dis3l2.23-27 TUTase activity is required for the Lin28-mediated inhibition of let-7 biogenesis, and RNAi-mediated knockdown of Zcchc11 has been shown to inhibit tumorigenesis of human breast, ovarian, melanoma, prostate, and liver cancer cells in mouse xenograft assays.28,29 Moreover, Zcchc11 knockdown inhibited the lung metastasis of liver cancer cells.29 In addition, Zcchc11 expression was found to be up-regulated in primary human liver and colorectal tumors compared with matched normal tissues.29 Taken together, emerging evidence supports that Zcchc6/11 is an attractive target for therapeutic intervention in Lin28 over-expressing cancers.
As a first step toward targeting the Lin28 oncogenic pathway in cancer, we developed a method for high-throughput screening of small molecules that can inhibit Zcchc11 TUTase activity. This assay uses recombinant Zcchc11 expressed and purified from E.coli, synthetic RNA substrate, and simple luciferase readout to measure TUTase activity. Using this approach we screened in duplicate libraries containing ∼15,000 compounds including known bioactive molecules as well as natural products to identify small molecules that can selectively inhibit Zcchc11 in vitro. Several of these ‘hits’ from the primary screen were validated using secondary assays as specific inhibitors of Zcchc11 activity in vitro. Our results demonstrate the feasibility of screening for TUTase inhibitors and present a relatively simple platform that can be exploited for future drug discovery efforts aimed at restoring let-7 expression in cancer.
Results
Purification and characterization of recombinant Zcchc11
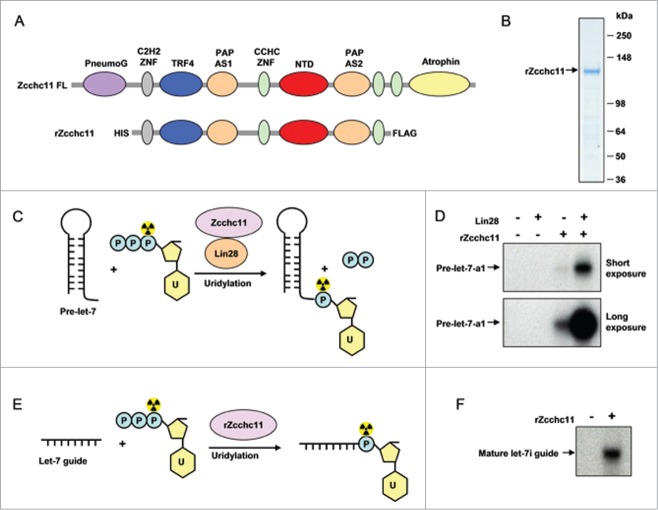
Zcchc11 is a large (∼185 kDa), multi-domain containing protein, making it difficult to express and purify in large quantities in recombinant form. Our previous research identified a minimal fragment of Zcchc11, containing a TRF4 domain, a catalytic nucleotidyl transferase domain (NTD), 2 PAP-associated domains, and 3 zinc finger domains, that together with Lin28, is necessary and sufficient for pre-let-7 uridylation (Fig. 1A).25 We therefore subcloned the cDNA encoding this minimal region of Zcchc11 with a FLAG tag epitope sequence at the 3′ end into the prokaryotic expression vector pETDuet-1. After expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) at 20°C overnight, the recombinant Zcchc11 (rZcchc11) protein was isolated by anti-FLAG affinity purification. The FLAG-beads were washed extensively, and rZcchc11 was eluted from the beads using FLAG-peptide. The isolated (∼130 kDa) rZcchc11 protein was free of major contaminants, and enzymatically active as shown by its ability to catalyze the Lin28-mediated uridylation of pre-let-7 (Fig. 1B–D). Importantly, this minimal fragment could be expressed and purified in the relatively large quantities required for assay design, optimization, and high-throughput screening.
Figure 1.
Purification of rZcchc11. (A) Schematic representation of Zcchc11 and the minimal region of Zcchc11 (rZcchc11) that is required for Lin28 mediated uridylation of pre-let-7. (B) SDS-PAGE and coomassie blue staining analysis of purified rZcchc11. (C and D) Uridylation of pre-let-7a1 by Lin28 and rZcchc11. (E and F) Uridylation of mature let-7i guide RNA by rZcchc11.
Our most recent work uncovered a novel function of Zcchc11, which preferentially uridylates the mature let-7 guide RNA in vitro and in cells in a Lin28-independent manner.30 We furthermore experimentally defined a sequence motif present in a small subset of mature miRNAs that confers this preferential uridylation activity.30 Consistent with these findings, rZcchc11 was capable of uridylating mature let-7 RNA in vitro (Fig. 1E,F). Since this assay is simpler, does not require Lin28, and is more amenable to scale-up, we favored developing this strategy for our high-throughput screening of Zcchc11 activity.
