Abstract
The tRNA modification field has a rich literature covering biochemical analysis going back more than 40 years, but many of the corresponding genes were only identified in the last decade. In recent years, comparative genomic-driven analysis has allowed for the identification of the genes and subsequent characterization of the enzymes responsible for N6-threonylcarbamoyladenosine (t6A). This universal modification, located in the anticodon stem-loop at position 37 adjacent to the anticodon of tRNAs, is found in nearly all tRNAs that decode ANN codons. The t6A biosynthesis enzymes and synthesis pathways have now been identified, revealing both a core set of enzymes and kingdom-specific variations. This review focuses on the elucidation of the pathway, diversity of the synthesis genes, and proposes a new nomenclature for t6A synthesis enzymes.
Keywords: comparative genomics, modified nucleosides, translation, tRNA, t6A, universal proteins
Introduction
tRNAs (tRNAs) are the central adaptors in the translation process responsible for decoding mRNAs. tRNAs harbor numerous post-transcriptional modifications that fine-tune their function. To date, more than 90 modifications1 have been identified in tRNA, and most organisms devote more genetic information to modifying tRNAs than to the tRNAs themselves.2 While modifications have been shown to affect different aspects of tRNA metabolism and shape interactions of tRNA molecules with the rest of the translation apparatus,3-5 most modifications to the anticodon-stem-loop (ASL) are required for accurate decoding.6,7 The diversity of tRNA modifications and how modifications affect function has been the topic of recent reviews,8-10 and for a summary of modifications to the ASL in Escherichia coli see the recent review by Helm and Alfonzo11 and Table 4 in de Crécy-Lagard, et al.10 In this review, we address the enzymes responsible for the formation of N6-threonyl-carbamoyl-adenosine (t6A) and its derivatives. This complex modification of adenosine is located at position 37, next to the anticodon (t6A37), and is one of the few universal modifications of the ASL.12
The hypermodified base t6A is present in nearly all ANN decoding tRNAs and has been studied in vitro and in vivo for more than 40 y.13-19 Since the first discovery of the modification by Schweizer, et al. in 1969,18 sporadic studies established the basic requirements for the synthesis of this universal modification, identifying the requirement for ATP, threonine and carbonate,15,17,20-22 but fell short of elucidating the multi-step path to its formation. Subsequent studies in which native E. coli tRNAfMet (harboring an unmodified A37) and yeast tRNAiMet transcripts were converted to t6A37 after microinjection into Xenopus laevis oocytes demonstrated that the formation of t6A occurred in the oocyte cytoplasm and used a conserved machinery.13,14 These studies also demonstrated that A37 and U36 were strict determinants for t6A formation, and that A38 enhances the efficiency of modification of A37 to t6A37.13 Finally, structural studies showed that t6A enhances anticodon-codon base-pairing by cross-strand base-stacking of the t6A base with the first position of the codon,23 and influences the structure of the ASL by preventing across the loop base-pairing between U33-A37, as well as stacking of bases A37 and A38.23-27
Only in the last 5 y have the t6A biosynthesis enzymes and the pathways been elucidated, revealing both a core set of enzymes and kingdom-specific variations (Fig. 1).28-33 Elucidation of this multi-step pathway, which requires the formation of an activated carbon dioxide intermediate, and the diversity of the enzymes required for synthesis are the focus of this review.
Figure 1.
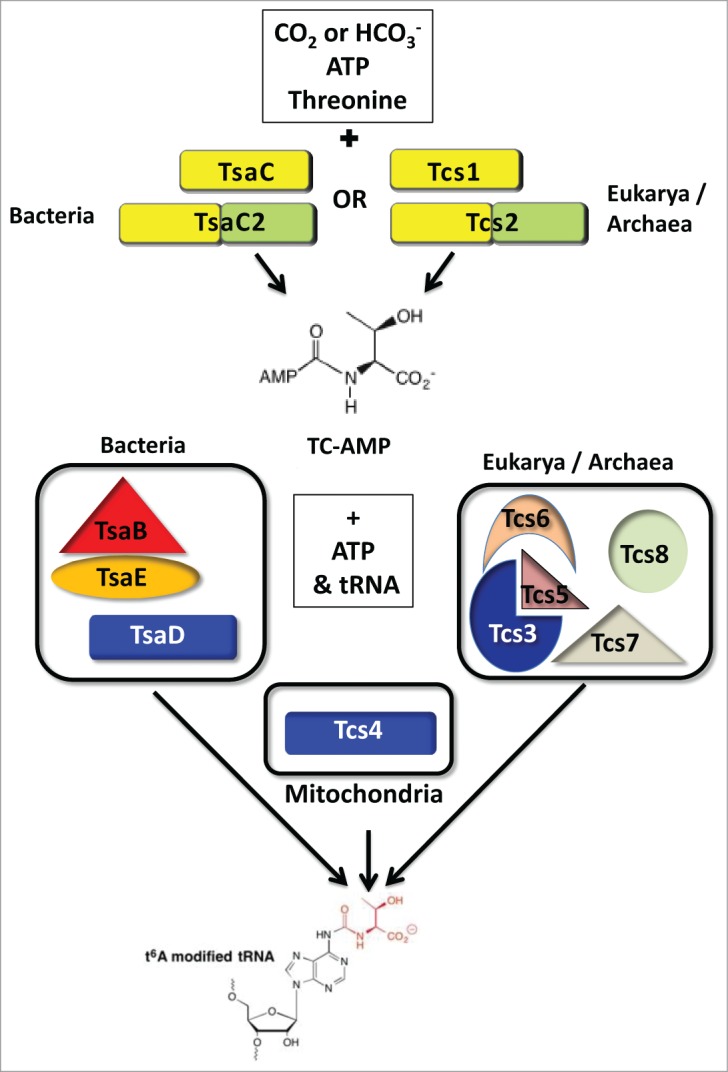
Diversity in the synthesis of the universal tRNA modification t6A. Two types of enzymes families, TsaC (YrdC) and TsaC2 (Sua5) in Bacteria and Tcs1 (YrdC) and Tcs2 (Sua5) in Eukarya and Archaea, catalyze the formation of TC-AMP. TsaC2 and Tcs2 contain a TsaC-domain plus an additional C-terminal Sua5-domain. To transfer threonylcarbamoyl (TC) to tRNA, Bacteria require TsaBDE, while Archaea and Eukarya use the KEOPS complex composed of Tcs3 (Kae1), Tcs5 (Bud32), Tcs6 (Pcc1) and Tcs7 (Cgi121) proteins. Tcs8 (Gon7) is found exclusively in Fungi. Mitochondria use the nuclear encoded Tcs4 (Qri7) for transfer of the TC to tRNA. Colors represent homology and correspond with Figure 2.
