Abstract
Developmental plasticity allows for the remarkable morphological specialization of individuals into castes in eusocial species of Hymenoptera. Developmental trajectories that lead to alternative caste fates are typically determined by specific environmental stimuli that induce larvae to express and maintain distinct gene expression patterns. Although most eusocial species express two castes, queens and workers, the ant Cardiocondyla obscurior expresses diphenic females and males; this provides a unique system with four discrete phenotypes to study the genomic basis of developmental plasticity in ants. We sequenced and analyzed the transcriptomes of 28 individual C. obscurior larvae of known developmental trajectory, providing the first in-depth analysis of gene expression in eusocial insect larvae. Clustering and transcription factor binding site analyses revealed that different transcription factors and functionally distinct sets of genes are recruited during larval development to induce the four alternative trajectories. In particular, we found complex patterns of gene regulation pertaining to sphingolipid metabolism, a conserved molecular pathway involved in development, obesity, and aging.
Keywords: developmental plasticity, Cardiocondyla obscurior, RNAseq, caste determination, sphingolipid, transcription factor binding site
Introduction
Developmental plasticity is a core biological phenomenon through which a genotype can produce a variety of phenotypes in response to different environmental cues (West-Eberhard 2003; Gilbert and Epel 2009). This responsiveness requires a sensitivity to (external) environmental input and affects most, if not all, complex developmental processes (Gilbert 2005). Furthermore, in a fluctuating environment, developmental plasticity can induce phenotypic variation of different magnitude that can be either nonadaptive (e.g., teratogenesis; Hamdoun and Epel 2007) or adaptive (e.g., predator-induced defenses in Daphnia; Boersma et al. 1998) (West-Eberhard 1989; DeWitt et al. 1998; Gilbert 2001; Ghalambor et al. 2007; Beldade et al. 2011). One specific form of developmental plasticity called polyphenism allows for the generation of two or more distinct phenotypes and underlies the evolutionary success of eusocial insects (ants, bees, wasps, and termites). Eusocial insect colonies are founded on the principle of division of labor (Hölldobler and Wilson 1990), where morphologically and behaviorally specialized individuals are produced from the same genotype due to gene-by-environment interactions. In this way, the ontogeny of a superorganism (i.e., a eusocial insect colony) parallels the differentiation of cells or organs in a multicellular organism (Bourke 2011). Such polyphenism is a prime example of the flexibility of an organism’s genetic makeup (Schlichting and Pigliucci 1998; West-Eberhard 2003).
The stable differentiation of individual eusocial insect larvae into distinct adult castes is known to be affected by external stimuli (e.g., temperature, pheromones, nutrition, and tactile interactions; Hölldobler and Wilson 2009; Penick and Liebig 2012), but we are only beginning to understand how such stimuli are transduced into neuronal, endocrine, and metabolic signals that ultimately regulate gene expression to control caste fate. This molecular cascade is best resolved for the honeybee Apis mellifera, where development of the queen caste is induced by feeding larvae with “royal jelly”, a special glandular secretion produced by worker bees. Components of this secretion, including Royalactin, Major Royal Jelly proteins, and a fatty acid Histone Deacetylase inhibitor, likely activate epidermal growth factor receptor (EGFR) signaling in the larval fat body (Kamakura 2011), which in turn activates downstream pathways that directly or indirectly affect developmental trajectories (e.g., insulin-like signaling, PI3K/TOR/S6K, Ras/Raf/MAPK; Patel et al. 2007; Kamakura 2011; Mutti et al. 2011; Wolschin et al. 2011; Badisco et al. 2013). Experimental gene knock-downs, as well as comparative transcriptome and methylome studies, performed mostly in honeybees, have revealed extensive caste-dependent changes in gene expression and epigenetic regulation (Kucharski et al. 2008; Elango et al. 2009; Bonasio et al. 2012; Shi et al. 2012; Simola, Ye, et al. 2013) for a wide range of genes, for example, hexamerins (Hoffman and Goodisman 2007; Hunt et al. 2007; Cameron et al. 2013), cytochrome P450s, spliceosomal genes (Cameron et al. 2013), the hypoxia pathway (Azevedo et al. 2011), and several other metabolic and developmental genes (Evans and Wheeler 2001; Barchuk et al. 2007; Hoffman and Goodisman 2007; Li et al. 2010; Begna et al. 2011).
The mechanisms regulating caste polyphenism in the evolutionarily independent lineage of ants are expected to be more sophisticated, owing to generally greater diphenism between queens and workers and the presence of extensive worker subcaste differentiation or queen polyphenism in some species (Hölldobler and Wilson 1990; Ruppell and Heinze 1999; Johnson and Linksvayer 2010). Furthermore, caste diphenism may be accompanied by striking morphological variation among males (Kugler 1983; Yamauchi et al. 1991; Heinze and Hölldobler 1993; Oettler et al. 2010).
Though transcriptomes (Feldmeyer et al. 2013), methylomes (Bonasio et al. 2010, 2012), and chromatin structure (Simola, Ye, et al. 2013) have been compared among different adult castes of ants, detailed studies about the molecular regulation of gene expression during larval ontogeny have not yet been conducted. This is partly due to an inherent difficulty in most ant species, as an individual’s caste-fate remains elusive until a late larval instar or the pupal stage. Yet, it is hypothesized that many of the key differences between adult castes emerge during larval development. Therefore, characterizing genes that are involved in this initial phase of caste divergence will help to unravel the ontogenetic and evolutionary underpinnings of caste determination and differentiation in eusocial insects.
Here we used Cardiocondyla obscurior, a minute myrmicine ant, as a model to study genome-wide gene expression variation among larvae of known caste fate within a replicated experimental design based on individual whole larval samples. Colonies of C. obscurior can be reared easily in the laboratory, and in this controlled setting we are able to induce larval development along the desired caste-fate trajectory in a reproducible manner.
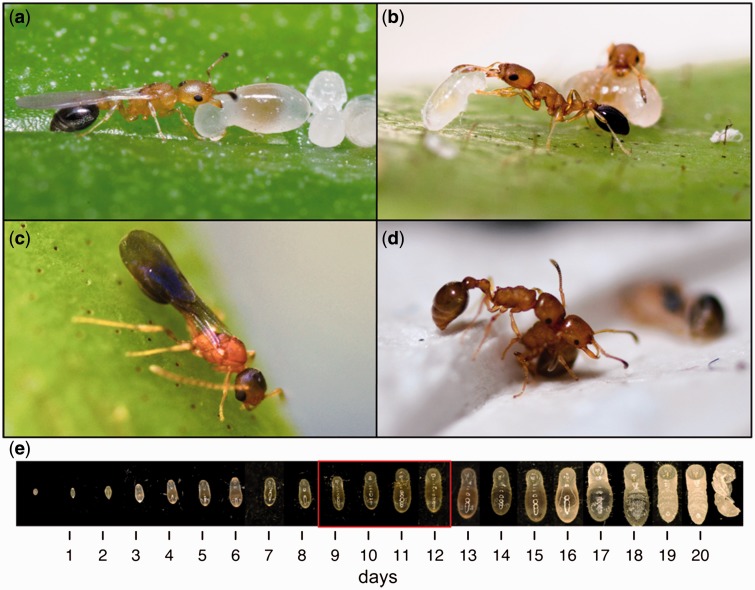
Furthermore, C. obscurior offers a particularly promising system to unravel the mechanisms underlying developmental plasticity in both female and male sexes. In addition to female queens (QU) and workers (WO), C. obscurior colonies regularly express wingless males (ergatoid [literally “workerlike”] males, EM) and occasionally also winged disperser males under laboratory conditions (WM) (fig. 1, table 1) (Heinze and Hölldobler 1993; Cremer and Heinze 2003; Schrempf and Heinze 2006; Cremer et al. 2011). The phenotypes of these two male forms differ substantially in many aspects (table 1). Winged males resemble typical males of other ant species in morphology, physiology and behavior, whereas ergatoid males have smaller eyes, a slender thorax, smaller brains, an unusually long life span and—unique for males of social Hymenoptera—a life-long spermatogenesis (Kugler 1983; Heinze and Hölldobler 1993). EMs also feature enlarged, sickle-shaped mandibles that are deployed in lethal fights with rival males over reproductive rights (Kinomura and Yamauchi 1987; Stuart et al. 1987).
Fig. 1.
Phenotypes of Cardiocondyla obscurior. Fertilized, diploid eggs develop either into (a) queens (QU) or (b) workers (WO), whereas unfertilized eggs remain haploid and develop into males of either the (c) winged (WM) or (d) wingless, ergatoid phenotype (EM). (e) Development of a C. obscurior worker larva from egg to pupa. The red box indicates the approximate sampling period for RNAseq experiments.
Table 1.