Development of an assay that monitors Zcchc11 activity
To develop this TUTase assay for high-throughput screening we required a non-radioactive detection method to monitor Zcchc11 activity. For this we decided to measure the levels of pyrophosphate (PPi) that is generated by Zcchc11-mediated nucleotide polymerization. The detection of pyrophosphate (PPi) can be achieved using a commercially available PPiLight assay, which converts the PPi level into luciferase intensity. Thus, the Zcchc11 enzymatic activity could be measured by simply monitoring the luciferase signal that is converted from the PPi generated by Zcchc11 catalysis (Fig. 2A). To establish and optimize this luciferase assay to monitor Zcchc11 activity, we incubated synthetic let-7 guide RNA with rZcchc11 and PPiLight substrate, in the presence or absence of UTP. We found this assay to be highly responsive to the UTP concentration in these reactions with changes in relative luciferase activity accurately reflecting the differential TUTase activity with ∼30 fold higher activity with 50 μM UTP compared to the background signal obtained in the absence of UTP (Fig. 2B). The luciferase activity in these reactions was found to also be dependent on the concentration of the mature let-7 guide RNA (Fig. 2B), further confirming that the detected luciferase signal is generated by the Zcchc11-catalyzed uridylation of mature let-7. Moreover, we further showed that rZcchc11 induces the luciferase activity in a dose-dependent manner (Fig. 2B), confirming that the intensity of the luciferase signal accurately reflects Zcchc11 enzymatic activity. Most importantly, this assay is highly sensitive, with 50 ng of rZcchc11 sufficient for a luciferase signal that is about 30 fold above the background. We therefore next tested the suitability of this luciferase-based assay for high-throughput applications.
Figure 2.
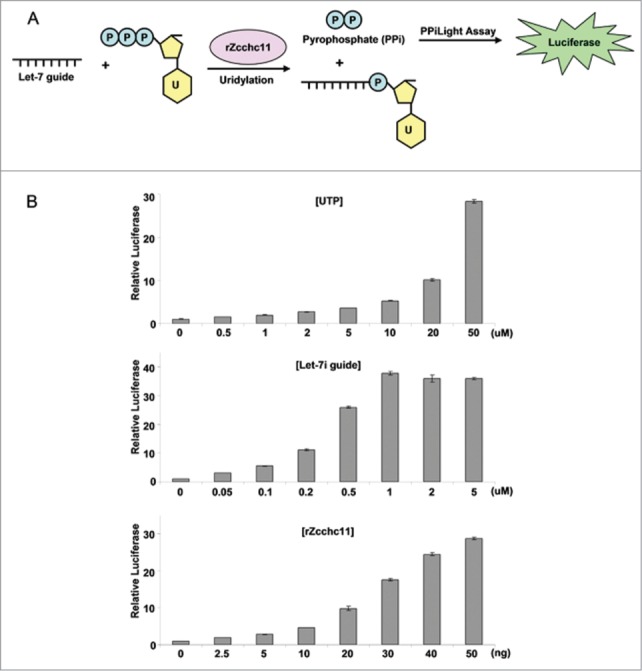
Optimization of PPi light assay to measure Zcchc11 activity. (A) Schematic demonstration of the PPi light assay. The PPi generated by rZcchc11-mediated uridylation is converted into luciferase signal by the PPiLight™ Inorganic Pyrophosphate Assay kit. (B) Optimization of PPi light assay for high throughput screening. Included (as indicated) is a titration of UTP, let-7i guide RNA, and rZcchc11 for the optimization of PPi light assay.
High-throughput screening for TUTase inhibitors
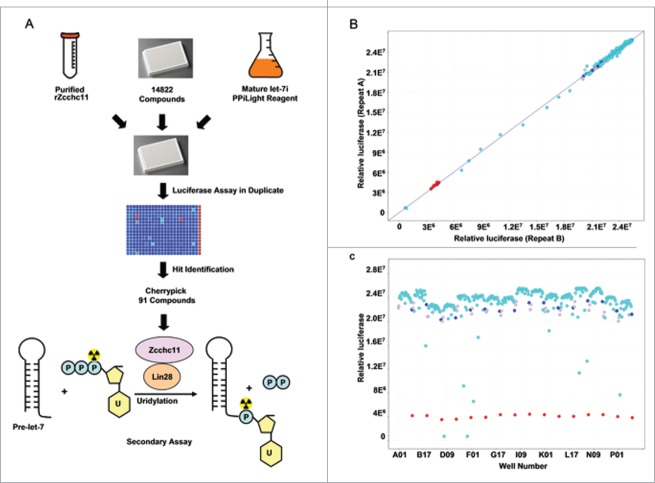
With the highly sensitive luciferase assay to monitor Zcchc11 enzymatic activity in place, we scaled up our system for the high-throughput screening of small molecule libraries to identify Zcchc11 inhibitors. As shown in Figure 3A, rZcchc11 and reaction buffer (containing let-7 RNA, UTP and PPi substrate) were subsequently added to 384 well plates by liquid handling robots together with the individual chemicals. After incubation, the luciferase signals were measured for each well to screen the small molecule compounds that can inhibit Zcchc11. The screening was performed in duplicate for each compound and the luciferase signals were highly correlated between the replicates (Fig. 3B). Compounds that reproducibly decreased the luciferase signal >2 -fold were considered as hits (Fig. 3C). In total we screened 14,822 compounds in duplicate, including 8,881 known bioactive compounds and 5,941 partially purified natural products. Based on the screening results, we cherry-picked 91 strong hits that can inhibit the luciferase >8 -fold for secondary screening to identify small molecule inhibitors of Zcchc11.
