Abstract
The innate immune system is the first line of defense against microbial pathogens, but tight regulation of gene expression is necessary to prevent the detrimental effects of unrestrained activation. Although the functions of most long noncoding RNAs (lncRNAs; >200 nucleotides) are unknown, many have been shown to regulate diverse cellular activities. Recent reports by us and others have suggested that lncRNAs may also play critical roles in transcriptional regulation of gene expression during innate immune responses. Following engagement of Toll-like receptors, lncRNAs form functional RNA–protein complexes that recruit activators or remove repressors of transcription, leading to rapid expression of inflammatory mediators. These discoveries suggest that lncRNAs may contribute to the gene regulatory networks that govern host–pathogen interactions.
Keywords: Innate immunity, vaccine design, non-coding RNAs, epigenetic regulation, Toll-like receptors
Innate Immunity and Toll-Like Receptors
The innate immune response is a phylogenetically ancient and conserved system that protects against external and internal “danger signals.” In vertebrates, this system not only serves as a rapidly mobilized first line of defense against invading pathogens, but also influences the development of the slower but more sophisticated adaptive immune response.1 Cells of the innate immune system must recognize and react to diverse classes of microbial pathogens that vary extensively in their molecular composition, structure, and life cycle. Pathogen detection is mediated by a group of pattern recognition receptors (PRRs) that recognize conserved structures called pathogen-associated molecular patterns (PAMPs). PRRs may be present on the host cell surface or intracellularly and, in most cases, they trigger intracellular signaling pathways that culminate in transcription of pro-inflammatory and anti-microbial molecules. Multiple classes of PRRs are known, including the membrane-associated Toll-like receptors (TLRs), C-type lectin receptors, the cytosolic RIG-I-like receptors, NOD-like receptors and AIM2-like receptors. Among these, TLRs are the best characterized and most extensively studied PRRs.1
TLRs are a family of type I transmembrane proteins composed of three domains: an extracellular leucine-rich domain for PAMP detection, a transmembrane domain, and an intracellular domain, termed the Toll/IL-1 receptor (TIR) domain. Upon receptor engagement, the TIR domains interact with adaptor proteins such as MyD88, TIR domain-containing adaptor protein-inducing IFN-β (TRIF), TIR-associated protein (TIRAP), and TRIF-related adaptor molecule (TRAM),2 which activate a cascade of kinase-mediated signaling events. TLR signaling ultimately leads to activation of NFκB, IRF, and AP-1 transcription factors that control expression of pro-inflammatory cytokines such as tumor necrosis factor-α (TNFα), interleukin (IL)-1, and the interferons (IFNs), all of which play central roles in anti-microbial defense.1 Accumulating evidence suggests that TLRs not only guard against exogenous threats but also recognize endogenous structures and molecules released from damaged or dead cells, known as damage-associated molecular patterns (DAMPs).3 This observation indicates that TLR-mediated signaling plays an important role not only in defense against infectious diseases but also in non-infective inflammatory responses to tissue/cellular damage.
Uncontrolled cytokine production is at the heart of many autoimmune and chronic inflammatory diseases, and as such, negative regulation of the innate immune response is critical for maintaining immune homeostasis and limiting the detrimental effects of excessive stimulation. Indeed, inappropriate or sustained activation of TLRs have been associated with several autoimmune disorders. TLR signaling can be negatively regulated at the transcriptional and post-transcriptional levels. Transcriptional regulation is achieved through expression of inhibitor proteins,4 recruitment of chromatin-modifying enzymes,5,6 prevention of transcription factor binding,7 and promotion of target mRNA decay.8 At the post-transcriptional level, TLR signaling is regulated by dissociation of adaptor complexes, proteasome-mediated degradation of signaling proteins, and miRNA-mediated post-transcriptional gene silencing.9,10
In recent years, noncoding RNAs have emerged as major regulators of gene expression. Several small (∼22 nucleotide) microRNAs (mRNAs), such as miR-14611 and miR-155,12 have been shown to regulate not only the innate immune response but also immune cell development and adaptive immunity.10 However, little is known of the immune-modulating functions of other noncoding RNAs. In the following sections, we discuss the emerging interest in long noncoding RNAs (lncRNAs) as regulators of innate immunity.