Discovery of the First t6A Synthesis Genes
The first enzyme of the t6A pathway was discovered in 2009 when it was found that a universal protein family, YrdC/Sua5 (COG0009), was involved in t6A modification.31 Based on the assumption that because t6A was universally conserved the t6A biosynthetic enzymes would also be universally conserved, this work used comparative genomic analysis to focus on universally conserved protein families of unknown function. At the time of this study, 9 universally conserved protein families were of unknown function. Of these, the YrdC/Sua5 family was judged the most likely candidate for involvement in t6A biosynthesis due to: 1) its similarity to HypF, which catalyzes a carbamoylation reaction34 similar to a putative step in t6A biosynthesis proposed in 1974 by both Elkins and Keller15 and Körner and Söll;17 2) mutations in the yeast yrdC ortholog SUA5 led to translation defects (initiation at non-AUG codons);35 and 3) E. coli YrdC was found to bind RNA and tRNA.36 The involvement of the YrdC/Sua5 family in t6A synthesis was experimentally validated using E. coli and Saccharomyces cerevisiae.31
In E. coli, yrdC is essential, but SUA5 can be deleted from S. cerevisiae, although the growth of the mutant is severely compromised. tRNAs analyzed from this mutant were devoid of t6A. The levels of t6A could be restored through complementation with SUA5Sc, yrdCEc, ywlCBs (Bacillus subtilis SUA5 homolog), and yrdCMm (Methanococcus maripaludis, an archaeal yrdC homolog). The essentiality phenotype of a E. coli yrdC deletion could be complemented by expressing orthologs from yeast, B. subtilis, and M. maripaludis in trans.31 Analysis of tRNAs in the complemented strains confirmed the presence of t6A.31 This work identified the first gene family involved in t6A synthesis, established that its function is universally conserved, and that members of the family could bind ATP, but because purified YrdC alone was not sufficient to produce t6A in tRNA transcripts in vitro, it also suggested that additional enzymes were needed for t6A synthesis, or that the role of YrdC/Sua5 family was indirect.
A second protein family involved with t6A synthesis, YgjD/Kae1/Qri7 (COG0533), was discovered in 2011.30 Like YrdC/Sua5, the YgjD/Kae1/Qri7 family of proteins is universally conserved, and also exhibited similarity to HypF, which harbors a fusion of YrdC-like and YgjD-like domains. Kae1 had first been described as a member of the KEOPS complex (Kinase, putative Endopeptidase and Other Proteins of Small size)37 also known as EKC (Endopeptidase-like Kinase Chromatin-associated complex),38 and had been proposed to be involved in a variety of phenomena unrelated to RNA modification.37,38 A phylogeny of this family revealed that yeast harbored 2 members of the family.39 The first, Kae1 had homologs in other eukaryotes and archaea, and the second, Qri7 was targeted to the mitochondria and was part of the bacterial YgjD clade.39
The hypothesis that the YgjD/Kae1/Qri7 family was involved in t6A synthesis was confirmed by extracting tRNAs from S. cerevisiae kae1Δ and showing they were devoid of t6A and that t6A levels could be restored by complementation with either ygjDEc or a version of QRI7Sc designed to remain in the cytoplasm. These results also indicated that members of the YgjD/Kae1/Qri7 family were isofunctional for t6A synthesis, at least in yeast. To test if YgjD/Kae1/Qri7 were isofunctional in E. coli, a PTET::ygjD strain was constructed (ygjD is only expressed when anhydrotetracycline, aTc, is added). Only the expression of the ygjD gene from E. coli allowed complementation of the essentiality phenotype in the absence of aTc. In contrast to the YrdC/Sua5 complementation results, expression of the KAE1Sc and QRI7Sc genes from yeast, the PRPKMm from Methanococcus maripaludis (PRPK is a fusion of Kae1-Bud32 in Archaea) or the B. subtilis ygjDBs did not complement the essentiality phenotype of the absence of ygjD.
While the protein families TsaC/Sua5 and Kae1/Qri7/TsaD were found to be strictly required for the biosynthesis of t6A, and a homolog of at least one member of each family is found in all domains of life (Fig. 2),30,31 YrdC and YgjD failed to produce t6A in vitro with transcript or t6A-deficient tRNA purified from yeast sua5Δ,30 suggesting that the biosynthetic machinery for t6A biosynthesis required more than these 2 proteins. Over the last 2 years, a flurry of papers have reported the identification of these missing proteins, and elucidated the complete bacterial,28,33 eukaryotic/archaeal,29,40-42 and mitochondrial32,43,44 biosynthetic pathways to t6A.
Figure 2.

Distribution of genes for biosynthesis of t6A and derivatives. Representative organisms from each domain of life were used to build a taxonomic tree in iToL (http://itol.embl.de).73,74 Filled circles indicate presence of genes. Genes for formation of ct6A are collapsed into a single column.
Synthesis of t6A Varies With Domains of Life
The observation that the YrdC/Sua5 family members were functionally interchangeable between domains31 while YgjD/Kae1/Qri7 were not30 lead to a model in which t6A biosynthesis occurred in 2 steps with kingdom, species, or organelles specific partners for the second step.
Bacteria
The identity of the remaining enzymes in bacterial t6A synthesis was predicted from 3 pieces of evidence. First, YgjD was shown to form an association network with YeaZ (a paralog of YgjD) and YjeE, based on physical interaction between the proteins and physical clustering of the genes;30,45 second, like YrdC and YgjD, YeaZ and YjeE were essential in E. coli;46 and third, complementation of the E. coli yjgD- essentiality phenotype required expression of both B. subtilis ygjD and yeaZ genes, suggesting that a YgjD/YeaZ interaction was necessary for t6A synthesis.30 Notably, it's been shown that only YeaZ-YgjD pairs from closely related organisms form complexes in vitro.47 The final evidence that YeaZ and YjeE were the missing proteins in t6A bacterial synthesis was provided by in vitro reconstitution experiments,28 which demonstrated that recombinant YrdC, YgjD, YeaZ, and YjeE proteins from E. coli28 were collectively both necessary and sufficient to generate t6A in reactions with threonine, bicarbonate, ATP, and either E. coli tRNAThr or tRNALys transcripts, or unfractionated tRNA from yeast sua5Δ. Notably, t6A formation was not observed when a tRNA transcript corresponding to tRNAGln from Methanothermaobacter thermautotrophicus, which does not naturally contain t6A, or a 17-mer corresponding to an unmodified ASL of E. coli tRNALys were used as a substrates.28 While the former was consistent with the natural lack of t6A in this tRNA, the latter was surprising since this ASL had previously been show to bind specifically to E. coli YrdC.28 The t6A synthesis pathway was subsequently reconstituted using the B. subtilis enzymes YwlC (an ortholog of yeast Sua5), and YdiBCE (orthologs of E. coli YjeE, YeaZ, and YgjD, respectively),33 demonstrating the universality of these enzymes in bacteria. With the newly established enzymatic role for YeaZ, YrdC, YgjD, and YjeE (and their orthologs) in the biosynthesis of threonylcarbamoyl-6-adenosine (t6A), these enzymes were renamed TsaB, TsaC, TsaD, and TsaE, respectively (Fig. 1).28
Archaea and Eukarya