Figure 3.
High-throughput screening of Zcchc11 inhibitors. (A) Flow chart of the high throughput screening stratgey. 91 of 14,822 screened compounds were cherry picked for secondary assay. (B) A representative screening plate showing the correlation of luciferase reading between 2 repeats. (C) A representative screening plate showing the luciferase reading of each well. B and C were generated with visualization software Vortex. Red: positive control; Dark blue: Negative control; Light blue: screening samples; Gray: empty well.
Validation of TUTase inhibitors
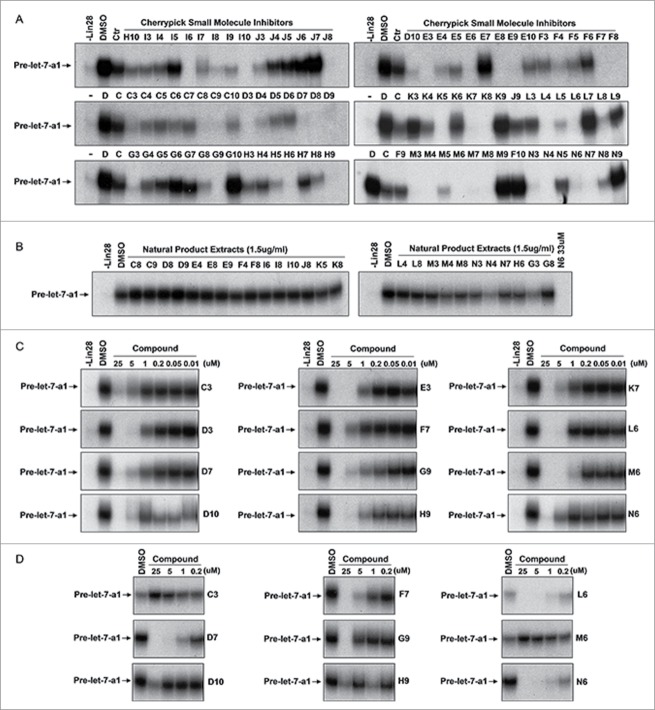
We next performed a secondary assay to verify the 91 ‘hits’ from our screen as TUTase inhibitors. For this we utilized the radioactive assay that monitors the Lin28-mediated uridylation of pre-let-7 by Zcchc11 (Fig. 1D). We found that 39 out of the 91 cherry-picked compounds could substantially block pre-let-7 uridylation at the same concentration that was used for the initial screening (33.3 μM for known bioactive compounds, 50 μg/ml for natural product extracts) (Fig. 4A). To help identify the most potent compounds we diluted the 39 compounds to select those that can inhibit the Zcchc11 activity at lower concentrations. Since most of the natural product extracts had little inhibitory effect at the concentration of 1.5 μg/ml (Fig. 4B), we subsequently focused on the known bioactive compounds to study their functions in inhibiting Zcchc11 activity. Dose curves were performed on 12 compounds. As shown in Figure 4C, the apparent IC50 of those bioactive compounds ranges from 0.2 to 2 μM. To further confirm this result, we re-purchased all 9 commercially available cherry-picked compounds and found that except for compounds C3 and M6, other compounds inhibited Zcchc11 at similar concentrations as the cherry-picked compounds provided by the screening facility (Fig. 4D).
Figure 4.
Secondary assay for TUTase inhibitors. (A) 91 cherry-picked compounds were analyzed with the Lin28 dependent secondary assay to determine their functions in regulation of Zcchc11 activity in uridylation of pre-let-7-a1. Concentrations used for the assay: known bioactive compounds: 33 μM; natural product extracts: 50 μg/ml. B and C: Test of diluted natural product extracts (1.5 μg/ml) (B) and known bioactive compounds (C) in uridylation assay. (D) Test of re-purchased compounds in inhibiting Lin28 and Zcchc11-mediated uridylation of pre-let-7a1.