The Functions of LncRNAs
The discovery that RNA molecules are transcribed from both coding and noncoding regions of the genome, including promoters, introns, and intergenic regions, indicated that the eukaryotic transcriptome is far more complicated than once imagined.13 Although the vast majority of lncRNAs, operationally defined as being more than 200 nucleotides in length and lacking in putative coding regions,13 have not yet been ascribed a function, they are increasingly implicated in the regulation of numerous cellular processes such as X-chromosome inactivation,14 regulation of p53 pathway,15 stem cell self-renewal and differentiation,16,17 epidermal differentiation,18 erythroid development,19 DNA damage response,20 chromosome architecture,21 and maintenance of active chromatin state.22 The low abundance and lack of sequence conservation of lncRNAs initially suggested they were of little biological significance.23 However, later efforts have made it clear that these molecules have great potential to advance our understanding of cell regulatory mechanisms in health and disease.
To identify mammalian lncRNAs with the highest probability of functional significance, Guttman et al.24 took advantage of their previous finding that chromatin at actively transcribed genes is enriched in trimethylated lysine 4 of histone H3 (H3K4me3) at the promoters and trimethylated lysine 36 of histone H3 (H3K36me3) along the actively transcribed regions.25 They analyzed genome-wide chromatin state maps and identified ∼1600 ncRNAs encoded in intergenic regions, which they named large intergenic RNAs (lincRNAs). These RNAs showed clear evidence of sequence conservation, albeit less than protein-coding genes,24 and a later study indicated that the conserved regions of many lncRNAs are often restricted to a single short sequence.26
Transcriptional Regulation by LncRNAs
LncRNAs regulate gene expression through multiple mechanisms, including chromatin remodelling, epigenetic regulation, transcription, mRNA splicing, RNA decay, and enhancer functions. However, most lncRNAs with known functions act at the transcriptional level by forming RNA–protein complexes (RNP). The three major routes for lncRNA-mediated transcriptional control are (1) RNP formation with chromatin-modifying proteins, (2) RNP formation with non-chromatin-binding nuclear proteins, and (3) RNP-independent enhancer-like activity (Fig. 1, Table 1). HOTAIR is an example of a lncRNA that regulates gene transcription through chromatin modification. HOTAIR is transcribed from the HOXC locus and interacts with Ezh2 and Suz12, subunits of the methyltransferase Polycomb repressor complex 2 (PRC2)27 PRC2 is primarily responsible for H3K27me3, a marker of closed chromatin. HOTAIR binding recruits PRC2 to gene loci and thus facilitates gene silencing; indeed, the HOTAIR–PRC2 RNP has been reported to silence hundreds of genes in the human genome.28 Not surprisingly, aberrant lncRNA function is linked to several human diseases, including cancer. For example, HOTAIR overexpression in cancer cells plays an important role in allowing expression of genes associated with invasiveness and metastasis.28 Binding of lncRNAs to chromatin modifiers can also activate transcription. One example is the lncRNA HOTTIP, which forms an RNP with WDR5, an adaptor protein for the MLL family of H3K4 methylases. Binding of HOTTIP to WDR5 has been shown to be required for H3K4 methylation and maintenance of pluripotency in mouse embryonic stem cells.29
Figure 1.
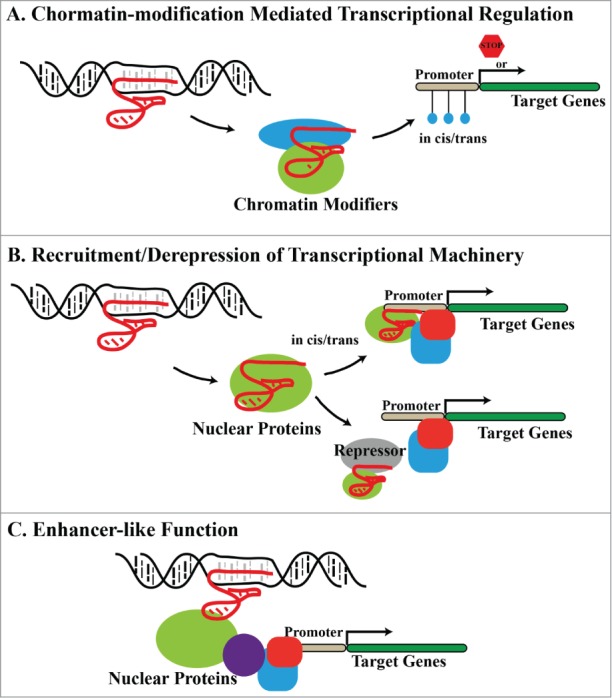
LncRNA-mediated transcriptional regulation. Long noncoding RNAs (lncRNAs; red solid lines) regulate gene transcription through three main mechanisms: (A) Interaction with and recruitment of chromatin-modifying enzymes (e.g., histone methylases, acetylases, and deacetylases) to the target gene locus. Modulation of the chromatin state by these enzymes leads to activation or repression of local genes. (B) Interaction with other RNA-binding factors such as hnRNPs to form RNA–protein complexes (RNPs). RNPs can either promote transcription by recruiting key proteins to the target gene promoters or derepress gene transcription by binding to existing gene repressors. (C) LncRNAs also have enhancer functions and help to change the chromatin architecture and recruit transcriptional machinery proteins to adjacent target gene locus to promote its transcription.