General Features of the Four Different Adult Phenotypes Expressed in Cardiocondyla obscurior.
| QU | WO | WM | EM | |
|---|---|---|---|---|
| Sex | Female | Female | Male | Male |
| Winged | Yes | No | Yes | No |
| Average life span (weeks) | 24 | 8 | 2 | 6 |
| Average size (mm) | ∼2 | ∼1.7 | ∼2.1 | ∼1.7 |
| Fertility | High | Sterile | Low | Higha |
| Individuals per colony | 5 | 28 | 0b | 1 |
| Number of ocelli | 3 | 0 | 3 | 0 |
| Antennae | Elbowed, long scape | Elbowed, long scape | Short scape | Elbowed, long scape |
| Gaster pigmentation | Dark | Dark | Dark | Light |
| Eye size | Medium | Medium | Large | Small |
| Mandible type | Normal | Normal | Reduced | Enlarged |
aLife long spermatogenesis.
bWinged males are only produced under lab conditions.
To deepen our understanding of the mechanisms underlying polyphenism in ants, we characterized the developmental transcriptomes of C. obscurior larvae by high throughput mRNA sequencing (RNAseq) using seven early third instar larvae from each of four known developmental trajectories (caste fate), rendering this study the first analysis of genome-wide gene expression differences of individual ant larvae. By analyzing gene expression patterns based on caste fate, we identified the genes that are putatively functional during the early stages of caste divergence.
Transcriptional regulation of gene expression is achieved through many factors, including transcription factors (TFs), DNA methylation, histone posttranslational modifications and histone variants, and noncoding RNAs. Among these factors, TFs are perhaps the most well studied and are established regulators of development and phenotypic plasticity in eukaryotes (Carroll 2000; Gilbert and Epel 2009; Latchman 2010). By binding to canonical and often evolutionarily conserved binding sites (transcription factor binding sites [TFBSs]) typically located in intergenic sequence proximal to genes (promoters) and in tandem groups of TFBSs distal to genes (enhancers), TFs can coordinately regulate transcription for groups of target genes, thereby driving specific developmental pathways. It was previously shown that there are significant evolutionary changes in the genome-wide TFBS landscape between solitary and eusocial insect genomes, suggesting that the regulation of gene transcription by TFs is a major mechanism facilitating caste differentiation in eusocial insects (Simola, Wissler, et al. 2013). In support of this, we found enrichment of different TFBSs in promotor regions of genes exhibiting differential expression by caste fate, thereby relating specific TFs to early developmental divergence of ant castes.
Sphingolipids are versatile structural compounds of cell membranes and have gained increasing interest in cancer research and developmental biology for their activity as second messengers affecting highly conserved molecular pathways, thus regulating cell growth, proliferation, differentiation, and apoptosis (Spiegel and Merrill 1996; Adachi-Yamada et al. 1999; Lebman and Spiegel 2008; Gault et al. 2010; Hamel et al. 2010; Kraut 2011; Pepperl et al. 2013; Sasamura et al. 2013; Zhu et al. 2013). Intriguingly, the sphingolipid metabolism is tightly interwoven with several pathways implicated in caste differentiation (TOR [Zhu et al. 2013], Wnt [Pepperl et al. 2013], and Notch/EGF [Baker and Thummel 2007]) and is also involved in the regulation of CytP450 expression (Merrill et al. 1999) and JH metabolism (Yang et al. 2010). These interactions led us to analyze expression patterns for genes involved in sphingolipid metabolism. We found differential regulation in all expressed key genes (Walls et al. 2013), indicating that developmental plasticity in ants is in part regulated by changes in sphingolipid homeostasis.
Together, our findings confirm that significant gene expression differences emerge early in larval development, priming larvae for the vast morphological reorganization during metamorphosis as pupae. Furthermore, such changes likely are controlled by a specific set of TFs and prominent in developmental toolkit genes (e.g., Wnt, EGFR, Notch), metabolic pathways (e.g., sphingolipids, oxidative phosphorylation), and cell-cycle processes (e.g., ribosomal and proteasomal proteins).
Results
To assess gene expression differences at an early stage of caste fate divergence, we performed high throughput sequencing of mRNA complements (RNAseq) from 28 individual and morphologically indistinguishable early third instar larvae, sampled shortly after their commitment to each of the four mature phenotypes (fig. 1e) (Schrempf and Heinze 2006). By sequencing seven independent biological replicates per treatment, our experimental design explicitly incorporates interindividual biological variation (Auer and Doerge 2010) (fig. 2a–c), which increases the sensitivity and specificity of our analyses (Liu et al. 2014).
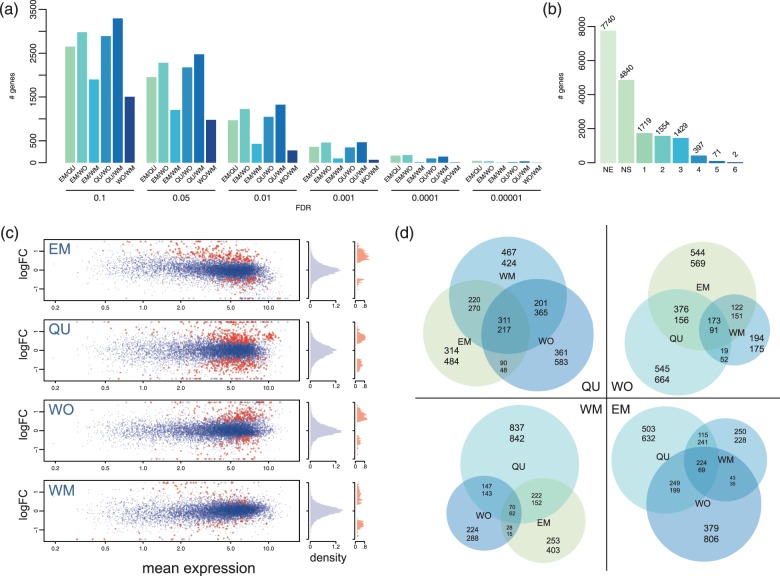
Fig. 2.
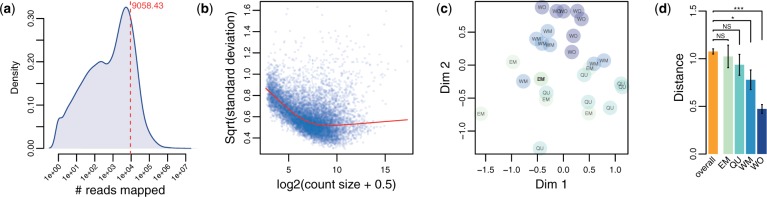
(a) Distribution of raw read coverage per gene. The red dashed line indicates mean read count per gene. (b) Genewise mean–variance relationship suggesting high levels of biological variation between samples (Law et al. 2014). (c) MDS plot for the top 500 most variable genes. (d) Mean Euclidean distances (±SE) of MDS-scaled samples between all samples (overall) and within each caste fate.
Differential Gene Expression and Gene Set Enrichment Analyses
The current official gene set of C. obscurior includes annotations for 17,552 protein-coding genes (Schrader et al. 2014). In our larval RNAseq data, 10,012 of these genes (57%) were expressed with a mean raw coverage of 9,058.43 reads per gene (fig. 2a) and high-level biological variation as suggested by the mean–variance relationship (fig. 2b, Law et al. 2014). To assess whether transcriptome samples cluster according to their developmental trajectory, we performed a multidimensional scaling (MDS) analysis with limma (Law et al. 2014) of the 500 most variable genes across all samples (fig. 2c) and calculated mean Euclidean distances between MDS-scaled data points (fig. 2d). This calculation of distance is conservative, as it relies on a lower dimensional projection of the data that do not account for the total variance. Compared with mean distances between pairs of all 28 samples, distances between EM and QU samples did not differ significantly (t-tests, EM: df = 22.2, P = 0.67, QU: df = 22.58, P = 0.23). However, among WM and WO samples distances were significantly smaller (t-test, WM: df = 22.95, P = 0.01, WO: df = 36.18, P < 0.0001). In addition, we found strong overlap between groups in the MDS analysis, in particular for QU and EM.
Using a false discovery rate (FDR) of 0.05 (fig. 3a), 5,172 genes showed differential expression in at least one of the six pairwise comparisons (EM/QU, EM/WO, EM/WM, QU/WO, QU/WM, and WO/WM; fig. 3b). As we were interested in phenotype-specific gene expression profiles, we retrieved the subsets of differentially expressed genes (at FDR < 0.05) that showed consistent changes in one group compared with the three others (fig. 3c and supplementary tables S1–S4, Supplementary Material online), for example, 311 genes showed significantly higher and 217 genes showed significantly lower expression in QU in each of the three pairwise comparisons with WO, EM, and WM (fig. 3d).
Fig. 3.