The TsaE and TsaB proteins in Bacteria have no homologs in Eukarya or Archaea. The identification of the additional components of t6A biosynthesis in these last 2 kingdoms came from the fact that Kae1 was part of the KEOPS/EKC complex. The other subunits of the KEOPS/EKC complex (Bud32, Cgi121, Pcc1, plus the fungal specific Gon7) were tested for a potential role in t6A synthesis, first genetically then in vitro. Mutation of PCC1 and BUD32, but not in CGI121, in yeast eliminated t6A on tRNAIle reportedly by a primer extension assay.41 However, the primer extension method reported by the Sternglanz laboratory has not been repeated by others. (de Crécy-Lagard laboratory, Goldberg laboratory, and Glavic laboratory, personal communication). Indeed, as recently shown by the Alfonzo laboratory, reverse-transcriptase bypassed t6A to stop downstream at m3C32.48 Analysis of bulk tRNA from a pcc1–4 allele by LC-MS/MS found t6A was reduced 30%.42 HPLC and LC-MS/MS analysis of bulk tRNAs purified from whole gene deletions in S. cerevisiae found that the bud32Δ and gon7Δ strains were devoid of t6A, while t6A levels in the pcc1Δ and cgi121Δ strains were reduced 30% and 60%, respectively, versus wild-type (Thiaville and de Crécy-Lagard, unpublished data). In the halophilic Archaea Haloferax volcanii, the kae1‑bud32 (gene fusion) and cgi121 are essential, precluding a direct genetic test of their role in t6A biosynthesis, and deletion of pcc1 had only a small decrease (∼16%) in total t6A content.40,49
Confirmation that the KEOPS/EKC complex was responsible for t6A formation came with in vitro reconstitution experiments. It was shown that both the KEOPS/EKC complex from Pyrococcus abyssi (Kae1, Bud32, Pcc1, and Cgi121), reconstituted from the individual genes expressed in E. coli, as well as the S. cerevisiae KEOPS complex (Kae1, Bud32, Pcc1, Cgi121, and Gon7), genes expressed in E. coli as a synthetic operon,50 can form t6A in vitro, when combined with Sua5 from yeast or Archaea.29
Mitochondria
Yeast mitochondrial tRNAs contain t6A,51 and while none the subunits of the KEOPS/EKC complex or Sua5 have paralogs targeted to the mitochondria,52,53 the Kae1 homolog Qri7 was found to be targeted to the mitochondria in yeast,39,53 Caenorhabditis elegans,39 human,54 rat,54 and Arabidopsis thaliana.54 It was subsequently demonstrated that the nuclear encoded Sua5 can localize to both the cytoplasm and to the mitochondria in yeast through the use of alternative translation initiation at 2, in-frame AUG sites.44 Translation from the first AUG encoded a mitochondrial signal peptide, and Sua5 was localized to the mitochondria. Sua5 translated from the second AUG remained in the cytoplasm.44 Co-expression of both Sua5 and Qri7 in E. coli complemented the TsaD essentiality when the expression of Qri7 alone did not,44 suggesting that Qri7 could substitute for the KEOPS complex or the TsaBDE proteins. In addition, expression of QRI7 in the cytoplasm of a bud32Δ yeast strain restored growth defects.32 This was confirmed when it was demonstrated that a minimal system comprised of only Sua5 and Qri7 is sufficient to synthesize t6A in vitro.32,44
Thus, the enzyme families TsaC/Sua5 and TsaD/Kae1/Qri7 are shared in all organisms, and Bacteria additionally require TsaBE, while Archaea and Eukarya use the other components of the KEOPS complex. Interestingly, although TsaBDE, KEOPS, and Qri7 are functional analogs, only the TsaD/Kae1/Qri7 protein is shared among the 3 systems (Fig. 2), suggesting that this protein family along with the TsaC/Sua5 family were part of the ancestral t6A synthesis core present in the last universal common ancestor (LUCA).39
Mechanistic Analysis of the t6A Synthesis Machineries
Experiments probing the role of ATP in bacterial t6A formation demonstrated that both AMP and ADP were products, and that ATP consumption could be uncoupled from RNA modification, with TsaC being the source of AMP (in a threonine dependent process) and TsaD/TsaB/TsaE together producing ADP.28 These observations were consistent with earlier mechanistic hypotheses30 in which the ATP requirement in t6A biosynthesis was rationalized on the presumed need for 2 activated acyl intermediates during the course of t6A formation (i.e. acyl-phosphate and/or acyl-adenylate), the first a phosphocarboxy species (e.g. carboxyphosphate or carboxyadenylate; Fig. 3, intermediate I) activated for transfer to the nitrogen of either threonine or adenosine-37, and the second an N-carboxyphospho species activated for transfer to the remaining component (threonine or adenosine-37;Fig. 3, intermediate III). However, closer scrutiny of the TsaC reaction in the B. subtilis system revealed that the product was threonylcarbamoyl-adenylate (TC-AMP, Fig. 1),33 an intermediate already activated for condensation with adenosine-37 of tRNA, thus obviating ADP formation as part of the activation steps proposed to be necessary in the biosynthesis of t6A. Furthermore, AMP formation by TsaC was shown to arise exclusively from hydrolysis of TC-AMP,33 and that PPi was the other product of the TsaC reaction, implying that formation of TC-AMP itself proceeds through an unusual direct carboxylation of threonine by CO2 or HCO3- (Fig. 4).
Figure 3.
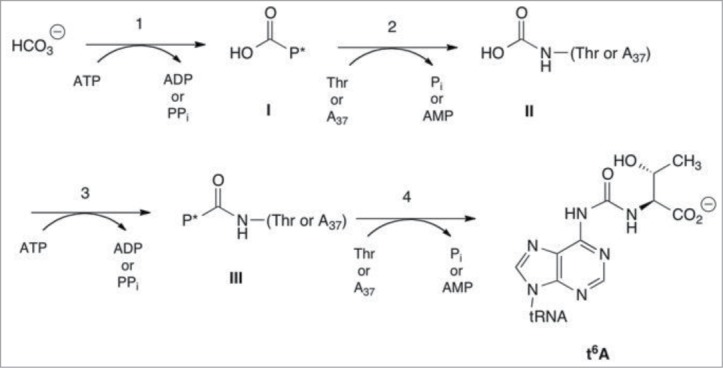
Early mechanistic proposal for the formation of t6A. P* refers to an activated acylphosphate species, either a simple acyl monophosphate or an acyl AMP.
Figure 4.
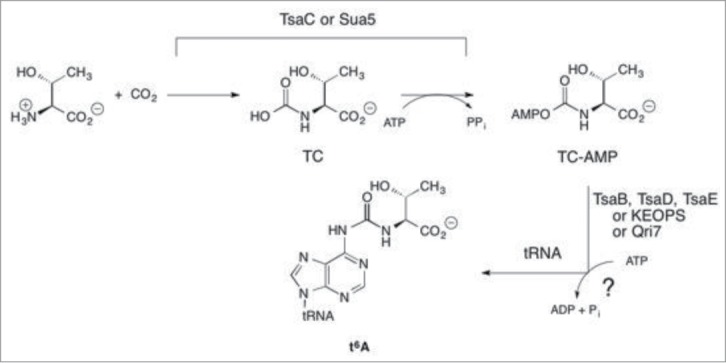
Stepwise formation of t6A illustrating the intermediates in the pathway.
The role of ADP production in t6A biosynthesis remains cryptic; in the bacterial system, t6A can be generated without formation of ADP by supplying purified TC-AMP to a reaction containing TsaD/TsaB/TsaE in the absence of ATP,33 while in Archaea and Eukarya formation of t6A appears to require the reaction of ATP to ADP,55 although it does not appear to serve a direct role in the reaction. In Archaea, Pcc1, Kae1, and Bud32 are minimally required to produce t6A in vitro,55 with Kae1 comprising the catalytic subunit responsible for condensing TC-AMP with tRNA. Bud32 was shown to be an ATPase in the presence of Kae1, while it autophosphorylates when it is in complex with Cgi121. Thus, Kae1 apparently modifies the phosphotransferase activity of Bud32 and switches it from a kinase to an ATPase.55 It's unclear what the specific role of this ATPase activity is, as neither Bud32 or Cgi121 participate directly in the t6A reaction. Cgi1321 appears to regulate activity by acting as an effector, where it's binding significantly changes the conformation of Bud32.55
Overall, the chemistry of t6A formation bears similarities to the TobZ system,56 an O-carbamoyltransferase comprised of a TsaC-like domain fused to a Kae1-like domain that carries out the carbamoylation of tobramycin to form nebramycin-5′. In the TobZ reaction, the TsaC domain catalyzes the conversion of carbamoylphosphate to carbamoyladenylate, while the Kae1 domain condenses the latter with tobramycin. Likewise, in t6A formation the TsaC/Sua5 homologs generate an adenylated intermediate, which then is condensed with tRNA by the TsaD/Kae1/Qri7 proteins. A notable difference in the systems is that in TobZ the initial substrate is carbamoyl phosphate, which undergoes a phosphotransfer reaction to generate carbamoyl adenylate. This interchange of phosphoryl moieties is chemically not necessary for the subsequent condensation of the carbamoyl group with tobramycin, as both species are activated for the coupling reaction. In t6A biosynthesis, the initial substrates for TsaC/Sua5 are threonine and CO2/HCO3-, which react first to form N-carboxythreonine followed by reaction with ATP to form TC-AMP.