Table 1.
Known lncRNA-interacting proteins
| LncRNAs | Proteins | Mode of Action | Function | Reference |
|---|---|---|---|---|
| Firre | hnRNP-U | Nuclear architecture, transcriptional regulation | Embryonic stem cell self-renewal | 21 |
| HOTAIR | Ezh2, Suz12 | Chromatin modification, transcriptional regulation | Cancer metastasis | 28, 51 |
| HOTTIP | WDR5 | Chromatin modification, transcriptional regulation | Hox gene expression | 22 |
| LincRNA-p21 | hnRNP-K | Transcriptional regulation | p53-mediated gene repression | 15 |
| LincRNA-p21 | HuR | Translational control | Gene suppression in HeLa cells | 52 |
| LincRNA-Cox2 | hnRNP-A/B, A1/B1 | Transcriptional regulation | Innate immunity | 31 |
| NEAT1 | SFPQ | Transcriptional regulation | Innate immunity | 34 |
| NeST | WDR5 | Chromatin modification, transcriptional regulation | Innate immunity | 33 |
| RepA | Ezh2 | Transcriptional regulation | XIST induction | 14 |
| RMST | Sox2 | Transcriptional regulation | Neurogenesis | 53 |
| TINCR | Stau1 | RNA stabilization | Epidermal differentiation | 18 |
| THRIL | hnRNP-L | Transcriptional regulation | Innate immunity | 30 |
| TUNA | PTBP1, hnRNP-K, and NCL | Transcriptional regulation | Embryonic stem cell self-renewal and neural differentiation | 17 |
| Lnc-DC | STAT3 | Phosphorylation and transcriptional regulatioin | Innate and Adaptive Immunity | 45 |
LncRNAs also modulate gene transcription by interacting with nuclear proteins other than chromatin modifiers. The heterogeneous ribonucleoproteins (hnRNPs) are a large family of proteins that were once thought to regulate gene expression exclusively through pre-mRNA processing and splicing. However, accumulating evidence suggests that many can associate with lncRNAs and function by recruiting transcriptional machinery to the target gene promoters. Examples of such RNPs are LincRNA-p21–hnRNP-K,15,17 THRIL–hnRNP-L,30 Firre–hnRNP-U,21 and LincRNA-Cox2–hnRNP-A/B.31 Finally, lncRNAs can function as enhancers of gene expression without binding to other cellular factors. In this case, active transcription of the lncRNA leads to increased expression of neighboring protein-coding genes through in cis and in trans regulation (Table 1).32
LncRNAs Are Novel Regulators of Innate Immunity
Several key discoveries over the past several years have identified key roles for lncRNAs in regulating the mammalian innate immune response (Fig. 2).30,31,33,34 These findings have served as an impetus for a more thorough examination of how lncRNAs may influence not only the innate response but also the more sophisticated adaptive immune response as well as immune cell development.
Figure 2.