(a) Number of differentially expressed genes for the six possible pairwise comparisons at different FDR cutoffs. (b) Summary of differential gene expression calls with six pairwise comparisons at an FDR of 0.05, NE, not expressed; NS, no significant difference in expression; 1–6; Number of genes with significant expression differences in 1–6 pairwise comparisons. (c) MA plots showing the mean expression across all 28 samples on the x-axis and the log-fold change for one over three other groups. Each dot represents one expressed gene, red dots show genes included in the compiled gene sets. Density plots show frequency distribution of all genes (blue) and of genes in the set genes (red). (d) Venn diagrams comparing phenotype-specific expression differences in WM, EM, QU, and WO. Each Venn diagram shows the number of genes with significantly higher (upper numbers) or lower expression (lower numbers) at an FDR < 0.05 in one, two, or three pairwise comparisons per caste fate. For each caste fate, sets of genes differentially expressed in all three pairwise comparisons (i.e., the central intersection) were used for enrichment analyses.
In the different gene sets, we assigned Drosophila orthologs to 69.70% (QU), 74.24% (WO), 62.46% (EM), and 53.79% (WM) of the genes using a reciprocal best alignment approach. When orthology was not resolved clearly, we assigned the closest homolog (BLASTp e value < 1e-10). We then performed functional enrichment analyses using Gene Ontology (GO) terms (see supplementary material, Supplementary Material online) and DAVID’s (Database for Annotation, Visualization and Integrated Discovery) Functional Annotation Clustering (v6.7), which combines enrichment scores from different resources (e.g., GO terms, protein-protein interactions, homologies) based on common biological themes. DAVID analysis returned 25 distinct Annotation Clusters on the 152 EM-specific genes and fewer clusters for the other gene sets (QU 12 [199 genes], WO 5 [53], WM 0 [0]) (table 2). In QU, most clusters are associated with cell-cycle processes (e.g., mitosis, DNA replication, nucleosome organization; table 2a). Furthermore, we found clusters associated with transcript splicing and diacylglycerol kinase activity. The functionally clustered gene set in QU contains 12 stronger expressed RpS, 14 RpL (13 overexpressed), and 4 mitochondrial ribosomal protein genes, which are significantly downregulated in QU. Furthermore, seven different histone genes (four H2A, one H1/H5, one H2b, and one H3) and the proteasome subunits α3, -6, -7 as well as β2, -4, -6, and -7 are expressed to a significantly lower level in the QU gene set (supplementary table S1, Supplementary Material online). As histone occupancy generally inhibits TF binding to DNA (Boyle et al. 2008; Neph et al. 2012; Wang et al. 2012), these results are consistent with a model of caste differentiation in which queen destined larvae increase chromatin accessibility to increase gene expression and cell proliferation.
Table 2.
Functional Enrichment of Genes Significantly Differentially Expressed in (a) Queens (QU), (b) Workers (WO), and (c) Ergatoid Males (EM).
| Cluster | Enrichment Score | Description |
|---|---|---|
| (a) QU | ||
| AC 1 | 14.85 | Mitosis 1 |
| AC 2 | 10.29 | Mitosis 2 |
| AC 3 | 6.20 | DNA replication |
| AC 4 | 3.95 | Protein–DNA interaction |
| AC 5 | 3.76 | Nucleotide binding |
| AC 6 | 3.65 | Proteasome |
| AC 7 | 3.38 | Mitosis 3 |
| AC 8 | 3.15 | Nucleosome organization |
| AC 9 | 2.83 | Prereplicative complex |
| AC 10 | 2.71 | Spindle organization |
| AC 11 | 2.71 | Splicing |
| AC 12 | 2.56 | Kinase activity |
| (b) WO | ||
| AC 1 | 11.34 | Oxidative phosphorylation |
| AC 2 | 6.69 | Energy generation |
| AC 3 | 4.01 | ATP metabolism |
| AC 4 | 2.86 | Oxidoreductase activity |
| AC 5 | 2.46 | Valine, leucine, and isoleucine degradation |
| (c) EM | ||
| AC 1 | 11.70 | Neurogenesis |
| AC 2 | 10.12 | Organogenesis |
| AC 3 | 8.51 | Cellular morphogenesis |
| AC 4 | 7.47 | Respiratory system development |
| AC 5 | 6.61 | TF activity |
| AC 6 | 5.83 | Imaginal disc differentiation |
| AC 7 | 5.54 | Exocrine System development |
| AC 8 | 5.46 | Limb & Wing morphogenesis |
| AC 9 | 5.06 | Organismal development & organogenesis |
| AC 10 | 4.87 | Axon guidance |
| AC 11 | 4.68 | Wnt pathway |
| AC 12 | 4.67 | Eye morphogenesis |
| AC 13 | 4.64 | Cell adhesion involved in morphogenesis |
| AC 14 | 4.47 | Homeobox |
| AC 15 | 4.00 | Cell–cell junction |
| AC 16 | 3.80 | EGF signaling |
| AC 17 | 3.60 | Gliogenesis |
| AC 18 | 3.28 | Compound eye pigmentation |
| AC 19 | 3.02 | Genitalia development |
| AC 20 | 2.92 | Vesicle formation |
| AC 21 | 2.84 | Immunoglobulin-like proteins |
| AC 22 | 2.64 | Frizzled |
| AC 23 | 2.52 | Metal ion binding |
| AC 24 | 2.42 | Gene expression |
| AC 25 | 2.19 | Apoptosis |
Note.—Annotations by DAVID; AC, Annotation Cluster; ES = −log(penrichment).
In WO, each of the five Annotation Clusters is associated with energy generation in mitochondria (e.g., oxidative phosphorylation, ATP metabolism, or valine, leucine, and isoleucine degradation; table 2b). We found stronger expression of five mRp genes (L20, L21, L32, S14, and S28) and three proteasome genes (α6, β3, and β7). In addition, four different cytochrome oxidase C subunits (IV, Vb, VIb, and VIIc) were expressed significantly higher in WO (supplementary table S2, Supplementary Material online).
Many of the 20 functional clusters in EM are associated with basic development (e.g., neurogenesis, imaginal disc development, exocrine system development; table 2c). In addition, we found clusters indicating substantial changes in core molecular pathways (e.g., Wnt, EGFR) (Hurlbut et al. 2007). For example, six of eight annotated Wnt homologs (wg, Wnt10, 3 homologs of Wnt2, Wnt5, and a homolog of Wnt6) in the C. obscurior genome are significantly overexpressed in EM. In addition, orthologs of the Wnt receptors fz2 and fz4, the coreceptor arr, and associated genes like stan, arm, nkd, and four different Cadherins (ft, Cad87A, Cad74A, and a homolog to Cad96Ca) are upregulated. Apart from Wnt pathway genes, we found differential regulation in various genes involved in Notch (N, Dl, H, Ser, shg, and heph) or EGFR signaling (Egfr, Hh, and Ptc) and also in three Toll-receptor homologs (Toll-6 and two Tollo homologs). In addition, differential expression in EM was abundant in TFs, for example, abd-b, castor, twist, Sp1, vielfältig, jumeau, jim, and homeobox genes (e.g., dll, en, smo) (see supplementary table S3, Supplementary Material online).
There were no significant Annotation Clusters for WM. However, some individual candidate genes might be of particular interest in the WM-specific gene set. For instance a homolog to Edem1, a gene coding for an alpha-mannosidase affecting life span in Drosophila melanogaster and Caenorhabditis elegans (Liu et al. 2009), is strongly downregulated in WM. In addition, we found differential expression in well-characterized developmental genes such as headcase, visgun, eyegone, as well as homologs to engrailed, prospero, AGO3, and capricious (supplementary table S4, Supplementary Material online).
Together, the enrichment analyses revealed strong functional differences between the gene sets, suggesting that development of the discrete phenotypes is regulated by distinct developmental programs in cell division (QU), energy generation (WO), and developmental toolkit genes (EM).
TF Binding Site Enrichment
As TFs function as important regulators of developmental plasticity, we tested whether gene expression may be regulated by distinct sets of TFs for each caste fate (fig. 4). Significant binding sites (TFBSs) for 59 factors were predicted by scanning 2-kb promoter sequences using available position weight matrix models. Caste-biased gene sets were subsequently analyzed for enrichment of each TFBS model individually using two measures (enrichment probability pE and presence probability pP). In the QU gene set, 13 TFBSs were either significantly enriched (pP < 0.01, Bonferroni corrected) or overrepresented (pP < 0.01, Bonferroni corrected), with five TFBSs being significant for both measures. Of eight significant TFBSs in the WO gene set, six were significant in both tests. The highest number of significantly enriched TFBSs was found in EM, with 19 TFBSs being significantly enriched (pE < 0.01, Bonferroni corrected) or overrepresented (pP < 0.01, Bonferroni corrected). Of these, ten TFBSs were significant in both tests. In contrast, we did not find any TFBSs to be significantly overrepresented in the WM gene set and only one TFBS (OVO) was significantly enriched (pE < 0.01, Bonferroni corrected).