We then considered the possible mechanism of action for those validated TUTase inhibitors identified in our screen. Since many of the verified hit compounds contain thiol groups that might be interacting with Cysteines in rZcchc11 (Fig. 5A), we further determined their inhibitory functions in reducing conditions. We first compared the inhibitory function of compound N6 in regular buffer or reducing conditions in which the reaction buffer was supplemented with 100 mM 2-Mercaptoethanol (β-ME). As shown in Figure 5B, addition of β-ME completely blocked the ability of compound N6 to inhibit Zcchc11 uridylation activity. We similarly tested all other validated hit compounds and found that only compounds L6 and F7 can inhibit uridylation in the reducing conditions, suggesting that most of the identified compounds inhibit Zcchc11 function through thiol-dependent mechanisms (Fig. 5B). Then we picked the apparent best inhibitory compounds (N6, L6, and F7) and performed dose-curve experiments to determine the IC50 for each compound. Serial dilution of the compounds revealed that the IC50 of compounds N6, L6 and F7 in regulating Lin28-mediated pre-let-7 uridylation is 0.38, 0.35, and 0.82 μM, respectively (Fig. 5C). To determine whether those compounds specifically inhibit Zcchc11 activity, we tested the activity of these compounds in inhibiting the uridylation activity of Cid1, the Schizosaccharomyces pombe Poly(U) Polymerase.31-34 As shown in Figure 5D, compounds N6 and L6 did not inhibit PUP activity, while compound F7 did inhibit PUP activity albeit at a higher concentration that for Zcchc11. These data support that compounds N6 and L6 selectively inhibit Zcchc11 but not a related TUTase.
Figure 5.
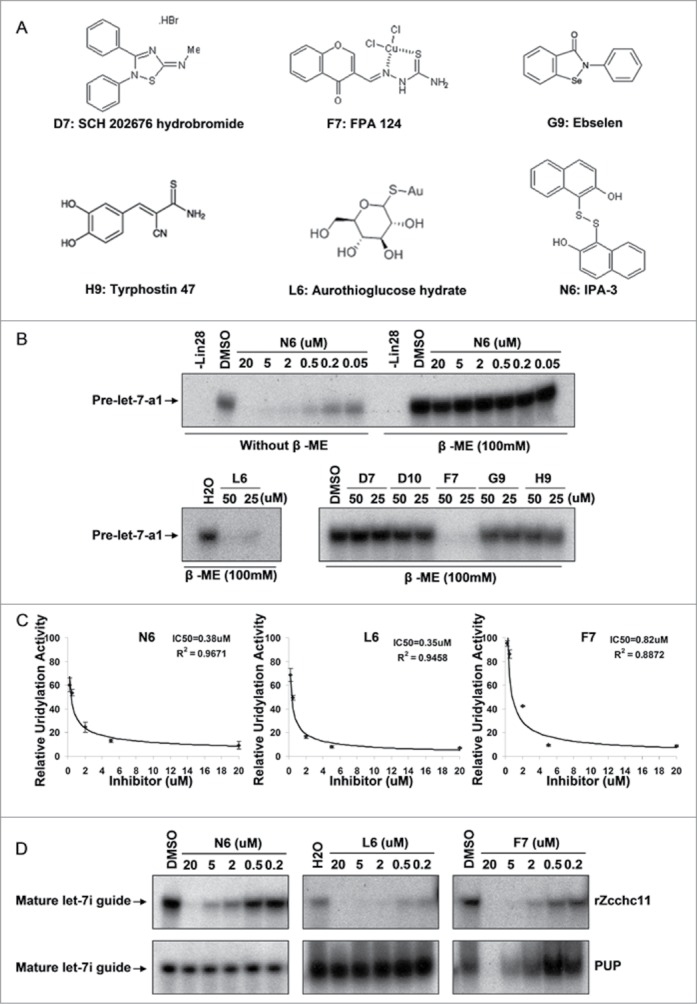
Mechanisms and specificity of Zcchc11 inhibition. (A) Structure of the identified compounds. (B) Test of compound inhibition in reducing conditions with 100 mM β-ME. (C) IC50 calculation from dose curve experiments with compounds N6, L6 and F7 in regulating Zcchc11 mediated uridylation. Signal intensitiy of each band was calculated by Image J and the inhibitory curves generated by Excel. (D) Specificity of compounds N6, L6 and F7 in regulating Zcchc11 activity in uridylation of mature let-7i guide RNA. Upper panel: roles of compounds in inhibiting rZcchc11 activity; Lower panel: roles of compounds in inhibiting PUP activity.