Structural Organization of t6A Biosynthetic Proteins
The highly reactive nature of TC-AMP is not compatible with a freely diffusible intermediate in the biosynthesis of t6A, and argues for the evolution of systems in which this intermediate is instead channeled, as in TobZ,56 from its site of production in TsaC/Sua5 to a second active-site (presumably in TsaD/Kae1/Qri7) where it undergoes reaction with tRNA. A number of observations are consistent with this proposal. First is the fact that the KEOPS complex from both Eukarya and Archaea is known to be a stable quaternary complex.29 Second, while the bacterial proteins do not form an isolable complex analogous to KEOPS, they do interact with one another as demonstrated by the analysis of the E. coli proteins in pull-down experiments, which demonstrated binding of TsaC to both TsaB and TsaD,28 and binding of TsaB to both TsaE and TsaD.28,45 Furthermore, the ability of B. subtilis YdiD (TsaD homolog) to complement the ΔtsaD essentiality phenotype in E. coli was dependent on co-expression of YdiB (TsaB homolog),30 while the ability of Qri7 to complement this phenotype was dependent on co-expression of Sua5.44 These observations are consistent with the requirement for specific physical interactions between these proteins necessary for function.
Additionally, crystallographic analysis has shown that protein-protein associations are conserved across the systems. For example, the crystal structure of Qri7 shows that dimerization is required for Qri7 function, and the dimerization surfaces for Qri7 are used by the archaeal/eukaryotic Kae1 binding to Pcc1 and the bacterial TsaD binding to TsaB.32 Interestingly, although the Pcc1 subunit of KEOPS/EKC shares no sequence similarity to Qri7 or TsaD, Pcc1 engages Kae1 in a manner surprisingly similar to dimerization of Qri7 and TsaD-TsaB.32
Thus, in all 3 systems the ability of the constituent proteins to physically interact with one another appears to be a requirement for t6A biosynthesis.
Naming Convention
The literature is polluted with a variety of names for each t6A synthesis protein and even for the complexes. With the defined enzymatic and biological function now established it is appropriate to unify the t6A nomenclature. For all Bacteria, we recommend the following suggestions, in agreement with Ken Rudd (Curator of EcoGene, U. of Miami) and published in Deutsch, et al., of TsaB, TsaC, TsaD and TsaE, to replace YeaZ, YrdC, YgjD, and YjeE, respectively. Additionally, Sua5 in bacteria should be renamed TsaC2. TsaC2 is defined as a protein containing both a TsaC and the additional C-terminal Sua5 domain. For Eukarya and Archaea, the use of Tcs (threonyl-carbamoyl synthesis) is recommended (in yeast, TSA1 and TSA2 are in use in yeast for thioredoxin). We recommend the following nomenclature: Tcs1 (YrdC), Tcs2 (Sua5), Tcs3 (Kae1), Tcs4 (Qri7), Tcs5 (Bud32), Tcs6 (Pcc1), Tcs7 (Cgi121), and Tcs8 (Gon7). A summary of the new and old names, as well as recommended functional descriptions can be found inTable 1. Additionally, we recommend naming the bacterial TsaBDE complex as well as the archeal/eukaryotic KEOPS/EKC complex to Threonyl-carbamoly Transferase Complex (TCTC), which will be in keeping with nomenclature of other members of the carbamoyl transferase family. The TCTC family can be further subdivided into bacterial (bTCTC), archaeal (aTCTC), and eukaryal (eTCTC).
Table 1.
Proposed names and functional roles for t6A synthesis genes.
| New Name | Old Names | Function |
|---|---|---|
| Bacteria | ||
| TsaB | YeaZ / YdiC | tRNA adenosine(37) threonylcarbamoyltransferase complex, dimerization subunit type 1 |
| TsaC | YrdC | L-threonylcarbamoyladenylate synthase (EC 2.7.7.87) type 1 |
| TsaC2 | Sua5 / YwlC | L-threonylcarbamoyladenylate synthase (EC 2.7.7.87) type 2 |
| TsaD | YgjD / YdiE | tRNA adenosine(37) threonylcarbamoyltransferase complex, transferase subunit |
| TsaE | YgjE / YdiB | tRNA adenosine(37) threonylcarbamoyltransferase complex, ATPase subunit type 1 |
| Archaea / Eukaryotes | ||
| Tcs1 | YrdC | L-threonylcarbamoyladenylate synthase (EC 2.7.7.87) type 1 |
| Tcs2 | Sua5 | L-threonylcarbamoyladenylate synthase (EC 2.7.7.87) type 2 |
| Tcs3 | Kae1 / gcp / OSGEP | tRNA adenosine(37) threonylcarbamoyltransferase complex, transferase subunit |
| Tcs4 | Qri7 / OSGEPL1 | tRNA adenosine(37) threonylcarbamoyltransferase, mitochondrial |
| Tcs5 | Bud32 | tRNA adenosine(37) threonylcarbamoyltransferase complex, ATPase subunit type 2 |
| Tcs6 | Pcc1 | tRNA adenosine(37) threonylcarbamoyltransferase complex, dimerization subunit type 2 |
| Tcs7 | Cgi121 | tRNA adenosine(37) threonylcarbamoyltransferase complex, regulator subunit |
| Tcs8 | Gon7 | tRNA adenosine(37) threonylcarbamoyltransferase complex, fungal specific subunit |
Distribution of the t6A Synthesis Genes Vary in Different Organisms
Annotation for the first enzyme of t6A synthesis, in bacteria, is complicated by the fact that 2 forms are found (the TsaC or TsaC2) and that 50% of the genomes analyzed harbor a TsaC paralog, YciO, that does not have the same function and does not contain the conserved KRSN tetrad.31,57 We reannotated all members of the COG0009 family (in 9176 bacterial genomes and all contained a TsaC or a TsaC2: 6745 contain TsaC (73%), 2846 contain TsaC2 (31%), and 859 (9%) contained both. In addition, 54% contained YciO (Fig. 2, and http://tinyurl.com/t6A-bacteria). To date, no clear pattern (phylogenetic or lifestyle) has emerged in terms of presence of TsaC or TsaC2, in any given genome and the functional differences between the 2 are not understood. Most bacteria contain both TsaB and TsaE; however, TsaE can be lost in symbiotic or intracellular bacteria, such as Wolbachia or Mycoplasmas (e.g., Mycoplasma genitalium and Mycoplasma pneumoniae). To date only 2 bacteria, Mycoplasma haemofelis and Mycoplasma suis strain Illinois, are missing both TsaB and TsaE58 (Fig. 2 and Table 2). It seems these organisms harbor a mitochondrial like minimal t6A synthesis system (unless another unidentified protein has been recruited).
Table 2.