Fig. 4.
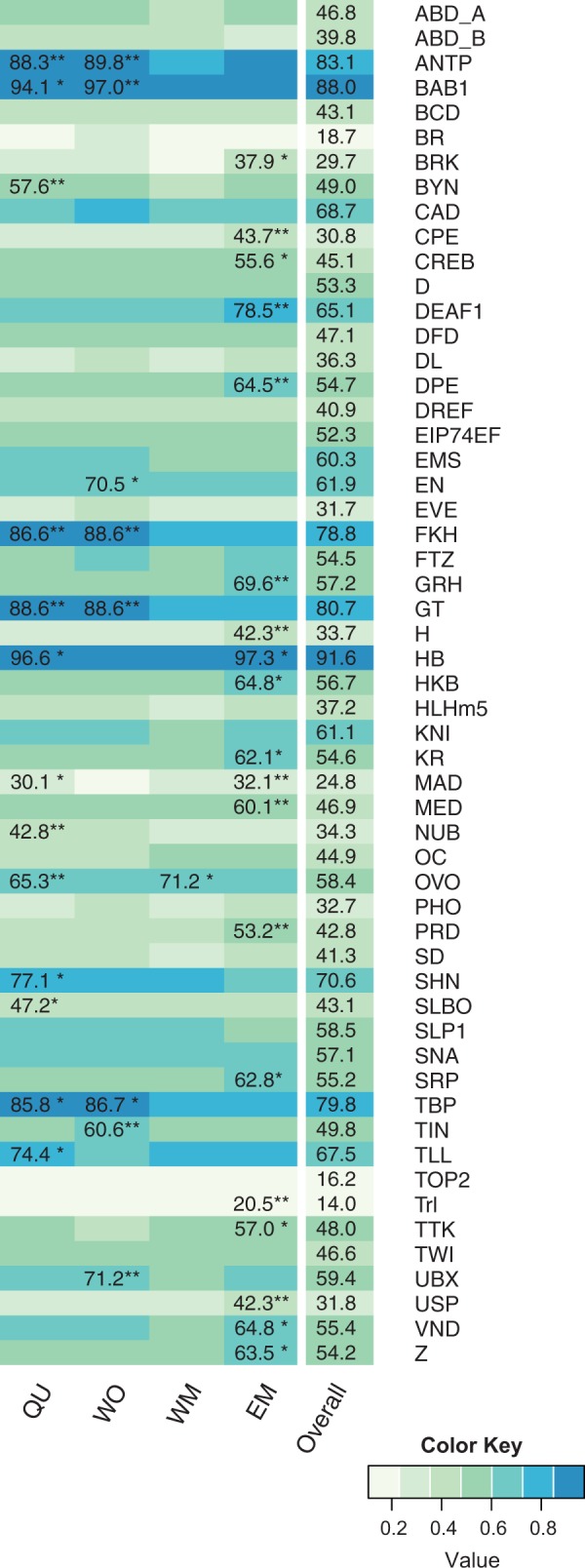
Transcription factor binding site enrichment for caste specific gene sets. The heat map shows the proportion of genes in the different gene sets that have at least one of the respective TFBSs in their promoter sequence. Numbers for significantly enriched/overrepresented TFBSs give the percentage of genes having the respective TFBS in their promotor. Asterisks indicate whether we found significant enrichment (pE < 0.01, asterisk at first position), overrepresentation (pP < 0.01, asterisk at second position), or both (two asterisks).
Sphingolipids
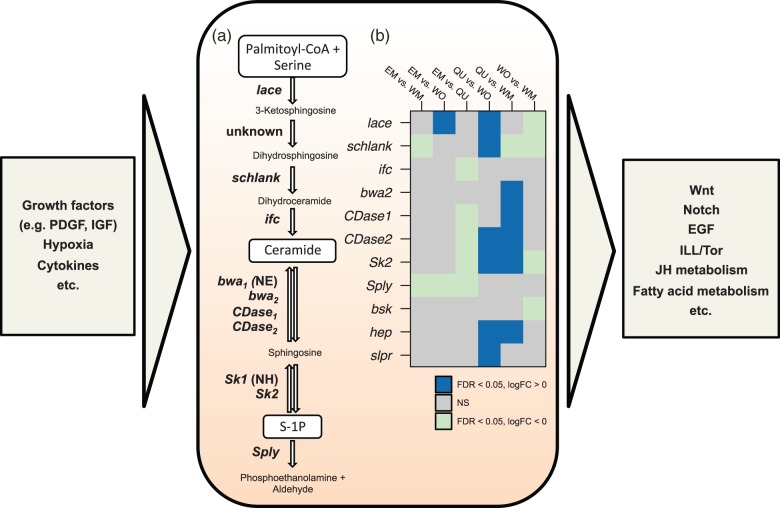
Based on previous evidence suggesting that sphingolipid metabolism regulates pathways involved in caste differentiation, we analyzed expression of sphingomyelin cycle genes in our data set. Interestingly, we found significant expression differences in all eight key genes (Walls et al. 2013) involved in the metabolic cycle (fig. 5 and supplementary table S5, Supplementary Material online). Lace, which codes for a serine-palmitoyltransferase responsible for de novo synthesis of sphingolipids, was weakly expressed in WO. Following the formation of dihydrosphingosine, the dihydroceramide synthase schlank (significant differences in expression: WM>WO, WM>QU, WM>EM, and QU>WO) and the sphingosine desaturase infertile crescent (ifc) (QU>EM) catalyze the production of the bioactive lipid ceramide. The conversion of ceramide to sphingosine involves different ceramidases coded by brain washing (bwa, with two homologs in C. obscurior. Bwa1: not expressed, bwa2: QU>WM), and ceramidase (CDase), which has two homologs in C. obscurior of which both show stronger expression in QU (CDase1: QU>EM, QU>WM and CDase2: QU>EM, QU>WM, QU>WO). Sphingosine is then converted to sphingosine-1-phosphate (S-1P) through the activity of a kinase coded by Sk2 (QU>EM, QU>WM, and QU>WO). The membrane-bound S1P-lyase Sply, which is downregulated in EM, converts S-1P to an aldehyde and phosphoethanolamine, thus removing S-1P irreversibly from the cycle.
Fig. 5.
Schematic representation of the sphingomyelin cycle (central box) and its involvement with other developmental pathways and mechanisms (left and right boxes). (a) The central metabolism in sphingolipid biosynthesis (Walls et al. 2013) with the involved genes (left of arrows) and metabolic products (between arrows). (b) Differential expression of key genes of the sphingomyelin cycle. The heat map shows significant (FDR < 0.05) positive (dark blue) or negative (light blue) log fold changes for each of the six pairwise comparisons in sphingomyelin cycle and downstream genes (basket [bsk], hemipterous [hep], slipper [slpr]). CDase2 and bwa2 are homologs to the Drosophila CDase and bwa genes, respectively. NH, no homolog identified in C. obscurior; NE, not expressed; NS, no significant difference.
In addition to the eight key genes of the sphingomyelin cycle, we also analyzed expression of downstream genes affected by cellular sphingolipid levels. For example, we found differences in slipper (QU>WO), hemipterous (QU>WO, QU>WM) and basket (EM>WO, WM>WO), three genes that form a ceramide-sensitive MAP k3 → JNK pathway involved in stress response and apoptosis in Drosophila (Kraut 2011).
Discussion
The pronounced differences between queen and worker castes make ants and other social insects compelling models to study the regulation of developmental plasticity. However, genetic and molecular studies focusing on the divergence during larval and pupal development are still rare (Abouheif and Wray 2002; Ometto et al. 2011; Chen et al. 2012; Berens et al. 2014). Studies on queen determination in different ant species have revealed differences in developmental switch points in caste differentiation during larval development, suggesting that the extent of queen–worker diphenism correlates with the timing of developmental divergence (Abouheif and Wray 2002; Penick et al. 2012). Furthermore, as shown in A. mellifera, caste determination might be a progressive process not controlled by a single fixed time point in development (Cameron et al. 2013). In the polyphenic ant C. obscurior, queen/worker and winged/ergatoid male determination has been studied in cross-fostering experiments and by juvenile hormone analog application(Schrempf and Heinze 2006; Du et al. 2007), indicating that in both males and females, caste fate is determined late in second larval instar development. In this study, we characterized gene expression during larval development shortly after the mature phenotype has been determined with high likelihood to unravel the genetic framework directing specific morphological differentiation into four distinct adult phenotypes.
Gene Expression Patterns
The great number of genes found to be differentially expressed in our comparison shows that distinct phenotypes adopt largely different gene expression profiles already during larval development, preceding morphological differentiation that occurs during metamorphosis. Therefore, the specific morphologies of queens, workers, ergatoid males, and winged males likely depend on gene expression patterns that are pre-established, as opposed to emerging first during pupation.