Test of Zcchc11 inhibitors in cells
We next determined whether the Zcchc11 inhibitors we identified could block TUTase activity in cells. Since uridylated pre-let-7 is rapidly degraded by Dis3l2, the levels of this RNA decay intermediate is very low in steady state cellular RNA samples. Therefore to more reliably detect the relative levels of uridylated pre-let-7 and to determine the inhibitory effect of Zcchc11 inhibitors, we tested the TUTase inhibitors in Dis3l2 knockdown mESCs in which the uridylated pre-let-7 is stabilized (Fig. 6A). As a control, we depleted Zcchc11 by siRNA in the Dis3l2 knockdown cells. As shown in Figure 6B, knockdown of Zcchc11 results in the dramatic decrease of the uridylated pre-let-7, confirming that the in vivo uridylation of pre-let-7 is mediated by Zcchc11. Treatment of the Dis3l2 knockdown cells with the screened compounds revealed that one of the compounds, compound L6 (Aurothioglucose hydrate), can inhibit the Zcchc11-mediated uridylation of pre-let-7 in vivo (Fig. 6C). Overall, these results support that compound L6 inhibits Zcchc11 enzymatic activity in cells.
Figure 6.
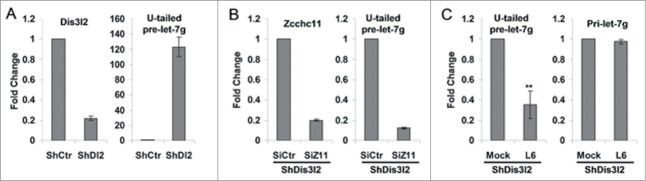
Test of Zcchc11 inhibitors in cells. (A) Dis3l2 knockdown (left panel) in V6.5 mESCs results in accumulations of uridylated pre-let7 (U-tailed pre-let-7, right panel). (B) Zcchc11 knockdown (left) in Dis3l2 depleted cells decreases uridylated pre-let7 level (U-tailed pre-let-7, right panel). (C) Treatment of Dis3l2 depleted mESCs with compound L6 (500 μM) results in decreased level of U-tailed pre-let-7, while have little effect on pri-let-7 expression. **, P < 0.01.
Discussion
The aberrant Lin28/TUTase/let-7 pathway is frequently found in cancers and can drive oncogenesis in vivo. Lin28A/B expression is associated with advanced disease in hepatocellular carcinoma (HCC), chronic myeloid leukemia (CML), Wilms' tumor, ovarian carcinoma, and germ cell tumors.16,22,35-44 Moreover, Lin28A/Lin28B expression is associated with poor clinical outcome and patient survival in HCC, ovarian cancer, Neuroblastoma, and Medulloblastoma.16,18,35,45 Therefore, identification of small molecule drugs that specifically inhibit the Lin28/let-7 pathway might prove to be a powerful approach for cancer therapy. However, there are currently no known pharmacological agents capable of targeting this pathway. Since TUTase activity is required for the Lin28 mediated repression of let-7 biogenesis, and is likely to be highly “druggable,” targeting TUTase activity might represent a promising strategy to inhibit the Lin28 oncogenic pathway in cancers. In addition, the TUTases were recently reported to uridylate miRNA and mRNA independent of Lin28,30,46,47 suggesting that TUTases are significant players in regulating posttranscriptional gene expression through uridylation. Therefore, potent TUTase inhibitors might not only be used for drug development against the Lin28 oncogenic pathway, but could also be used to study uridylation-mediated RNA decay. For this purpose, we developed an in vitro assay suitable for high-throughput screening to identify small molecules that can specifically inhibit Zcchc11 TUTase activity.
In this study, we designed, optimized, and applied a luciferase-based assay for the high-throughput screening of chemical libraries to identify small molecule inhibitors of Zcchc11, a critical enzyme involved in the Lin28-meidated control of let-7 miRNA biogenesis. We screened 14,822 compounds in duplicate and identified 91 compounds that can inhibit the Zcchc11 enzyme activity. To exclude possible false positive compounds, we performed a secondary assay to identify bona fide inhibitors of Lin28-mediated pre-let-7 uridylation. This led to our discovery of compounds that specifically inhibit the Zcchc11 activity in vitro. Inspection of the molecular structures of these validated inhibitory compounds revealed the presence of reactive thiol groups in most of them. This supports the possibility that these compounds inhibit the TUTase by covalently binding to Cysteines in Zcchc11. Considering the rZcchc11 fragment used in these studies contains 37 Cysteine residues, identifying those that are preferentially bound by each of the compounds remains a challenge. One possibility however is that these inhibitory compounds bind to certain Cysteine(s) within one or more of the Zinc fingers to interfere with Zcchc11 binding its RNA substrate. The inclusion of 2-Mercaptoethanol (β-ME) in the reaction buffer will help refine our high-throughput screening assay by excluding this class of inhibitors as positive hits.