Homologs of t6A biosynthetic genes in Bacteria.
| Organism | TsaC | TsaC2 (Sua5) | TsaB | TsaD | TsaE |
|---|---|---|---|---|---|
| E. coli K12 | b3282 | b1807 | b3064 | b4168 | |
| Vibrio cholerae O1 El Tor | VC0054 | VC1079 | VC1989 | VC0521 | VC0343 |
| Caulobacter crescentus NA1000 | CCNA_03501 | CCNA_00057 | CCNA_00069 | CCNA_03648 | |
| Mycoplasm gentialium G37 | MG259* | MG208 | MG046 | N.P. | |
| Mycoplasm pulmonis | MYPU_6130# | MYPU_1190 | MYPU_1180 | MYPU_1200 | |
| Bacillus subtilis subsp. subtilis str. 168 | BSU36950 | BSU05920 | BSU05940 | BSU05910 | |
| Haemophilus influenzae Rd | HI0656 | HI0388 | HI0530 | HI0065 | |
| Acinetobacter baylyi APD1 | ACIAD0208 | ACIAD0677 | ACIAD1332 | ACIAD2376 | |
| Salmonella Typhii TY2 | STY4395 | STY1950 | STY3387 | STY4714 | |
| Francisella novisida U112 | FTN_0158 | FTN_1148 | FTN_1565 | FTN_0274 | |
| Pseudomonas aeruginosa PA01 | PA0022 | PA3685 | PA0580 | PA4948 | |
| Burkholderia thailandensis E264 | BTH_I0669 | BTH_I2001 | BTH_II0616 | BTH_I0723 | |
| Stapholcoccus aureus subsp aureus MW2 | MW0860 | MW2040 | MW1975 | MW1973 | MW1976 |
M. gentialium MG259 is a TsaC/HemK fusion.
M. pulmonis TsaC (MYPU_6130) and HemK (MYPU_1060).
N.P.: Not Present.
Like Bacteria, all Eukarya and Archaea contain either a homolog of Tcs1 (TsaC/YrdC) or Tcs2 (TsaC2/Sua5). We have found one organism that has both, the fungi Pseudocercospora fijiensis CIRAD86, also known as Mycosphaerella fijiensis CIRAD86 (NCBI Taxonomic ID: 383855). As with bacteria, there is not a clear phylogenetic inheritance between organisms with Tcs1 or Tcs2 in Archaea or Eukarya, but a taxonomic relationship does exists in eukaryotes. Fungi exclusively contain Tcs2 (with P. fijiensis as an exception), while all Plants (including Chlamydomonas reinhardtii) and all Metazoans exclusively contain Tcs1. Of the 53 Archaea analyzed, 25 contain Tcs1 and 28 contain Tcs2. The only taxonomic relationship found is in the order Halobacteriales that exclusively contain Tcs1, Figure 2 and Table 3 (http://tinyurl.com/t6A-Arc-Euk).
Table 3.
Homologs of t6A biosynthetic genes in Archaea and Eukarya.
| Organism | Tcs1 YrdC) | Tcs2 (Sua5) | Tcs3 (Kae1) | Tcs4 (Qri7) | Tcs5 (Bud32) | Tcs6 (Pcc1) | Tcs7 (Cgi121) | Tcs8 (Gon7) |
|---|---|---|---|---|---|---|---|---|
| Haloferax volcanii DS2 | HVO_0253 | HVO_1895+ | HVO_1895 + | HVO_0652 | HVO_0013 | |||
| Homo sapiens | 1p34.3 | 14q11.2 | 2q32.2 | 20q13.2 | Xq28 | 2p24.3-p24.1 | ||
| Drosophila melanogaster | CG10438 | CG4933 | CG14231 | CG10673 | CG42498 | N.P. | ||
| Plasmodium falciparum | PFL0175c | PF3D7_1030600 PF3D7_0408900.1‡ | N.P. | MAL7P1.26 | N.P. | PFE0580w | ||
| Saccharomyces cerevisiae S228C | YGL169w | YKR038c | YDL104c | YGR262c | YKR095w-A | YML036w | YJL184w | |
| Schizosaccharomyces pombe | SPCC895.03c | SPBC16D10.03 | SPCC1259.10 | SPAP27G11.07c | SPAC4H3.13 | SPCC24B10.12 | SPAC6B12.18 | |
| Arabidopsis thaliana | AT5G60590 | AT4G22720 | AT2G45270 | AT5G26110 | AT5G53045 | AT4G34412 |
H. volcanii Tcs3 and Tcs5 occur as a gene fusion (HVO_1895).
P. falciparum PF3D7_0408900.1 (Tcs3b) targets to the apicoplast and is similar to Tcs3.
N.P.: Not Present.
All Archaea contain a single Tcs3 (Kae1) homolog, while Eukarya also contain a single Tcs3 homolog and also have a Tcs4 (Qri7) homolog (evolutionarily related to the bacterial TsaD), which will function in the organelles. In all genomes analyzed, both Tcs3 and Tcs4 were found in the nuclear genome and not in the organelle. Specifically, the human nuclear genome contains Tcs3 (OSGEP) for cytoplasmic t6A synthesis and Tcs4 (OSGEPL1) was shown to target to the mitochondria.54 Note to the reader, the Oberto, et al. paper incorrectly referred to OSGEPL as the Tcs4 homolog (instead of OSGEPL1), the human mitochondrial targeting protein. As an example for plants, Arabidopsis thaliana contains nuclear encoded Tcs3 (AT4G22720) and Tcs4 (AT2G45270). Tcs4At contains a strong chloroplast targeting signal, but has only been detected in the mitochondria.54 The human pathogen Plasmodium falciparum (causative agent of malaria) presents an interesting case for t6A synthesis, as the mitochondria utilize fully modified cytoplasmic tRNAs for mitochondrial translation (requirement for t6A machinery is unknown), and P. falciparum contains an apicoplast originating from secondary endosymbiosis of an alage.59 P. falciparum contains 2 nuclear encoded homologs of Tcs3 (Table 3): a Tcs3 that is similar to the yeast Tcs3, and an apicoplast-targeting Tcs3b, that is similar to Tcs3,60 but is phylogenetically distant from all known Tcs3 and from the bacterial TsaD (Thiaville and de Crécy-Lagard, unpublished). Tcs3 interacts with both Tcs5 (Bud32) and Tcs7 (Cgi121), and Tcs3b interacts with multiple proteins associated with the apicoplast ribosome (Mallari and Goldberg, personal communication). Tcs2 has not been detected in the apicoplast, and it is currently unknown how the first step in t6A synthesis occurs. (Mallari and Goldberg, personal communication).
Tcs5 (Bud32) is found in all Eukarya and Archaea sequenced to date. In the 53 Archaea analyzed, Tcs5 and Tcs3 are adjacent ORFs in 13 genomes and are fused in 25 genomes, demonstrating a strong functional linkage between the proteins of these genes. Tcs6 (Pcc1) and Tcs7 (Cgi121) are found in nearly all Archaea and Eukarya. Notable exceptions are the absence of Tcs6 in P. falciparum and the absence of Tcs7 in Drosophila melanogaster. Tcs8 (Gon7) is a fungal specific protein. Tcs8 is required for t6A formation in yeast (Thiaville and de Crécy-Lagard, unpublished), but the function of Tcs8 is currently unknown.