Furthermore, despite the apparent morphological similarities between alate and wingless phenotypes between sexes (e.g., female workers and ergatoid males), we did not find consistent overlap in gene expression profiles (fig. 3d) or TFBS enrichment (fig. 5) comparing these phenotypes. Assessing whether analogous morphological traits (e.g., winglessness) do in fact emerge by separate developmental routes however requires detailed studies on the vestigial imaginal discs of EM and WO.
Though an RNAseq experiment with whole body samples does not allow for a deeper analysis of tissue-specific expression profiles, it did allow for the identification of systemic gene expression changes. Furthermore, by sampling during early larval development when morphological differences have not yet emerged, we are able to minimize allometric bias in cell number or proportion of tissues.
Functional enrichment analysis revealed substantial ontogenetic differences between EMs and the other three phenotypes, driven by changes in pleiotropic and interconnected signaling networks (Notch, EGFR, Wnt, etc.). The general importance of Wnts in development and morphogenesis in arthropods is well-known (e.g., in CNS, eye, genitalia, imaginal discs, mouthparts; Murat et al. 2010) and it is also likely to be involved in wing loss in ants (Abouheif and Wray 2002; Rajakumar et al. 2012). The great number of differentially expressed Wnt, Notch, and EGFR pathway genes in EM thus indicates that changes in timing and/or orchestration of gene expression in such evolutionarily conserved developmental circuits underlie the evolution of the ergatoid male phenotype in Cardiocondyla.
In addition, absence of developmental toolkit pathways in DAVID analyses of the three other gene sets suggests that, during the particular period of sampling, basic developmental mechanisms experience differential regulation specifically in EMs. This is likely a consequence of strong sexual selection on EM-specific traits. EMs attack and kill young rivals upon their eclosion, resulting in an evolutionary arms-race over production of the first male in multiqueen colonies (Yamauchi et al. 2006; Suefuji et al. 2008). Consequently, selection is predicted to favor the early emergence of EMs; accordingly, EMs do exhibit reduced developmental time compared with the other male phenotype (Schrempf and Heinze 2006). Thus, phenotype-specific expression profiles in our analysis might emerge from alternative developmental rates that are set by upstream changes in developmental signaling pathways, for example, through Notch and Wnt signaling. In support of this, a recent study also found DNA methylation differences in Wnt genes between queen- and worker-destined honeybee larvae, indicating a prominent role for these networks in plastic development of eusocial insects (Shi et al. 2012).
The prevalence of genes involved in mitosis and nucleosome organization in QU implies specific changes in cell cycle and chromatin structure. Many mitosis-related genes are downregulated in QU, potentially reflecting a state of arrested cell division. In fact, restrained growth in queen larvae is known from honeybees, where queen-destined larvae remain smaller during early differentiation and only later outgrow their worker-destined siblings (Stabe 1930). Conversely, the high frequency of overexpressed ATP metabolic genes in the WO-specific gene set indicates an increased energy requirement in worker larvae during early third instar.
This is in contrast to studies done in A. mellifera that suggest an increase in energy generation in queen-destined larvae (Corona et al. 1999; Li et al. 2010; Begna et al. 2011; Cameron et al. 2013). However, in honeybees, growth rates of queen-destined larvae predominantly increase late in development and queens develop faster than workers (Page and Peng 2001). In C. obscurior (Schrempf and Heinze 2006) and likely other ants (Bowsher et al. 2007), queens develop at the same rate than workers but for a longer period. Thus, caste-specific energy requirements of larvae might not be conserved across these two distant taxa.
TF Binding Sites
Testing for enrichment of TFBSs in cis-regulatory regions, we again found specific differences between QU, WO, EM, and WM. The lack of a single TFBS enriched in all four phenotypes suggests that larval development is not controlled by common TFs, but instead that each developmental phenotype recruits its own set of TFs to produce the respective gene expression signatures. Yet, we found five TFBSs (antennapedia [ANTP], bric a brac 1 [BAB1], fork head [FKH], giant [GT], and TATA-binding protein [TBP]) with significant enrichment in both QU and WO, potentially revealing TFs specifically recruited to female larval development, for example, as shown for bab1 in Drosophila (Kopp et al. 2000). We did not find a similar overlap between the two male phenotypes, as WMs showed only one TFBS (OVO/Shavenbaby, a germline expressed factor shared with queens). How the interplay of different TFs directs gene expression and development in each caste remains elusive, in particular since significant overabundance of binding sites did not correlate with stronger TF gene expression in the respective larvae.
Sphingomyelin Cycle
We discovered significant differences in the expression of most sphingolipid metabolism genes comparing all four phenotypes, suggesting a role for the sphingomyelin cycle in caste differentiation. In addition, previous studies revealed that mutations in sphingolipid metabolism genes in Drosophila and Caenorhabditis produce phenotypes that strikingly resemble differences observed between castes in eusocial insects, in particular concerning fecundity and longevity. For example, loss of bwa decreases longevity and increases larval developmental time in Drosophila, and pharmacological inactivation of Bwa increased Juvenile Hormone acid methyltransferase activity (Yang et al. 2010). Sterility was observed in ifc deficient Drosophila (Phan et al. 2007). Deletion of sply leads to flight muscle degradation and apoptosis in reproductive organs (Herr et al. 2003); similar defects were found in Sk2 mutants (Herr et al. 2004). Schlank- mutant flies show defects in fat storage and larval growth, and corresponding mutants in C. elegans (hyl-1; lagr-1, hyl-1, and hyl-2) show significant lifetime extension (Tedesco et al. 2008; Menuz et al. 2009; Mosbech et al. 2013).
Similarly, the four phenotypes of C. obscurior are characterized by considerable differences in longevity, fecundity, larval development, and morphology (table 1). The distinct differential expression in most key genes in C. obscurior and their established regulatory functions and roles in Drosophila development strongly suggest previously unappreciated role for sphingolipids in caste fate differentiation in ants. We therefore propose to extend the current model of caste determination and differentiation in eusocial insects to include sphingolipids as important mediators of developmental plasticity.
Conclusion
Eusocial insect castes illustrate the concept of developmental conversion (Smith-Gill 1983), in that plastic responses in development are not continuous reaction norms proportional to the environmental stimulus, but threshold dependent discrete phenotypes. Previous studies on caste differentiation and developmental plasticity in eusocial insects already showed that evolutionarily conserved developmental pathways are involved in this process and that the initial environmental signal is transduced to substantially alter gene expression profiles during development. In accordance, our comparison of gene expression patterns in early larvae of C. obscurior revealed that a large number of genes are differentially expressed between larvae of different caste-fate and that many developmental pathways and TFs appear to be involved in caste differentiation. Caste differentiation can be considered as active, regulatory plasticity involving versatile developmental and physiological changes rather than passive plasticity, a (largely nonadaptive) direct response to environmental influences (Schlichting and Pigliucci 1995, 1998; Whitman and Agrawal 2009; Forsman 2014). Eusocial insects evolved from monophenic ancestors and the worker–queen diphenism thus likely evolved from passive plasticity expressed by the solitary ancestors (Hunt and Amdam 2005). This transition from putatively non- or even mal-adaptive plasticity to highly adaptive plasticity is expected to occur, if the ancestral plasticity produces phenotypes that are close enough to a new phenotypic optimum for directional selection to act upon (Ghalambor et al. 2007), allowing for the evolution of a stable queen–worker diphenism as we see it today. In Cardiocondyla, a similar mechanism might have also enabled the evolution of male diphenism, in that the extant male phenotypes evolved from a single, merely passively plastic winged male phenotype (Tsuji et al. 1994). Intriguingly, the extent of male phenotypic plasticity is highly variable in the genus Cardiocondyla, with several species producing ergatoid males exclusively, others producing both morphs (Oettler et al. 2010), and some species additionally producing intermorphs between winged and wingless males (Cremer et al. 2002; Yamauchi et al. 2005; Heinze et al. 2013). Hence it appears that phenotypic plasticity in males evolved under species-specific selection regimes in Cardiocondyla, potentially rendering this genus a showcase for the role of genetic accommodation in the evolution of novelty (West-Eberhard 1989, 2005; Pfennig et al. 2010; Moczek et al. 2011; Schlichting and Wund 2014).
Materials and Methods
Organism
Cardiocondyla obscurior is a cosmotropical tramp ant species with a presumed native range in Southeast Asia (Seifert 2003). Our experimental colonies used in this study are derived from colonies that were originally collected in an introduced population in Brazil (permitted by the Brazilian Ministry of Science and Technology [RMX 004/02]) and thereafter propagated in the lab for several years. Experimental colonies were reared at 12 h 28 °C light/12 h 23 °C dark in plastered petri dishes and fed twice a week with parts of cockroaches or Drosophila and honey-soaked shreds of paper. Water was provided ad libitum. All animal treatment guidelines applicable to ants under international and German law have been followed.