When tested in cells, one of the identified compounds (Aurothioglucose hydrate) can block the uridylation of pre-let-7 in vivo. Aurothioglucose hydrate has been used clinically to treat rheumatoid arthritis and has been reported to inhibit PKCiota-Par6 interaction and suppress the growth of non-small-cell lung cancer in vitro and in vivo.48 Our data reveal a novel function of Aurothioglucose hydrate in inhibiting Zcchc11 activity. Although the motivation for this study was to find small molecules that can inhibit the TUTase to elevate let-7 level in Lin28-expresisng cells, the treatment of mouse P19 (embryonal carcinoma) cells, mouse embryonic stem cells (ESCs), and a small panel of human cancer cell lines did not lead to elevated let-7 expression (data not shown). This might be due to several reasons including: (1) Due to the general cytotoxic effects of several of the compounds, we were only able to treat cells with relatively low concentrations of compound that might not be high enough to induce let-7 upregulation. (2) Some of the compounds might have poor cell permeability and stability in vivo. (3) The reducing environment in the cell cytoplasm might partially block the function of the sensitive compounds. Therefore, further chemical modifications will be required to reduce cytotoxic effect and increase specificity of the identified lead compounds in targeting the Zcchc11 activity.
In summary, we established a luciferase based high-throughput screening methodology to identify small molecule compounds that can specifically inhibit the Zcchc11 TUTase activity in vitro and in vivo. Our study represents a proof-of-concept that may be further exploited by more extensive high-throughput screening and/or a screening strategy focused on libraries of nucleotide analogs as a first step toward therapeutic targeting of the TUTase in the oncogenic Lin28/let-7 pathway.
Materials and Methods
Plasmids
For rZcchc11 expression, a fragment of Zcchc11 was PCR amplified and subcloned using with the following primers: Sal I-mZ11-F: ACGCgtcgacTCTCAGAAGCAGCAGACTCA; Not I-FLAG-mZ11-R:ATAAGAATgcggccgcCTTGTCGTCATCGTCTTTGTAGTCTCTTTTCCTTTTGGGACAGTCT to the prokaryotic expression vector pETDuet-1. The FLAG-Lin28 plasmid was previously described.25
Immunoprecipitation and recombinant protein purification
Immunoprecipitation and recombinant protein purification were performed as previously described.25 Briefly, Flag-Lin28 expression plasmids were transfected into HEK293T cells with Lipofectamine 2000 (Invitrogen). 48 hours later, whole cell lysate was collected for the purification of Flag-Lin28 with anti-Flag M2 beads (Sigma). For recombinant Zcchc11 protein purification, rZcchc11 was induced with 500 μM IPTG overnight at 20°C. The FLAG tagged rZcchc11 proteins were then purified using anti-Flag M2 beads according to the manufacture's instructions.
In vitro uridylation assay
In vitro uridylation assay was performed as previously described.25 0.01 μM unlabeled synthetic pre-let-7a1 or mature let-7i guide RNA (Dharmacon) were incubated with purified proteins and buffer containing [α-32P] UTP at 37°C for 1h. A limited amount of (125 nM) [α-32P] UTP was added in the uridylation system, therefore the pre-let-7a1 or mature let-7i are predominantly monouridylated in these reaction conditions. The products were then analyzed on 15% denaturing polyacrylamide gels and radioactively labeled RNAs were detected by autoradiography. For IC50 calculation, band intensities were quantified with ImageJ. Where indicated reactions were supplemented with 100 mM 2-Mercaptoethanol (β-ME).
Gene knockdown and Real-Time PCR
The shRNA and siRNA mediated gene knockdown experiment was performed as previously described.26 Total RNA samples were collected for real-time PCR assay to detect the relative gene expression. To detect the poly-uridylated pre-let7g, small RNAs (<200 bp) were first isolated with mirVana miRNA Isolation Kit (Life Technologies), then same amount of isolated small RNA samples were used for the reverse transcription with oligodA12 and subsequent real-time PCR analysis.26
PPi light assay
The PPi light assay was performed with the PPiLight™ Inorganic Pyrophosphate Assay kit (Lonza). Briefly, reaction system containing 50 ng rZcchc11 protein, 50 μM UTP, 0.75 μM let-7i synthetic guide RNA, 4 units of RNasin (Promega), and PPi substrates was incubated at 37°C for 30 mins. Luciferase intensity was then measured using luminometer (BioTek).