Derivatives of t6A
Currently, there are 3 known derivatives of t6A: ct6A (cyclic t6A), m6t6A (N6-methyl-N6-threonycarbamoyladenosine), and ms2t6A (2-methylthio-N6-threonycarbamoyladenosine).1
A new twist in the t6A field was recently discovered with the identification of cyclic form of t6A (ct6A), a cyclized active ester of t6A with an oxazolone ring.61 Renumber starting here throughout the end of the manuscript. TcdA (previously CsdL in E. coli) catalyzes an ATP-dependent dehydration of t6A to ct6A; this reaction is performed by Tcd1 (YHR003c) and by Tcd2 (YKL027w) in yeast.61 The harsh treatment for preparing tRNAs for LC-MS/MS analysis had masked the presence of the true arrangement of t6A, and ct6A appears to help tRNALys decode the noncognate codons AGA and UAG.61 At least for E. coli, ct6A appears to occur on all t6A-modified tRNAs.61 Unlike TsaB, C, D, and E, TcdA is not essential for E. coli (minor growth defect), and is not universally conserved in bacteria (Fig. 2).61 Whether this represents the final functional form of t6A, or if this a species-specific solution for a particular problem has not been addressed. Additionally, Tcd1 and Tcd2 localize to the outer membrane, not to the surface of yeast mitochondria,62 and mutations in either render cells mitochondrial deficient.61 How yeast cytoplasmic tRNAs would be converted to ct6A is currently unknown. Also, neither ct6A nor homologs of TcdA have been found in Archaea, and ct6A does not occur in humans61 (Fig. 2).
The second known derivative of t6A, m6t6A, was initially thought to only occur in E. coli on the 2 tRNAThr(GGU) species that decode ACC and ACU.63 The limited distribution of m6t6A is confounded by the small number of organisms in which tRNAs have been sequenced.7 m6t6A is formed by TsaA (E. coli YaeB was recently identified as the gene responsible for TsaA activity, and renamed TrmO)64 by transferring a methyl group from S-adenosylmethionine (AdoMet) to tRNAThr(GGU) containing t6A.63 TrmO has a unique single-sheeted β-barrel structure and does not belong to any known classes of methyltransferases, representing a novel category of AdoMet-dependent methyltransferase (Class VIII). Interestingly, t6A is required for the formation of the m6 moiety at position 37 of tRNAThr(GGU), and in ΔtrmO, tRNAThr(GGU) A37 will be modified to ct6A, suggesting that t6A is a common precursor to both m6t6A and ct6A.64 m6t6A slightly improves translational efficiency at the codon ACY.63,64 TrmO is widely distributed throughout life and cross-kingdom functional analysis was performed to show the activity was conserved.64 (Fig. 2)
The third known derivative of t6A, ms2t6A, is found only on tRNALys(UUU) in a subset of organisms.1 Particularly, ms2t6A is found in B. subtilis, some Archaea, and in human, but not in E. coli. YqeV (MtaB) in B. subtilis and Cdkal1 in humans are responsible for the insertion of the sulfur moiety and methylation at position 2 of the adenosine containing t6A.65-66 MtaB has been shown to increase the accuracy of decoding lysine codons.66,67 Loss of the Cdkal1 homolog in mice is correlated to increase Type 2 diabetes.67,68 It is not clear if ct6A is the base to form ms2t6A, or like m6t6A, ms2t6A is formed from t6A. (Fig. 2).
Concluding Remarks
The biosynthesis of t6A is just one example of a “rediscovery” of tRNA modifications in the genomic era, which has allowed for the discovery of globally unknown genes for enzyme reactions that were discovered more than 40 y earlier. In Bacteria, the 4 genes involved in t6A biosynthesis, due to their prokaryotic-specific essentiality and because tsaB and tsaE are found only in bacteria, had been identified as potential antibacterial and inhibitor targets prior to the discovery of their role in t6A synthesis was even established.45,69-72 The unique Tcs3b found in P. falciparum also presents itself as an attractive anti-malarial target. For these proteins to be viable targets, it is critical to understand their distribution profile and potential range of action as well as the mechanisms underlying the essentiality phenotypes to predict resistance mechanisms. Clearly, one should use caution in designing drugs targeting TsaB and TsaE in Mycoplasmas spp. since the genes are absent. Caution would also be needed for drugs targeting TsaC, due to the possibility of cross reactivity in humans, although TsaC2 and TsaD may be viable options.
The discovery of the t6A pathways now allows us to address more systematically the causes of the pleiotropic phenotypes caused by the absence of t6A synthesis enzymes. Are these due to mistranslation of target proteins, to a role of t6A as a determinant for other components of the translation apparatus, or to a role of t6A or of t6A synthesis proteins in other processes than translation? Indeed, the recent discovery of a molecule similar to the t6A nucleoside in dauer signaling in nematodes73 opens an unforeseen role for t6A derivatives in biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Basma El Yacoubi and Jennifer Thiaville for insightful comments and editing the manuscript. We thank Jeremy Mallari, Dan Goldberg, Diego Rojas-Benítez, and Álvaro Glavic for sharing results ahead of publication.
Funding
This work was supported by the National Institutes of Health (grant number R01 GM70641 to V. de C.-L.). P.C.T. was funded in part by the Chateaubriand Fellowship from the French Embassy in the United States.
References
- 1. Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Björk GR. Biosynthesis and function of modified Nucleosides. In: Söll D, Rajbhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. ASM Press, Wahsington, DC; 1995; 165-205. [Google Scholar]
- 3. Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem 2002; 277:16391-5; PMID:11861649; http://dx.doi.org/ 10.1074/jbc.M200253200 [DOI] [PubMed] [Google Scholar]
- 4. Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 2001; 20:4863-73; PMID:11532950; http://dx.doi.org/ 10.1093/emboj/20.17.4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krüger MK, Pedersen S, Hagervall TG, Sørensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol 1998; 284:621-31; PMID:9826503; http://dx.doi.org/ 10.1006/jmbi.1998.2196 [DOI] [PubMed] [Google Scholar]
- 6. Agris PF, Vendeix FAP, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 2007; 366:1-13; PMID:17187822; http://dx.doi.org/ 10.1016/j.jmb.2006.11.046 [DOI] [PubMed] [Google Scholar]
- 7. Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009; 37:D159-62; PMID:18957446; http://dx.doi.org/ 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/ 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA 2013; 4:35-48; PMID:23139145; http://dx.doi.org/ 10.1002/wrna.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; http://dx.doi.org/ 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 11. Helm M, Alfonzo JD. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical legoland. Chem Biol 2014; 21:174-85; PMID:24315934; http://dx.doi.org/ 10.1016/j.chembiol.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides : a guided tour. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin: Landes Bioscience; 2009. 1-18. [Google Scholar]
- 13. Morin A, Auxilien S, Senger B, Tewari R, Grosjean H. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: an in vivo study with Xenopus laevis oocytes. RNA 1998; 4:24-37; PMID:9436905 [PMC free article] [PubMed] [Google Scholar]
- 14. Grosjean H, De Henau S, Doi T, Yamane A, Ohtsuka E, Ikehara M, Beauchemin N, Nicoghosian K, Cedergren R. The in vivo stability, maturation and aminoacylation of anticodon-substituted escherichia coli initiator methionine tRNAs. Eur J Biochem 1987; 166:325-32; PMID:3301339; http://dx.doi.org/ 10.1111/j.1432-1033.1987.tb13518.x [DOI] [PubMed] [Google Scholar]