Larvae Sampling
Experimental colonies of C. obscurior were treated to exclusively produce only one of the four different phenotypes. Production of worker larvae (WO) was achieved by establishing colonies comprised one fertilized queen, one ergatoid male, and ten workers. Larvae produced within the first 2 months after colony foundation will develop into workers (Suefuji et al. 2008). Queen larvae (QU) were produced by treating eggs from worker-producing colonies (see above) with the juvenile hormone analog methoprene (Schrempf and Heinze 2006). Collected eggs were rinsed twice with 1 mg/ml methoprene dissolved in acetone and transferred to a queen-less colony of 20 workers. To produce males, we reared virgin queens that only produce haploid eggs, which develop either into either ergatoid or winged males. Under standard conditions (see above), colonies produce ergatoid males. Winged males are rare. However, stressful environmental conditions can induce their production (Cremer and Heinze 2003). We established colonies consisting of two unfertilized queens and ten workers and reared these at a constant 23 ° (colder than normal; see above). Some of these colonies ceased production of ergatoid males and exclusively produced winged males, especially in the later stage of their queens’ life (>30 weeks; Heinze and Schrempf 2012), which allowed us to sample winged male larvae (WM).
We sampled larvae that had chitinized mandibles, a characteristic feature of the third instar (Schrempf and Heinze 2006), and were smaller than 800 µm (fig. 1e). All colonies that were used for sampling larvae were screened twice a week from 2 weeks before the first larva was sampled until 3 weeks after the last larva had been sampled to confirm the exclusive production of workers, queens, ergatoid males, or winged males. For RNAseq studies, sampled larvae were placed individually in 1.5-ml Eppendorf tubes, snap-frozen in liquid nitrogen, and stored at −80 °C.
Gene Expression Analysis with RNAseq
Total RNA was extracted from 28 individuals during the early third larval instar with the RNeasy Plus Micro kit (Qiagen), yielding 27–270 ng per individual larvae.
Starting from 20 ng input RNA, double strand, unstranded, multiplexed cDNA libraries for single-end sequencing of the 28 separate samples were generated. Briefly, cDNA was generated by reverse transcription using pseudorandom and oligo-dT priming. Using single-primer isothermal linear amplification, double-stranded DNA was generated and amplified (Ovation RNAseq system V2, NuGEN). After clean up with QIAgen’s MinElute cleanup kit, 1.5 µg cDNA was fragmented to 100–300 bp by shearing (Covaris S2 AFA). Libraries from 150 ng cDNA were prepared with the Encore Rapid DR Multiplex System (NuGEN), quantified (KAPA library quantification kit), and distributed randomly across four different lanes (Auer and Doerge 2010). Sequencing was carried out at the in-house sequencing center (KFB, University of Regensburg, Germany) on an Illumina HiSeq1000. Raw sequencing reads have been deposited in the NCBI short read archive (accession no. SRX879674, SRX879676, SRX879678).
Sequencing reads produced from QU were published previously (Schrader et al. 2014; SRX692538). Sequencing yielded approximately 532 M raw reads that were filtered for adapter contamination (cutadapt; Martin 2011), parsed through quality filtration (Trimmomatic v0.27, options: LEADING:10 TRAILING:10 SLIDING:4:10 MINLEN:15), and mapped against the reference genome (Schrader et al. 2014) using the tophat2 (v2.0.8) and bowtie2 (v 2.1.0) package (Phan et al. 2007; Langmead and Salzberg 2012; Kim et al. 2013; –b2-sensitive, default settings). On average, a mapping rate of approximately 55% was obtained. De novo assembly of unmapped reads with velvet (Zerbino and Birney 2008) and subsequent BLAST analyses did not yield contigs of traceable origin, suggesting that the unmapped reads are of no biological significance. Indeed, similar mapping rates have been reported by other studies using the same or very similar library preparation protocols (Beane et al. 2011; Chen et al. 2011; Malboeuf et al. 2012; Burruel et al. 2013; Leal et al. 2013; Sun et al. 2013). Gene expression analysis was performed with limma (Smyth et al. 2002), DEseq (Anders and Huber 2010), and DEseq2 (Love et al. 2014) based on count tables produced with HTseq (Anders 2010; Yang et al. 2010) against the Cobs1.4 official gene set (Schrader et al. 2014). Even though limma was originally designed for microarray data, it outcompetes some recently published software in RNAseq analysis (Rapaport et al. 2013). In addition, it more extensively supports modeling multisample comparisons in the underlying generalized linear model, so that we chose limma for the analysis of gene expression differences in this particular experiment. Read counts were converted to log2 counts per million (library size normalization), quantile normalized and precision weighted with voom (Law et al. 2014) for subsequent modeling in limma.
Functional Clustering and Gene Set Enrichment
We performed a reciprocal BLAST analysis against Drosophila annotated proteins (r5.39) to retrieve the ortholog or closest homolog (BLASTp, e value cutoff 1e-10) for each C. obscurior gene. Caste-specific gene sets were tested for functional clustering by parsing lists of the respective Drosophila closest homologs/orthologs through DAVID 6.7 (http://david.abcc.ncifcrf.gov, last accessed February 22, 2015) at an EASE cutoff of 0.01 (Huang et al. 2008, 2009). Each retrieved Annotation Cluster received a descriptive name summarizing the annotation terms.
TF Enrichment Analysis
TFBS annotations were obtained in promotor regions (0–2 kb upstream) of all 17,552 annotated protein-coding genes for 59 different TFs using pwm_scan and a P value cutoff of 2e-04 (Levy and Hannenhalli 2002; Simola, Wissler, et al. 2013). Caste-specific gene sets were then analyzed for enrichment of each TF individually using two measures (enrichment probability pE and presence probability pP) to identify TFs likely to be involved in the regulation of the query gene set (Veerla et al. 2010). The enrichment probability pE was estimated by bootstrap analysis using the total number of occurrences of a given TFBS in the promotors of a given gene set. A background distribution of occurrences was generated by randomly sampling gene lists of the same length as the query list (105 iterations). The probability pE of obtaining the number of occurrences of a TFBS by chance in the query gene set was then calculated as the percentile of the observed number compared with the background distribution (1/105 ≤ pE ≤ 1). Similarly, the presence probability pP was also estimated by bootstrap by computing the proportion of genes in a gene set (query or background) that contains at least one occurrence of a given TFBS in their promoters.
Supplementary Material
Supplementary material and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Mary Jane West-Eberhard for her encouraging comments on the manuscript and two anonymous reviewers for providing helpful comments. This work was funded by the Deutsche Forschungsgemeinschaft (He1623/31). D.F.S. was supported by a Howard Hughes Medical Institute Collaborative Innovation Award (HCIA) #2009005 (J. Liebig, S. Berger, D. Reinberg).