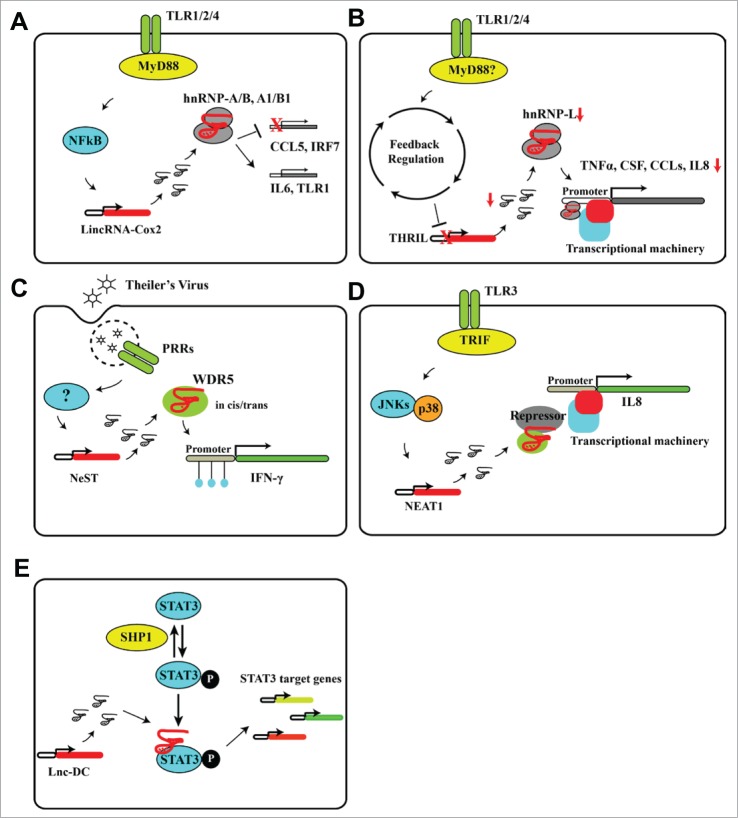
Regulation of innate immune responses by lncRNAs. TLR signaling induces expression of lncRNAs involved in innate immune regulation, including lincRNA-Cox2, NeST, THRIL, and NEAT1. (A) LincRNA-Cox2 interacts with hnRNP-A/B and hnRNP-A1/B1 to form RNPs that can activate (e.g., IL-6) or repress (e.g., CCL5) gene expression. (B) TLR signaling can activate endogenous feedback-regulation networks to limit the potentially damaging effects of excessive inflammation. This downregulates expression of the lncRNA THRIL, which is required for transcription of inflammatory genes. Downregulation of THRIL thus helps to restrain TLR-induced gene activation. (C) Viral infection upregulates expression of lncRNAs, including NeST. NeST interacts with the adaptor protein WDR5 and recruits the chromatin-modifying histone methyltransferase MLL to the target gene locus. Alterations in Figure 2 (See previous page). the local chromatin state can induce or repress expression of target genes such as IFN-γ. (D) Activation of TLR3 signaling induces expression of NEAT1. This lncRNA binds to transcriptional repressor proteins such as SFPQ and thus allows transcription of local inflammatory genes to proceed. (E) Lnc-DC binds with STAT3 to protect it from dephosphorylation induced by phophatases such as SHP1, which leads to increased phosphor-STAT3 and thus activates downstream gene expression.
The first demonstrations that lncRNAs can be induced in innate immune cells came from studies of TLR4-activated cells.24 Human dendritic cells treated with a TLR-4 agonist showed significant upregulation of ∼20 lincRNAs, most of which were located in a cluster associated with NFκB signaling. One of these, named lincRNA-Cox2 for its location upstream of the NFκB target gene COX2, was induced ∼1000-fold by TLR4 stimulation but was only weakly induced by TLR3 stimulation.24 Consistent with this, induction of lincRNA-Cox2 was later confirmed by Carpenter et al. to be MyD88- and NFκB-dependent.31 These investigators combined RNAi-mediated silencing and next-generation sequencing analysis to demonstrate that lincRNA-Cox2 could both activate and repress gene expression in mouse bone marrow-derived macrophages. For example, lincRNA-Cox2 suppressed IRF7 and CCL5 in resting cells but was required for induction of TLR1 and IL-6 in response to TLR2 stimulation with Pam3CSK4. Mechanistically, lincRNA-Cox2 mediates these effects by interacting with hnRNP-A/B and hnRNP-A1/B1 and recruiting transcriptional machinery to the target gene promoters.31
The lncRNA NeST was shown to control susceptibility to persistent Theiler's virus infection and resistance to Salmonella enterica Typhimurium infection in mice by regulating IFN-γ expression.33 NeST, formerly known as Tmevpg1, is located close to the IFN-γ gene on chromosome 10 of the mouse genome; a locus previously demonstrated by genetic mapping to be closely linked with susceptibility to Theiler's virus-induced neurological and inflammatory diseases.35,36 NeST had long been suspected to be involved in regulation IFN-γ expression,37 but the mechanism was not known. Gomez et al. showed that NeST acts in trans to increase IFN-γ expression by interaction with the WDR5/MLL adaptor protein and alteration of H3K3 methylation at the IFN-γ promoter. In addition, overexpression of NeST protected mice against lethal Salmonella infection. These data provided mechanistic insight into the physiological function of NeST in the response to viral and bacterial infection.