- 15. Elkins BN, Keller EB. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 1974; 13:4622-8; PMID:4609459; http://dx.doi.org/ 10.1021/bi00719a024 [DOI] [PubMed] [Google Scholar]
- 16. Sibler AP, Dirheimer G, Martin RP. Yeast mitochondrial tRNAIle and tRNAMetm: nucleotide sequence and codon recognition patterns. Nucleic Acids Res 1985; 13:1341-5; PMID:3889839; http://dx.doi.org/ 10.1093/nar/13.4.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Körner A, Söll D. N-(purin-6-ylcarbamoyl)threonine: biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett 1974; 39:301-6; PMID:4604806; http://dx.doi.org/ 10.1016/0014-5793(74)80135-3 [DOI] [PubMed] [Google Scholar]
- 18. Schweizer MP, Chheda GB, Baczynskyj L, Hall RH. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. a new component of transfer ribonucleic acid. Biochemistry 1969; 8:3283-9; PMID:4897334; http://dx.doi.org/ 10.1021/bi00836a023 [DOI] [PubMed] [Google Scholar]
- 19. Ishikura H, Yamada Y, Murao K, Saneyoshi M, Nishimura S. The presence of N-; [9-(β-D-ribofuranosyl)purin-6-ylcarbamoyl]threonine in serine, methionine and lysine transfer RNA's from escherichia coli. Biochem Biophys Res Commun 1969; 37:990-5; PMID:4902784; http://dx.doi.org/ 10.1016/0006-291X(69)90229-0 [DOI] [PubMed] [Google Scholar]
- 20. Chheda GB, Hong CI, Piskorz CF, Harmon GA. Biosynthesis of N-(purin-6-ylcarbamoyl)-L-threonine riboside. incorporation of L-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem J 1972; 127:515-9; PMID:4561775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Powers DM, Peterkofsky A. Biosynthesis and specific labeling of N-(purin-6-ylcarbamoyl)threonine of escherichia coli transfer RNA. Biochem Biophys Res Commun 1972; 46:831-8; PMID:4550699; http://dx.doi.org/ 10.1016/S0006-291X(72)80216-X [DOI] [PubMed] [Google Scholar]
- 22. Powers DM, Peterkofsky A. The presence of N-(purin-6-ylcarbamoyl)threonine in transfer ribonucleic acid species whose codons begin with adenine. J Biol Chem 1972; 247:6394-401; PMID:4561935 [PubMed] [Google Scholar]
- 23. Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol 2004; 11:1186-91; PMID:15558052; http://dx.doi.org/ 10.1038/nsmb861 [DOI] [PubMed] [Google Scholar]
- 24. Weissenbach J, Grosjean H. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA arg III on codon-anticodon and anticodon-anticodon interactions. a thermodynamic and kinetic evaluation. Eur J Biochem 1981; 116:207-13; PMID:6788546; http://dx.doi.org/ 10.1111/j.1432-1033.1981.tb05320.x [DOI] [PubMed] [Google Scholar]
- 25. Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 2000; 39:13396-404; PMID:11063577; http://dx.doi.org/ 10.1021/bi0013039 [DOI] [PubMed] [Google Scholar]
- 26. Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005; 44:8078-89; PMID:15924427; http://dx.doi.org/ 10.1021/bi050343f [DOI] [PubMed] [Google Scholar]
- 27. Lescrinier E, Nauwelaerts K, Zanier K, Poesen K, Sattler M, Herdewijn P. The naturally occurring N6-threonyl adenine in anticodon loop of schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res 2006; 34:2878-86; PMID:16738127; http://dx.doi.org/ 10.1093/nar/gkl081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deutsch C, El Yacoubi B, de Crécy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 2012; 287:13666-73; PMID:22378793; http://dx.doi.org/ 10.1074/jbc.M112.344028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perrochia L, Crozat E, Hecker A, Zhang W, Bareille J, Collinet B, van Tilbeurgh H, Forterre P, Basta T. In vitro biosynthesis of a universal t6A tRNA modification in archaea and eukarya. Nucleic Acids Res 2013; 41:1953-64; PMID:23258706; http://dx.doi.org/ 10.1093/nar/gks1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crécy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 2011; 30:882-93; PMID:21285948; http://dx.doi.org/ 10.1038/emboj.2010.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crécy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 2009; 37:2894-909; PMID:19287007; http://dx.doi.org/ 10.1093/nar/gkp152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan LCK, Mao DYL, Neculai D, Strecker J, Chiovitti D, Kurinov I, Poda G, Thevakumaran N, Yuan F, Szilard RK, et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res 2013; 41:6332-46; PMID:23620299; http://dx.doi.org/ 10.1093/nar/gkt322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lauhon CT. Mechanism of N6-threonylcarbamoyladenosine (t(6)A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry 2012; 51:8950-63; PMID:23072323; http://dx.doi.org/ 10.1021/bi301233d [DOI] [PubMed] [Google Scholar]
- 34. Rosano C, Zuccotti S, Stefani M, Bucciantini M, Ramponi G, Bolognesi M. Crystallization and preliminary X-ray characterization of the acylphosphatase-like domain from the Escherichia coli hydrogenase maturation factor HypF. Acta Crystallogr D Biol Crystallogr 2002; 58:524-5; PMID:11856843; http://dx.doi.org/ 10.1107/S0907444901021874 [DOI] [PubMed] [Google Scholar]
- 35. Hampsey M, Na JG, Pinto I, Ware DE, Berroteran RW. Extragenic suppressors of a translation initiation defect in the cyc1 gene of saccharomyces cerevisiae. Biochimie 1991; 73:1445-55; PMID:1666843; http://dx.doi.org/ 10.1016/0300-9084(91)90177-3 [DOI] [PubMed] [Google Scholar]
- 36. Teplova M, Tereshko V, Sanishvili R, Joachimiak A, Bushueva T, Anderson WF, Egli M. The structure of the yrdc gene product from escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci 2000; 9:2557-66; PMID:11206077; http://dx.doi.org/ 10.1110/ps.9.12.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 2006; 124:1155-68; PMID:16564010; http://dx.doi.org/ 10.1016/j.cell.2005.12.044 [DOI] [PubMed] [Google Scholar]
- 38. Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle J-C, Ilan L, Hofmann K, Namane A, Mann C, Libri D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J 2006; 25:3576-85; PMID:16874308; http://dx.doi.org/ 10.1038/sj.emboj.7601235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oberto J, Breuil N, Hecker A, Farina F, Brochier-Armanet C, Culetto E, Forterre P. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res 2009; 37:5343-52; PMID:19578062; http://dx.doi.org/ 10.1093/nar/gkp557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naor A, Thiaville PC, Altman-Price N, Cohen-Or I, Allers T, de Crécy-Lagard V, Gophna U. A genetic investigation of the KEOPS complex in halophilic archaea. PLoS One 2012; 7:e43013; PMID:22927945; http://dx.doi.org/ 10.1371/journal.pone.0043013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin E V, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 2011; 30:873-81; PMID:21183954; http://dx.doi.org/ 10.1038/emboj.2010.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Daugeron MC, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res 2011; 39:6148-60; PMID:21459853; http://dx.doi.org/ 10.1093/nar/gkr178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tominaga T, Kobayashi K, Ishii R, Ishitani R, Nureki O. Structure of saccharomyces cerevisiae mitochondrial qri7 in complex with AMP. Acta Crystallogr 2014; 70:1009-14; PMID:25084372; http://dx.doi.org/ 10.1107/S2053230X14014046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thiaville PC, El Yacoubi B, Perrochia L, Hecker A, Prigent M, Thiaville JJ, Forterre P, Namy O, Basta T, de Crécy-Lagard V. Cross kingdom functional conservation of the core universally conserved threonylcarbamoyladenosine tRNA synthesis enzymes. Eukaryot Cell 2014; 13:9:1222-31; PMID:25038083; http://dx.doi.org/ 10.1128/EC.00147-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Handford JI, Ize B, Buchanan G, Butland GP, Greenblatt J, Emili A, Palmer T. Conserved network of proteins essential for bacterial viability. J Bacteriol 2009; 191:4732-49; PMID:19376873; http://dx.doi.org/ 10.1128/JB.00136-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of escherichia coli K-12 in-frame, single-gene knockout mutants: the keio collection. Mol Syst Biol 2006; 2:2006.0008; PMID:16738554; http://dx.doi.org/ 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rajagopala SV, Yamamoto N, Zweifel AE, Nakamichi T, Huang HK, Mendez-Rios JD, Franca-Koh J, Boorgula MP, Fujita K, Suzuki K, et al. The escherichia coli K-12 ORFeome: a resource for comparative molecular microbiology. BMC Genomics 2010; 11:470; PMID:20701780; http://dx.doi.org/ 10.1186/1471-2164-11-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spears JL, Gaston KW, Alfonzo JD. Analysis of tRNA editing in native and synthetic substrates. Methods Mol Biol 2011; 718:209-26; PMID:21370051; http://dx.doi.org/ 10.1007/978-1-61779-018-8_13 [DOI] [PubMed] [Google Scholar]