References
- Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada T, Gotoh T, Sugimura I, Tateno M, Nishida Y, Onuki T, Date H. De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Mol Cell Biol. 1999;19:7276–7286. doi: 10.1128/mcb.19.10.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. HTSeq: analysing high-throughput sequencing data with Python. 2010 doi: 10.1093/bioinformatics/btac166. Available from: http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo SV, Caranton OAM, de Oliveira TL, Hartfelder K. Differential expression of hypoxia pathway genes in honey bee (Apis mellifera L.) caste development. J Insect Physiol. 2011;57:38–45. doi: 10.1016/j.jinsphys.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Badisco L, Van Wielendaele P, Vanden Broeck J. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol. 2013;4:202. doi: 10.3389/fphys.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, Thummel CS. Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res. 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begna D, Fang Y, Feng M, Li J. Mitochondrial proteins differential expression during honeybee (Apis mellifera L.) queen and worker larvae caste determination. J Proteome Res. 2011;10:4263–4280. doi: 10.1021/pr200473a. [DOI] [PubMed] [Google Scholar]
- Beldade P, Mateus ARA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol. 2011;20:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- Berens AJ, Hunt JH, Toth AL. Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol Biol Evol. 2014;32:690–703. doi: 10.1093/molbev/msu330. [DOI] [PubMed] [Google Scholar]
- Boersma M, Spaak P, De Meester L. Predator-mediated plasticity in morphology, life history, and behavior of daphnia: the uncoupling of responses. Am Nat. 1998;152:237–248. doi: 10.1086/286164. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 2010;329:1068–1071. doi: 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke AF. Principles of social evolution. Oxford: Oxford University Press; 2011. [Google Scholar]
- Bowsher JH, Wray GA, Abouheif E. Growth and patterning are evolutionarily dissociated in the vestigial wing discs of workers of the red imported fire ant, Solenopsis invicta. J Exp Zool Part B: Molecular and Developmental Evolution. 2007;308:769–776. doi: 10.1002/jez.b.21200. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burruel V, Klooster KL, Chitwood J, Ross PJ, Meyers SA. Oxidative damage to rhesus macaque spermatozoa results in mitotic arrest and transcript abundance changes in early embryos. Biol Reprod. 2013;89:72. doi: 10.1095/biolreprod.113.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RC, Duncan EJ, Dearden PK. Biased gene expression in early honeybee larval development. BMC Genomics. 2013;14:903 doi: 10.1186/1471-2164-14-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. doi: 10.1016/s0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu Z, Gong S, Wu X, Taylor WL, Williams RW, Matta SG, Sharp BM. Genome-wide gene expression profiling of nucleus accumbens neurons projecting to ventral pallidum using both microarray and transcriptome sequencing. Front Neurosci. 2011;5:98. doi: 10.3389/fnins.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hu Y, Zheng H, Cao L, Niu D, Yu D, Sun Y, Hu S, Hu F. Transcriptome comparison between honey bee queen- and worker-destined larvae. Insect Biochem Mol Biol. 2012;42:665–673. doi: 10.1016/j.ibmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Corona M, Estrada E, Zurita M. Differential expression of mitochondrial genes between queens and workers during caste determination in the honeybee Apis mellifera. J Exp Biol. 1999;202:929–938. doi: 10.1242/jeb.202.8.929. [DOI] [PubMed] [Google Scholar]
- Cremer S, Heinze J. Stress grows wings: environmental induction of winged dispersal males in Cardiocondyla ants. Curr Biol. 2003;13:219–223. doi: 10.1016/s0960-9822(03)00012-5. [DOI] [PubMed] [Google Scholar]
- Cremer S, Lautenschläger B, Heinze J. A transitional stage between the ergatoid and winged male morph in the ant Cardiocondyla obscurior. Insectes Soc. 2002;49:221–228. [Google Scholar]
- Cremer S, Schrempf A, Heinze J. Competition and opportunity shape the reproductive tactics of males in the ant Cardiocondyla obscurior. PLoS One. 2011;6:e17323. doi: 10.1371/journal.pone.0017323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt TJ, Sih A, Wilson DS. Costs and limits of phenotypic plasticity. Trends Ecol Evol. 1998;13:77–81. doi: 10.1016/s0169-5347(97)01274-3. [DOI] [PubMed] [Google Scholar]
- Du Y, Schrempf A, Heinze J. Environmental determination of the male morph in the ant Cardiocondyla obscurior (Hymenoptera: Formicidae) Eur J Entomol. 2007;104:243–246. [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0001. research0001.1–0001.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer B, Elsner D, Foitzik S. Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Mol Ecol. 2013;23:151–161. doi: 10.1111/mec.12490. [DOI] [PubMed] [Google Scholar]
- Forsman A. Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity. 2014 doi: 10.1038/hdy.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol. 2007;21:394–407. [Google Scholar]
- Gilbert SF. Ecological developmental biology: developmental biology meets the real world. Dev Biol. 2001;233:1–12. doi: 10.1006/dbio.2001.0210. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Mechanisms for the environmental regulation of gene expression. Birth Defects Res C. 2005;72:291–299. doi: 10.1002/bdrc.20026. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Epel D. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland (MA): Sinauer Associates. 2009 [Google Scholar]
- Hamdoun A, Epel D. Embryo stability and vulnerability in an always changing world. Proc Natl Acad Sci U S A. 2007;104:1745–1750. doi: 10.1073/pnas.0610108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel S, Fantini J, Schweisguth F. Notch ligand activity is modulated by glycosphingolipid membrane composition in Drosophila melanogaster. J Cell Biol. 2010;188:581–594. doi: 10.1083/jcb.200907116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Aumeier V, Bodenstein B, Crewe RM, Schrempf A. Wingless and intermorphic males in the ant Cardiocondyla venustula. Insectes Soc. 2013;60:43–48. [Google Scholar]
- Heinze J, Hölldobler B. Fighting for a harem of queens: physiology of reproduction in Cardiocondyla male ants. Proc Natl Acad Sci U S A. 1993;90:8412–8414. doi: 10.1073/pnas.90.18.8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Schrempf A. Terminal investment: individual reproduction of ant queens increases with age. PLoS One. 2012;7:e35201. doi: 10.1371/journal.pone.0035201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Creason MB, Phan VH, Saba JD, Harris GL. Characterization of the Drosophila sphingosine kinases and requirement for Sk2 in normal reproductive function. J Biol Chem. 2004;279:12685–12694. doi: 10.1074/jbc.M310647200. [DOI] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 2003;130:2443–2453. doi: 10.1242/dev.00456. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Goodisman MAD. Gene expression and the evolution of phenotypic diversity in social wasps. BMC Biol. 2007;5:23. doi: 10.1186/1741-7007-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B, Wilson EO. 1990 The ants. Cambridge (MA): Harvard University Press. [Google Scholar]
- Hölldobler B, Wilson EO. 2009 The superorganism. New York: W. W. Norton & Company. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JH, Amdam GV. Bivoltinism as an antecedent to eusociality in the paper wasp genus Polistes. Science. 2005;308:264–267. doi: 10.1126/science.1109724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JH, Kensinger BJ, Kossuth JA, Henshaw MT, Norberg K, Wolschin F, Amdam GV. A diapause pathway underlies the gyne phenotype in Polistes wasps, revealing an evolutionary route to caste-containing insect societies. Proc Natl Acad Sci U S A. 2007;104:14020–14025. doi: 10.1073/pnas.0705660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Linksvayer TA. Deconstructing the superorganism: social physiology, groundplans, and sociogenomics. Q Rev Biol. 2010;85:57–79. doi: 10.1086/650290. [DOI] [PubMed] [Google Scholar]
- Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473:478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomura K, Yamauchi K. Fighting and mating behaviors of dimorphic males in the ant Cardiocondyla wroughtoni. J Ethol. 1987;5:75–81. [Google Scholar]
- Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kraut R. Roles of sphingolipids in Drosophila development and disease. J Neurochem. 2011;116:764–778. doi: 10.1111/j.1471-4159.2010.07022.x. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Kugler J. The males of Cardiocondyla emery (Hymenoptera: Formicidae) with the description of the winged male of Cardiocondyla wroughtoni (Forel) Isr J Entomol. 1983;17:1–21. [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman DS. Eukaryotic transcription factors. 5th ed. Waltham (MA): Academic Press; 2010. [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Choo Y-M, Xu P, da Silva CS, Ueira-Vieira C. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc Natl Acad Sci U S A. 2013;110:18704–18709. doi: 10.1073/pnas.1316059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebman DA, Spiegel S. Thematic review series: sphingolipids. Cross-talk at the crossroads of sphingosine-1-phosphate, growth factors, and cytokine signaling. J Lipid Res. 2008;49:1388–1394. doi: 10.1194/jlr.R800008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mamm Genome. 2002;13:510–514. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- Li J, Wu J, Begna Rundassa D, Song F, Zheng A, Fang Y. Differential protein expression in honeybee (Apis mellifera L.) larvae: underlying caste differentiation. PLoS One. 2010;5:e13455. doi: 10.1371/journal.pone.0013455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication. Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-L, Lu W-C, Brummel TJ, Yuh C-H, Lin P-T, Kao T-Y, Li F-Y, Liao P-C, Benzer S, Wang H-D. Reduced expression of alpha-1,2-mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell. 2009;8:370–379. doi: 10.1111/j.1474-9726.2009.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, Pesko KN, Boutwell CL, DeVincenzo JP, Ebel GD, et al. Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res. 2012;41:e13. doi: 10.1093/nar/gks794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- Menuz V, Howell KS, Gentina S, Epstein S, Riezman I, Fornallaz-Mulhauser M, Hengartner MO, Gomez M, Riezman H, Martinou JC. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science. 2009;324:381–384. doi: 10.1126/science.1168532. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Nikolova-Karakashian M, Schmelz EM, Morgan ET, Stewart J. Regulation of cytochrome P450 expression by sphingolipids. Chem Phys Lipids. 1999;102:131–139. doi: 10.1016/s0009-3084(99)00081-x. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Sultan S, Foster S, Ledon-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW. The role of developmental plasticity in evolutionary innovation. Proc R Soc Lond B Biol Sci. 2011;278:2705–2713. doi: 10.1098/rspb.2011.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech M-B, Kruse R, Harvald EB, Olsen ASB, Gallego SF, Hannibal-Bach HK, Ejsing CS, Færgeman NJ. Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One. 2013;8:e70087. doi: 10.1371/journal.pone.0070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat S, Hopfen C, McGregor AP. The function and evolution of Wnt genes in arthropods. Arthropod Struct Dev. 2010:1–7. doi: 10.1016/j.asd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Mutti NS, Dolezal AG, Wolschin F, Mutti JS, Gill KS, Amdam GV. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J Exp Biol. 2011;214:3977–3984. doi: 10.1242/jeb.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettler J, Suefuji M, Heinze J. The evolution of alternative reproductive tactics in male Cardiocondyla ants. Evolution. 2010;64:3310–3317. doi: 10.1111/j.1558-5646.2010.01090.x. [DOI] [PubMed] [Google Scholar]
- Ometto L, Shoemaker D, Ross KG, Keller L. Evolution of gene expression in fire ants: the effects of developmental stage, caste, and species. Mol Biol Evol. 2011;28:1381–1392. doi: 10.1093/molbev/msq322. [DOI] [PubMed] [Google Scholar]
- Page RE, Peng C. Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp Gerontol. 2001;36:695–711. doi: 10.1016/s0531-5565(00)00236-9. [DOI] [PubMed] [Google Scholar]
- Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K, Amdam GV. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS One. 2007;2:e509. doi: 10.1371/journal.pone.0000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penick CA, Liebig J. Regulation of queen development through worker aggression in a predatory ant. Behav Ecol. 2012;23:992–998. [Google Scholar]
- Penick CA, Prager SS, Liebig J. Juvenile hormone induces queen development in late-stage larvae of the ant Harpegnathos saltator. J Insect Physiol. 2012;58:1643–1649. doi: 10.1016/j.jinsphys.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Pepperl J, Reim G, Lüthi U, Kaech A, Hausmann G, Basler K. Sphingolipid depletion impairs endocytic traffic and inhibits Wingless signaling. Mech Dev. 2013;130:493–505. doi: 10.1016/j.mod.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol Evol. 2010;25:459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Phan VH, Herr DR, Panton D, Fyrst H, Saba JD, Harris GL. Disruption of sphingolipid metabolism elicits apoptosis-associated reproductive defects in Drosophila. Dev Biol. 2007;309:329–341. doi: 10.1016/j.ydbio.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar R, San Mauro D, Dijkstra MB, Huang MH, Wheeler DE, Hiou-Tim F, Khila A, Cournoyea M, Abouheif E. Ancestral developmental potential facilitates parallel evolution in ants. Science. 2012;335:79–82. doi: 10.1126/science.1211451. [DOI] [PubMed] [Google Scholar]
- Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppell O, Heinze J. Alternative reproductive tactics in females: the case of size polymorphism in winged ant queens. Insectes Soc. 1999;46:6–17. [Google Scholar]
- Sasamura T, Matsuno K, Fortini ME. Disruption of Drosophila melanogaster lipid metabolism genes causes tissue overgrowth associated with altered developmental signaling. PLoS Genet. 2013;9:e1003917. doi: 10.1371/journal.pgen.1003917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD, Pigliucci M. Gene regulation, quantitative genetics and the evolution of reaction norms. Evol Ecol. 1995;9:154–168. [Google Scholar]
- Schlichting CD, Pigliucci M. 1998 Phenotypic evolution: a reaction norm perspective. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Schlichting CD, Wund MA. Phenotypic plasticity and epigenetic marking: an assessment of evidence for genetic accommodation. Evolution. 2014;68:656–672. doi: 10.1111/evo.12348. [DOI] [PubMed] [Google Scholar]
- Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K, Weichselgartner T, Kemena C, Stökl J, Schultner E, et al. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun. 2014;5:5495. doi: 10.1038/ncomms6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf A, Heinze J. Proximate mechanisms of male morph determination in the ant Cardiocondyla obscurior. Evol Dev. 2006;8:266–272. doi: 10.1111/j.1525-142X.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- Seifert B. The ant genus Cardiocondyla (Insecta: Hymenoptera: Formicidae)—a taxonomic revision of the C. elegans, C. bulgarica, C. batesii, C. nuda, C. shuckardi, C. stambuloffii, C. wroughtonii, C. emeryi, and C. minutior species groups. Ann Nat Hist Mus Wien Ser B Bot Zool. 2003:203–338. [Google Scholar]
- Shi YY, Yan WY, Huang ZY, Wang ZL, Wu XB, Zeng ZJ. Genomewide analysis indicates that queen larvae have lower methylation levels in the honey bee (Apis mellifera) Naturwissenschaften. 2012;100:193–197. doi: 10.1007/s00114-012-1004-3. [DOI] [PubMed] [Google Scholar]
- Simola DF, Wissler L, Donahue G, Waterhouse RM, Helmkampf M, Roux J, Nygaard S, Glastad KM, Hagen DE, Viljakainen L, et al. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 2013;23:1235–1247. doi: 10.1101/gr.155408.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Ye C, Mutti NS, Dolezal K, Bonasio R, Liebig J, Reinberg D, Berger SL. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 2013;23:486–496. doi: 10.1101/gr.148361.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Gill SJ. Developmental plasticity—developmental conversion versus phenotypic modulation. Am Zool. 1983;23:47–55. [Google Scholar]
- Smyth GK, Ritchie M, Thorne N, Wettenhall J, Shi W, Hu Y. 2002 Limma: linear models for microarray data. http://www.bioconductor.org/packages/release/bioc/vignettes/limma/inst/doc/;?>usersguide.pdf. [Google Scholar]
- Spiegel S, Merrill AH. Sphingolipid metabolism and cell growth regulation. FASEB J. 1996;10:1388–1397. doi: 10.1096/fasebj.10.12.8903509. [DOI] [PubMed] [Google Scholar]
- Stabe HA. The rate of growth of worker, drone and queen larvae of the honeybee, Apis mellifera Linn. J Econ Entomol. 1930;23:447–453. [Google Scholar]
- Stuart R, Francoeur A, Loiselle R. Lethal fighting among dimorphic males of the ant, Cardiocondyla wroughtonii. Naturwissenschaften. 1987;74:548–549. [Google Scholar]
- Suefuji M, Cremer S, Oettler J, Heinze J. Queen number influences the timing of the sexual production in colonies of Cardiocondyla ants. Biol Lett. 2008;4:670–673. doi: 10.1098/rsbl.2008.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Asmann YW, Nair A, Zhang Y, Wang L, Kalari KR, Bhagwate AV, Baker TR, Carr JM, Kocher J-P A, et al. Impact of library preparation on downstream analysis and interpretation of RNA-Seq data: comparison between Illumina PolyA and NuGEN ovation protocol. PLoS One. 2013;8:e71745. doi: 10.1371/journal.pone.0071745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco P, Jiang J, Wang J, Jazwinski SM, Johnson TE. Genetic analysis of hyl-1, the C. elegans homolog of LAG1/LASS1. Age. 2008;30:43–52. doi: 10.1007/s11357-008-9046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji N, Yamauchi K, Yamamura N. A mathematical model for wing dimorphism in male Cardiocondyla ants. J Ethol. 1994;12:19–24. [Google Scholar]
- Veerla S, Ringnér M, Höglund M. Genome-wide transcription factor binding site/promoter databases for the analysis of gene sets and co-occurrence of transcription factor binding motifs. BMC Genomics. 2010;11:145. doi: 10.1186/1471-2164-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls SM, Attle SJ, Brulte GB, Walls ML, Finley KD, Chatfield DA, Herr DR, Harris GL. Identification of sphingolipid metabolites that induce obesity via misregulation of appetite, caloric intake and fat storage in Drosophila. PLoS Genet. 2013;9:e1003970. doi: 10.1371/journal.pgen.1003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;30:249–278. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proc Natl Acad Sci U S A. 2005;102:6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman DW, Agrawal AA. 2009 What is phenotypic plasticity and why is it important. In: Whitman D, Ananthakrishnan NT, editors. Phenotypic plasticity of insects. Enfield (NH): Science Publishers. Vol. 10. p. 1–63. [Google Scholar]
- Wolschin F, Mutti NS, Amdam GV. Insulin receptor substrate influences female caste development in honeybees. Biol Lett. 2011;7:112–115. doi: 10.1098/rsbl.2010.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K, Asano Y, Lautenschläger B, Trindl A, Heinze J. A new type of male dimorphism with ergatoid and short-winged males in Cardiocondyla cf. kagutsuchi. Insectes Soc. 2005;52:274–281. [Google Scholar]
- Yamauchi K, Furukawa T, Kinomura K, Takamine H, Tsuji K. Secondary polygyny by inbred wingless sexuals in the dolichoderine ant Technomyrmex albipes. Behav Ecol Sociobiol. 1991;29:313–319. [Google Scholar]
- Yamauchi K, Ishida Y, Hashim R, Heinze J. Queen-queen competition by precocious male production in multiqueen ant colonies. Curr Biol. 2006;16:2424–2427. doi: 10.1016/j.cub.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Yang Q, Gong Z-J, Zhou Y, Yuan J-Q, Cheng J, Tian L, Li S, Lin X-D, Xu R, Zhu Z-R, Mao C. Role of Drosophila alkaline ceramidase (Dacer) in Drosophila development and longevity. Cell Mol Life Sci. 2010;67:1477–1490. doi: 10.1007/s00018-010-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shen H, Sewell AK, Kniazeva M, Han M. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. eLife. 2013;2:e00429. doi: 10.7554/eLife.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.