Recent work from our lab identified a previously undocumented lncRNA, THRIL, as a crucial regulator of TNFα induction upon TLR1/2 signaling in human cells.30 THRIL is located in the opposite strand next to the Bri3bp gene, with ∼450 bp overlapping with the Bri3bp mRNA 3′UTR. THRIL was found to regulate basal transcription and induction of TNFα expression by interacting with hnRNP-L. HnRNP-L is located in both the nucleus and cytoplasm. In the nucleus, hnRNP-L binds to and regulates the activities of Line-1 retrotransposon38; in the cytoplasm, it binds to and stabilizes target mRNAs response to stress stimuli.39 Our work revealed a novel role for the THRIL–hnRNP-L RNP complex as a transcriptional activator at the TNFα promoter. This role was consistent with earlier findings that hnRNP-L interacts with the multi-protein Mediator complex.40 Mediator is a well-documented multi-protein transcriptional co-activator that interacts with RNA polymerase II and other components of the transcriptional machinery.41 hnRNP-L also interacts with P-TEFb complex,42 an important elongation factor for gene transcription. This report showed that hnRNP-L bound with P-TEFb complex and Aire to regulate the expression of many autoantigen genes in the thymus and play a role in establishing immunological tolerance in T cells.42
NEAT1 is another lncRNA regulator of innate immune gene expression.34 NEAT1 is essential for formation of nuclear paraspeckles,43 which are ribonucleoprotein bodies in interchromatin space that regulates specific gene expression through retention of RNA and transcriptional proteins.44 NEAT1 expression is induced by binding of the synthetic dsRNA analog poly I:C to TLR3. Upon expression, NEAT1 binds with the DNA- and RNA-binding protein splicing factor, proline/glutamine-rich (SFPQ) and prevents SFPQ suppression of several immune-related genes downstream of TLR3-p38 pathway including IL-8. In addition to the synthetic ligand, infection of cells with the natural TLR3 stimulators influenza virus and herpes simplex virus also induced NEAT1 expression. This study therefore established that, unlike other lncRNAs identified so far, NEAT1 does not act directly at the target gene promoter but instead functions by binding and sequestering a transcriptional repressor protein, thereby releasing inhibition of target gene expression.
Lnc-DC is a newly identified lncRNA that regulates dendritic cell differentiation and T cell activation,45 which is a critical link between innate- and adaptive immune regulation.46 Distinct from other identified lncRNAs, lnc-DC functions through direct binding with STAT3, an important transcription factor that regulates many immune associated genes. Wang et al. provided evidence that lnc-DC interacts with STAT3 through its 3′-end segment, which prevents it from binding with phosphatases including SHP1. Thus, this interaction promotes the phosphorylated state of STAT3 and lead to increased STAT3 activity.
Finally, enhancers like RNAs have also been identified recently to regulate innate immune response.47 Collectively, these recent reports demonstrate that lncRNAs have an emerging role in regulating the innate immune responses by a variety of mechanisms. This is an emerging field and further work will undoubtedly continue to shed light on this exciting new function.
Concluding Remarks and Future Perspectives
The human immune system has evolved intricate and highly sophisticated mechanisms to guard against lethal infection from constantly mutating microbial pathogens. The studies described here suggest that lncRNAs may represent another component of the innate immune repertoire. The fact that lncRNAs do not show strong sequence conservation across species may represent a selection advantage for them to quickly adapt to new challenges by changing the nucleotide sequence and form new secondary structures to recruit protein factors. In the coming years, we will see many more examples of lncRNA-mediated regulation of host–pathogen interactions, and perhaps some unique functions beyond control of gene expression.