High-throughput screening
On the day of screening, 10 μl of BC150 buffer (20 mM Tris–HCl pH 7.9, 20% glycerol, 0.2 mM EDTA, 150 mM KCl) containing Zcchc11 (∼50ng per well) was added into 384-well, white, low volume plates (Corning) with Multidrop™ Combi nL Reagent Dispenser (Thermo Scientific). Compounds were immediately added into wells by pin-transfer robot (Seiko). Wells in column 23 contained DMSO (100 nl/well) and served as the negative controls. Column 24 also contained the positive control that can inhibit Zcchc11 activity (from collaborator, unpublished) at 33.3uM. After pin transfer, 20 μl of reaction buffer containing let-7i guide RNA, RNAsin, PPi substrate and UTP was added to the wells using Multidrop™ Combi nL Reagent Dispenser (Thermo Scientific). Plates were then incubated at room temperature for 60 mins, then the luciferase intensities quantified with the Envision plate reader (PerkinElmer). Z´ factor of the screening was higher than 0.8, as calculated following the instructions of ICCB at Harvard Medical School (http://iccb.med.harvard.edu/screening-information/overview-and-guidelines/#quantitative).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank William Lowry (UCLA) for sharing unpublished data and providing the positive control compound used in the screen. Thanks also to James Thornton for technical advice with Zcchc11 purification and TUTase activity assays. We are grateful to Jennifer Smith and other members of the ICCB-Longwood screening facility for input on assay design, equipment use, and data analysis.
Funding
S.L. is a Damon Runyon-Sohn Pediatric Fellow supported by the Damon Runyon Cancer Research Foundation (DRSG-7–13). R.I.G is supported by grants from the US National Cancer Institute (NCI) (R01CA163467), and The American Cancer Society (121635-RSG-11–175–01-RMC).
References
- 1.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10:704-14; PMID:19763153; http://dx.doi.org/ 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol 2010; 22:35-45; PMID:19907325; http://dx.doi.org/ 10.1097/CCO.0b013e328333dcbb [DOI] [PubMed] [Google Scholar]
- 3.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015; 15:321-33; PMID:25998712; http://dx.doi.org/ 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 2009; 8:843-52; PMID:19221491; http://dx.doi.org/ 10.4161/cc.8.6.7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barh D, Malhotra R, Ravi B, Sindhurani P. Microrna let-7: an emerging next-generation cancer therapeutic. Curr Oncol 2010; 17:70-80; PMID:20179807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007; 315:1576-9; PMID:17322030; http://dx.doi.org/ 10.1126/science.1137999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006; 29:903-6; PMID:16651716; http://dx.doi.org/ 10.1248/bpb.29.903 [DOI] [PubMed] [Google Scholar]
- 8.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 2008; 14:400-9; PMID:18674967; http://dx.doi.org/ 10.1016/j.molmed.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 9.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131:1109-23; PMID:18083101; http://dx.doi.org/ 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- 10.Barh D, Malhotra R, Ravi B, Sindhurani P. Microrna let-7: an emerging next-generation cancer therapeutic. Curr Oncol 2010; 17:70-80; PMID:20179807; http://dx.doi.org/ 10.3747/co.v17i1.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, et al.. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67:7713-22; PMID:17699775; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1083 [DOI] [PubMed] [Google Scholar]
- 12.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15:509-24; PMID:25027649; http://dx.doi.org/ 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 13.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008; 10:987-93; PMID:18604195; http://dx.doi.org/ 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- 14.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 2008; 320:97-100; PMID:18292307; http://dx.doi.org/ 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008; 32:276-84; PMID:18951094; http://dx.doi.org/ 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 16.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al.. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009; 41:843-8; PMID:19483683; http://dx.doi.org/ 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamano R, Miyata H, Yamasaki M, Sugimura K, Tanaka K, Kurokawa Y, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, et al.. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer 2012; 106:1415-23; PMID:22433967; http://dx.doi.org/ 10.1038/bjc.2012.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, Mestdagh P, van Sluis P, Valentijn LJ, van Nes J, et al.. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012; 44:1199-206; PMID:23042116; http://dx.doi.org/ 10.1038/ng.2436 [DOI] [PubMed] [Google Scholar]