- 49. Blaby IK, Phillips G, Blaby-Haas CE, Gulig KS, El Yacoubi B, de Crécy-Lagard V. Towards a systems approach in the genetic analysis of archaea: accelerating mutant construction and phenotypic analysis in haloferax volcanii. Archaea 2010; 2010:426239; PMID:21234384; http://dx.doi.org/ 10.1155/2010/426239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Collinet B, Friberg A, Brooks MA, van den Elzen T, Henriot V, Dziembowski A, Graille M, Durand D, Leulliot N, Saint André C, et al. Strategies for the structural analysis of multi-protein complexes: lessons from the 3D-repertoire project. J Struct Biol 2011; 175:147-58; PMID:21463689; http://dx.doi.org/ 10.1016/j.jsb.2011.03.018 [DOI] [PubMed] [Google Scholar]
- 51. Sibler AP, Dirheimer G, Martin RP. Codon reading patterns in saccharomyces cerevisiae mitochondria based on sequences of mitochondrial tRNAs. FEBS Lett 1986; 194:131-8; PMID:2416594; http://dx.doi.org/ 10.1016/0014-5793(86)80064-3 [DOI] [PubMed] [Google Scholar]
- 52. Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature 2003; 425:737-41; PMID:14562106; http://dx.doi.org/ 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 53. Huh WK, Falvo J V, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature 2003; 425:686-91; PMID:14562095; http://dx.doi.org/ 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 54. Haussuehl K, Huesgen PF, Meier M, Dessi P, Glaser E, Adamski J, Adamska I. Eukaryotic GCP1 is a conserved mitochondrial protein required for progression of embryo development beyond the globular stage in arabidopsis thaliana. Biochem J 2009; 423:333-41; PMID:19694617; http://dx.doi.org/ 10.1042/BJ20091023 [DOI] [PubMed] [Google Scholar]
- 55. Perrochia L, Guetta D, Hecker A, Forterre P, Basta T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res 2013; 41:9484-99; PMID:23945934; http://dx.doi.org/ 10.1093/nar/gkt720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parthier C, Görlich S, Jaenecke F, Breithaupt C, Bräuer U, Fandrich U, Clausnitzer D, Wehmeier UF, Böttcher C, Scheel D, et al. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew Chem Int Ed Engl 2012; 51:4046-52; PMID:22383337; http://dx.doi.org/ 10.1002/anie.201108896 [DOI] [PubMed] [Google Scholar]
- 57. Gerdes S, El Yacoubi B, Bailly M, Blaby IK, Blaby-Haas CE, Jeanguenin L, Lara-Núñez A, Pribat A, Waller JC, Wilke A, et al. Synergistic use of plant-prokaryote comparative genomics for functional annotations. BMC Genomics 2011; 12: 1:S2; PMID:21810204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, et al. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet 2014; 10:e1004363; PMID:24809820; http://dx.doi.org/ 10.1371/journal.pgen.1004363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci 2010; 365:749-63; PMID:20124342; http://dx.doi.org/ 10.1098/rstb.2009.0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mallari JP, Oksman A, Vaupel B, Goldberg DE. Kinase-associated endopeptidase 1 (Kae1) participates in an atypical ribosome-associated complex in the apicoplast of Plasmodium falciparum. J Biol Chem 2014; 289(43):300025-39; PMID: 25204654; doi: 10.1074/jbc.M114.586735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 2013; 9:105-11; PMID:23242255; http://dx.doi.org/ 10.1038/nchembio.1137 [DOI] [PubMed] [Google Scholar]
- 62. Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schönfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell 2006; 17:1436-50; PMID:16407407; http://dx.doi.org/ 10.1091/mbc.E05-08-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qian Q, Curran JF, Björk GR. The methyl group of the N6-methyl-N6-threonylcarbamoyladenosine in tRNA of escherichia coli modestly improves the efficiency of the tRNA. J Bacteriol 1998; 180:1808-13; PMID:9537379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kimura S, Miyauchi K, Ikeuchi Y, Thiaville PC, de Crécy-Lagard V, Suzuki T. Discovery of the β-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res 2014; 42:9350-65; PMID:25063302; http://dx.doi.org/ 10.1093/nar/gku618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Anton BP, Russell SP, Vertrees J, Kasif S, Raleigh EA, Limbach PA, Roberts RJ. Functional characterization of the YmcB and YqeV tRNA methylthiotransferases of Bacillus subtilis. Nucleic Acids Res 2010; 38:6195-205; PMID:20472640; http://dx.doi.org/ 10.1093/nar/gkq364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF, Douki T, Fontecave M, Mulliez E, Atta M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J Biol Chem 2010; 285:28425-33; PMID:20584901; http://dx.doi.org/ 10.1074/jbc.M110.106831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wei F, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. Deficit of tRNA(Lys) modification by cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 2011; 121:3598-608; PMID:21841312; http://dx.doi.org/ 10.1172/JCI58056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei FY, Tomizawa K. Functional loss of cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr J 2011; 58:819-25; PMID:21908934; http://dx.doi.org/ 10.1507/endocrj.EJ11-0099 [DOI] [PubMed] [Google Scholar]
- 69. Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, Fish R, Jamotte T, Curchod ML, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol 1998; 16:851-6; PMID:9743119; http://dx.doi.org/ 10.1038/nbt0998-851 [DOI] [PubMed] [Google Scholar]
- 70. Freiberg C, Wieland B, Spaltmann F, Ehlert K, Brötz H, Labischinski H. Identification of novel essential escherichia coli genes conserved among pathogenic bacteria. J Mol Microbiol Biotechnol 2001; 3:483-9; PMID:11361082 [PubMed] [Google Scholar]
- 71. Allali-Hassani A, Campbell TL, Ho A, Schertzer JW, Brown ED. Probing the active site of YjeE: a vital Escherichia coli protein of unknown function. Biochem J 2004; 384:577-84; PMID:15324301; http://dx.doi.org/ 10.1042/BJ20041082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lerner CG, Hajduk PJ, Wagner R, Wagenaar FL, Woodall C, Gu YG, Searle XB, Florjancic AS, Zhang T, Clark RF, et al. From bacterial genomes to novel antibacterial agents: discovery, characterization, and antibacterial activity of compounds that bind to HI0065 (YjeE) from haemophilus influenzae. Chem Biol Drug Des 2007; 69:395-404; PMID:17581233; http://dx.doi.org/ 10.1111/j.1747-0285.2007.00521.x [DOI] [PubMed] [Google Scholar]
- 73. Bose N, Ogawa A, von Reuss SH, Yim JJ, Ragsdale EJ, Sommer RJ, Schroeder FC. Complex small-molecule architectures regulate phenotypic plasticity in a nematode. Angew Chemie 2012; 51:12438-43; PMID:23161728; http://dx.doi.org/ 10.1002/anie.201206797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127-8; PMID:17050570; http://dx.doi.org/ 10.1093/bioinfor-matics/btl529 [DOI] [PubMed] [Google Scholar]
- 75. Letunic I, Bork P. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011; 39:W475-8; PMID:21470960; http://dx.doi.org/ 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]