In the meantime, a number of questions in the lncRNA field need to be addressed. One is how the specificity of lncRNA–protein interactions is determined. RNA secondary structure has been suggested to be crucial for recognition by the RNA-binding protein partner, with the lncRNA serving as a scaffold for assembly of the complex. It will be important to gather more data on lncRNA secondary structures before we can understand how to manipulate them to achieve gene-specific regulation. However, the relatively large size of these molecules makes it difficult to predict their structures within cells. Studies are already underway to address this problem and novel methods are being developed to systematically identify variations in lncRNA structure.48 In addition to secondary structure, the primary RNA sequence may play a critical role in localizing lncRNA–protein complexes to the appropriate genomic loci. Indeed, many lncRNAs discovered to date show preferences for specific loci.15,17,28,30 It is likely that new methodology, such as RNase-protected RNA sequencing, will be needed to fully understand the structure-function relationship and molecular mechanisms underlying lncRNA-mediated regulation of gene expression.
Another challenge for lncRNA research is to determine their coding potential. Although computational analysis suggests otherwise, the possibility that lncRNAs could code for short peptides cannot yet be ruled out. Indeed, ribosome-associated lncRNAs have been reported, and the ribosomal profiling pattern was shown to be more similar to mRNA-coding regions than 3′UTRs, suggesting that lncRNAs may indeed undergo translation.49 However, others have argued that this finding could be attributed to abnormally low ribosomal occupancy by mRNAs due to the presence of stop codons.50 In fact, the 5′UTRs of mRNAs, which are not translated, have ribosomal profiling patterns similar to those of coding sequences. Thus, ribosome occupancy may not be a good indicator of the translation potential of lncRNAs. Traditional in vitro translation assays are typically too insensitive to identify short peptides, suggesting the need for a more robust and convenient method for evaluating the coding potential of any given lncRNA.
Despite the rapid increase in our understanding of lncRNA function, this field is still in its infancy and many questions and challenges remain. Undoubtedly, technological and methodological advances will continue to clarify how lncRNAs influence diverse biological processes and contribute to the overall physiological and pathophysiological behavior of cells. A better understanding of the relationship between RNA structure, protein recruitment, and gene regulation may soon allow us to design novel therapeutics that can manipulate target gene expression for the treatment of many human diseases, including cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to our colleague Dr. R. Zhou and members of the Rana laboratory for helpful discussions. We apologize to our colleagues whose work we could not cite owing to space limitations.
Funding
This work was supported in part by grants from the US National Institutes of Health (to T.M.R).
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010; 11:373-84; PMID:20404851; http://dx.doi.org/ 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 2.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007; 7:353-64; PMID:17457343; http://dx.doi.org/ 10.1038/nri2079 [DOI] [PubMed] [Google Scholar]
- 3.Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol 2011; 11:807-22; PMID:22094985 [DOI] [PubMed] [Google Scholar]
- 4.Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, Koga R, Takeda K. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity 2006; 24:41-51; PMID:16413922; http://dx.doi.org/ 10.1016/j.immuni.2005.11.004 [DOI] [PubMed] [Google Scholar]
- 5.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009; 457:854-8; PMID:19212405; http://dx.doi.org/ 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitmore MM, Iparraguirre A, Kubelka L, Weninger W, Hai T, Williams BR. Negative regulation of TLR-signaling pathways by activating transcription factor-3. J Immunol 2007; 179:3622-30; PMID:17785797; http://dx.doi.org/ 10.4049/jimmunol.179.6.3622 [DOI] [PubMed] [Google Scholar]
- 7.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science 2007; 317:675-8; PMID:17673665; http://dx.doi.org/ 10.1126/science.1142953 [DOI] [PubMed] [Google Scholar]
- 8.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 2009; 458:1185-90; PMID:19322177; http://dx.doi.org/ 10.1038/nature07924 [DOI] [PubMed] [Google Scholar]
- 9.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol 2012; 33:449-58; PMID:22721918; http://dx.doi.org/ 10.1016/j.it.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 10.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 2010; 10:111-22; PMID:20098459; http://dx.doi.org/ 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- 11.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006; 103:12481-6; PMID:16885212; http://dx.doi.org/ 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol 2010; 185:6226-33; PMID:20937844; http://dx.doi.org/ 10.4049/jimmunol.1000491 [DOI] [PubMed] [Google Scholar]