- 19.Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS, Rustgi AK. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev 2013; 27:2233-45; PMID:24142874; http://dx.doi.org/ 10.1101/gad.224659.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbach A, Yermalovich A, Zhang J, Spina CS, Zhu H, Perez-Atayde AR, Shukrun R, Charlton J, Sebire N, Mifsud W, et al.. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 2014; 28:971-82; PMID:24732380; http://dx.doi.org/ 10.1101/gad.237149.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, Comerford SA, Ramezani S, Sun X, Parikh MS, et al.. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014; 26:248-61; PMID:25117712; http://dx.doi.org/ 10.1016/j.ccr.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009; 139:693-706; PMID:19878981; http://dx.doi.org/ 10.1016/j.cell.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 2009; 16:1021-5; PMID:19713958; http://dx.doi.org/ 10.1038/nsmb.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009; 138:696-708; PMID:19703396; http://dx.doi.org/ 10.1016/j.cell.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 25.Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012; 18:1875-85; PMID:22898984; http://dx.doi.org/ 10.1261/rna.034538.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013; 497:244-8; PMID:23594738; http://dx.doi.org/ 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 2013; 19:1632-8; PMID:24141620; http://dx.doi.org/ 10.1261/rna.040055.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piskounova E, Polytarchou C, Thornton JE, Lapierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B Inhibit let-7 MicroRNA Biogenesis by Distinct Mechanisms. Cell 2011; 147:1066-79; PMID:22118463; http://dx.doi.org/ 10.1016/j.cell.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu X, Meng Z, Liang W, Tian Y, Wang X, Han W, Lou G, Lou F, Yen Y, Yu H, Jove R, Huang W. miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene 2014; 33:4296-306; PMID:24056962; http://dx.doi.org/ 10.1038/onc.2013.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornton JE, Du P, Jing L, Sjekloca L, Lin S, Grossi E, Sliz P, Zon LI, Gregory RI. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res 2014; 42:11777-91; PMID:25223788; http://dx.doi.org/ 10.1093/nar/gku805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 2009; 16:616-23; PMID:19430462; http://dx.doi.org/ 10.1038/nsmb.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Tello P, Gabus C, Thore S. Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure 2012; 20:977-86; PMID:22608966; http://dx.doi.org/ 10.1016/j.str.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 33.Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol 2012; 19:782-7; PMID:22751018; http://dx.doi.org/ 10.1038/nsmb.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunde BM, Magler I, Meinhart A. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic acids research 2012; 40:9815-24; PMID:22885303; http://dx.doi.org/ 10.1093/nar/gks740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L, Katsaros D, Shaverdashvili K, Qian B, Wu Y, de la Longrais IA, Preti M, Menato G, Yu H. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur J Cancer 2009; 45:2212-8; PMID:19477633; http://dx.doi.org/ 10.1016/j.ejca.2009.05.003 [DOI] [PubMed] [Google Scholar]
- 36.Wang YC, Chen YL, Yuan RH, Pan HW, Yang WC, Hsu HC, Jeng YM. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis 2010; 31:1516-22; PMID:20525879; http://dx.doi.org/ 10.1093/carcin/bgq107 [DOI] [PubMed] [Google Scholar]
- 37.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 2009; 28:347-58; PMID:19153603; http://dx.doi.org/ 10.1038/emboj.2008.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006; 384:51-61; PMID:16971064; http://dx.doi.org/ 10.1016/j.gene.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 39.King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B Promotes Colon Cancer Progression and Metastasis. Cancer Res 2011; 71:4260-8; PMID:21512136; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji J, Wang XW. A Yin-Yang balancing act of the lin28/let-7 link in tumorigenesis. J Hepatol 2010; 53:974-5; PMID:20739081; http://dx.doi.org/ 10.1016/j.jhep.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys 2010; 76:5-8; PMID:20005451; http://dx.doi.org/ 10.1016/j.ijrobp.2009.08.028 [DOI] [PubMed] [Google Scholar]
- 42.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010; 29:2153-9; PMID:20101213; http://dx.doi.org/ 10.1038/onc.2009.500 [DOI] [PubMed] [Google Scholar]
- 43.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell 2010; 140:445-9; PMID:20178735; http://dx.doi.org/ 10.1016/j.cell.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 44.West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al.. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009; 460:909-13; PMID:19578360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, Sultan M, Stachurski K, Ryzhova M, Warnatz HJ, et al.. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014; 510:537-41; PMID:24847876; http://dx.doi.org/ 10.1038/nature13268 [DOI] [PubMed] [Google Scholar]
- 46.Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014; 159:1365-76; PMID:25480299; http://dx.doi.org/ 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones MR, Blahna MT, Kozlowski E, Matsuura KY, Ferrari JD, Morris SA, Powers JT, Daley GQ, Quinton LJ, Mizgerd JP. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS Genet 2012; 8:e1003105; PMID:23209448; http://dx.doi.org/ 10.1371/journal.pgen.1003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stallings-Mann M, Jamieson L, Regala RP, Weems C, Murray NR, Fields AP. A novel small-molecule inhibitor of protein kinase Ciota blocks transformed growth of non-small-cell lung cancer cells. Cancer Res 2006; 66:1767-74; PMID:16452237; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-3405 [DOI] [PubMed] [Google Scholar]