- 13.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al.; FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group). The transcriptional landscape of the mammalian genome. Science 2005; 309:1559-63; PMID:16141072; http://dx.doi.org/ 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750-6; PMID:18974356; http://dx.doi.org/ 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409-19; PMID:20673990; http://dx.doi.org/ 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477:295-300; PMID:21874018; http://dx.doi.org/ 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014; 53:1005-19; PMID:24530304; http://dx.doi.org/ 10.1016/j.molcel.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013; 493:231-5; PMID:23201690; http://dx.doi.org/ 10.1038/nature11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 2011; 25:2573-8; PMID:22155924; http://dx.doi.org/ 10.1101/gad.178780.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, Zhang X, Lu X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J 2013; 32:2833-47; PMID:24097061; http://dx.doi.org/ 10.1038/emboj.2013.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 2014; 21:198-206; PMID:24463464; http://dx.doi.org/ 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120-4; PMID:21423168; http://dx.doi.org/ 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, et al. Mouse transcriptome: neutral evolution of 'non-coding' complementary DNAs. Nature 2004; 431; http://dx.doi.org/ 10.1038/nature03016 [DOI] [PubMed] [Google Scholar]
- 24.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223-7; PMID:19182780; http://dx.doi.org/ 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448:553-60; PMID:17603471; http://dx.doi.org/ 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011; 147:1537-50; PMID:22196729; http://dx.doi.org/ 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006; 6:846-56; PMID:17060944; http://dx.doi.org/ 10.1038/nrc1991 [DOI] [PubMed] [Google Scholar]
- 28.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; http://dx.doi.org/ 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife (Cambridge) 2014; 3:e02046; PMID:24521543; http://dx.doi.org/ 10.7554/eLife.02046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 2014; 111:1002-7; PMID:24371310; http://dx.doi.org/ 10.1073/pnas.1313768111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013; 341:789-92; PMID:23907535; http://dx.doi.org/ 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010; 143:46-58; PMID:20887892; http://dx.doi.org/ 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 2013; 152:743-54; PMID:23415224; http://dx.doi.org/ 10.1016/j.cell.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 2014; 53:393-406; PMID:24507715; http://dx.doi.org/ 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 35.Bureau JF, Montagutelli X, Bihl F, Lefebvre S, Guénet JL, Brahic M. Mapping loci influencing the persistence of Theiler's virus in the murine central nervous system. Nat Genet 1993; 5:87-91; PMID:8220433; http://dx.doi.org/ 10.1038/ng0993-87 [DOI] [PubMed] [Google Scholar]
- 36.Bihl F, Brahic M, Bureau JF. Two loci, Tmevp2 and Tmevp3, located on the telomeric region of chromosome 10, control the persistence of Theiler's virus in the central nervous system of mice. Genetics 1999; 152:385-92; PMID:10224268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol 2003; 77:5632-8; PMID:12719555; http://dx.doi.org/ 10.1128/JVI.77.10.5632-5638.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res 2013; 41:575-85; PMID:23161687; http://dx.doi.org/ 10.1093/nar/gks1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature 2009; 457:915-9; PMID:19098893; http://dx.doi.org/ 10.1038/nature07598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell 2012; 45:459-69; PMID:22264826; http://dx.doi.org/ 10.1016/j.molcel.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010; 11:761-72; PMID:20940737; http://dx.doi.org/ 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giraud M, Jmari N, Du L, Carallis F, Nieland TJ, Perez-Campo FM, Bensaude O, Root DE, Hacohen N, Mathis D, et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc Natl Acad Sci U S A 2014; 111:1491-6; PMID:24434558; http://dx.doi.org/ 10.1073/pnas.1323535111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 2009; 35:467-78; PMID:19716791; http://dx.doi.org/ 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 2009; 186:637-44; PMID:19720872; http://dx.doi.org/ 10.1083/jcb.200906113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344:310-3; PMID:24744378; http://dx.doi.org/ 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]
- 46.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol 2000; 18:767-811; PMID:10837075; http://dx.doi.org/ 10.1146/annurev.immunol.18.1.767 [DOI] [PubMed] [Google Scholar]
- 47.IIott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 2014; 5:3979; PMID:24909122; http://dx.doi.org/ 10.1038/ncomms4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 2014; 505:706-9; PMID:24476892; http://dx.doi.org/ 10.1038/nature12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011; 147:789-802; PMID:22056041; http://dx.doi.org/ 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013; 154:240-51; PMID:23810193; http://dx.doi.org/ 10.1016/j.cell.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129:1311-23; PMID:17604720; http://dx.doi.org/ 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/ 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 2013; 51:349-59; PMID:23932716; http://dx.doi.org/ 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]