Abstract
Short hairpin RNAs (shRNAs) are widely used for gene knockdown by inducing the RNA interference (RNAi) mechanism, both for research and therapeutic purposes. The shRNA precursor is processed by the RNase III-like enzyme Dicer into biologically active small interfering RNA (siRNA). This effector molecule subsequently targets a complementary mRNA for destruction via the Argonaute 2 (AGO2) complex. The cellular role of Dicer concerns the processing of pre-miRNAs into mature microRNA (miRNA). Recently, a non-canonical pathway was reported for the biogenesis of miR-451, which bypasses Dicer and is processed instead by the slicer activity of AGO2, followed by the regular AGO2-mediated mRNA targeting step. Interestingly, shRNA designs that are characterized by a relatively short basepaired stem also bypass Dicer to be processed by AGO2. We named this design AgoshRNA as these molecules depend on AGO2 both for processing and silencing activity. In this study, we investigated diverse mechanistic aspects of this new class of AgoshRNA molecules. We probed the requirements for AGO2-mediated processing of AgoshRNAs by modification of the proposed cleavage site in the hairpin. We demonstrate by deep sequencing that AGO2-processed AgoshRNAs produce RNA effector molecules with more discrete ends than the products of the regular shRNA design. Furthermore, we tested whether trimming and tailing occurs upon AGO2-mediated processing of AgoshRNAs, similar to what has been described for miR-451. Finally, we tested the prediction that AgoshRNA activity, unlike that of regular shRNAs, is maintained in Dicer-deficient cell types. These mechanistic insights could aid in the design of optimised AgoshRNA tools and therapeutics.
Keywords: AgoshRNA, Argonaute 2, Dicer, RNA processing, shRNA
Introduction
RNA interference (RNAi) is an evolutionary conserved mechanism in which double stranded RNA (dsRNA) triggers sequence-specific gene silencing at the post-transcriptional level.1,2 The key players of this pathway are small non-coding RNAs and the largest class consists of the miRNAs.3 The primary miRNA transcripts are processed by the nuclear Microprocessor complex, consisting of the RNAse III-like enzyme Drosha and its dsRNA-binding partner DGCR8, into precursor miRNAs of ∼70 nucleotides (nt) that are characterized by a hairpin structure.4,5 This pre-miRNA is exported from the nucleus to the cytoplasm by Exportin5 and processed further by the RNase III-like Dicer enzyme–in association with the TAR RNA binding protein (TRBP) and the protein activator of PKR (PACT)–into miRNA duplexes of ∼20–24 base pairs (bp) with 2 nt 3′-overhangs. The miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) by association with Argonaute 2 (AGO2) protein.6,7 Depending on the thermodynamic properties of the miRNA duplex, preferentially one strand will be selected as mature miRNA in guiding the RISC complex to complementary mRNA targets.8,9
RNAi can also be triggered by gene constructs containing an RNA polymerase III cassette to express a short transcript that adopts a hairpin conformation of approximately 21 bp with a 2 nt 3′-overhang.10-12 Such shRNA molecules skip Drosha processing and enter the RNAi pathway at the Dicer processing step, resulting in the generation of an siRNA that triggers mRNA cleavage (Fig. 1A, top). The cellular miRNAs require Drosha and subsequently Dicer to yield the miRNA duplex (Fig. 1A, bottom).4,5 Besides these canonical shRNA/miRNA processing pathways, several alternative mechanisms have recently been described. Drosha-independent processing has been reported for several pre-miRNAs, namely mirtrons, tRNAZ and small nucleolar RNAs (snoRNAs).13-18 Although Dicer was considered to be essential for miRNA processing, recent evidence indicates that miR-451 is processed in a Dicer-independent, but AGO2-dependent manner.19-24 Among the pre-miRNAs, pre-miRNA-451 has as special features a short basepaired stem of only 17 bp and a relatively small loop of 4 nt and is processed by AGO2 at the 3′-strand of the hairpin between the 10th and 11th bp, yielding a ∼30 nt miRNA (Fig. 1B, bottom). Subsequent 3′-uridylation and trimming creates a ∼22–26 mature miR-451. A role for the poly(A)-specific ribonuclease (PARN) has recently been implied in the trimming reaction, but this modification is not necessary for miR-451 activity.25
Figure 1.

Canonical (Dicer-mediated) and non-canonical (AGO-2 mediated) paths for shRNA and miRNA processing (A) Canonical Dicer-mediated processing of shRNAs and pre-miRNAs. Top: a regular short hairpin RNA (shRNA) of 21 bp in length is processed by Dicer into siRNAs. Bottom: a regular pre-miRNA is processed by Dicer into the mature miRNA duplex. The guide strand (thick line) of the siRNA/miRNA duplex will subsequently be incorporated into RISC to induce RNAi silencing. (B) Non-canonical AGO2-mediated processing of AgoshRNAs and miR-451. Top: AgoshRNAs are hairpins of only 19 bp with a small terminal loop that bypass Dicer-processing and undergo cleavage by AGO2 between bp 10 and 11 on the 3′ side of the duplex (◂). Bottom: miR-451 is the founding member of a class of Dicer-independent, AGO2-processed miRNAs. A thick line represents the guide strand. Hairpin characteristics shared with the AgoshRNA design are the short stem and small loop. The miR-451 is processed further by tailing and trimming as indicated.
A similar non-canonical class of shRNAs was described more recently that is also processed in a Dicer-independent, AGO2-dependent manner (Fig. 1B, top). In analogy to miR-451, these so-called AgoshRNAs are typically shorter (17–19 bp) than regular shRNAs and have a small loop of 3–5 nt.26 We recently demonstrated the modulating role of a top G-U base pair, again similar to recent miR-451 findings.27,28 Other structural elements or sequence motifs that trigger non-canonical AGO2 processing remain largely unknown. Furthermore, it remains unclear whether the processed AgoshRNA is subject to uridylation and trimming upon AGO2 cleavage. In this study, we tested several aspects of AgoshRNA biology. The importance of a perfectly basepaired duplex around the AGO2 cleavage site was probed. We also tested the prediction that AgoshRNA products may have more defined ends than regular shRNA products and whether other typical miR-451 properties (trimming/tailing) occur during AgoshRNA processing. Furthermore, we confirmed the prediction that AgoshRNAs retain full activity in a Dicer-deficient cell line.
Results
Probing AGO2 processing by disruption of the basepaired cleavage site in AgoshRNAs
We previously mapped the AGO2 cleavage site in AgoshRNAs between bp 10 and 11 at the 3′ side of the 19 bp stem.26 This position is marked in the prototype wild-type (wt) AgoshRNA molecule shown in Figure 2 and mimics the well-studied AGO2 cleavage of regular siRNA-mRNA duplexes, which also occurs between bp 10 and 11.29-31 To test whether basepairing around the cleavage site is needed for AGO2-mediated processing, we designed 5 AgoshRNA mutants with a local destabilization near the center of the basepaired duplex (Fig. 2). Mutants 1 and 2 have a single nt substitution in the 3′ arm of the duplex to create a mismatch at bp position 11 or 10, respectively. The local stem destabilization was gradually increased in mutants 3, 4 and 5 that create a symmetric 4 nt, 6 nt and 8 nt internal loop, respectively. The predicted thermodynamic stability (ΔG in kcal/mol) of the hairpins is plotted below the structures, illustrating the gradual destabilization from −36.9 to −26.3 kcal/mol.
Figure 2.

Design of AgoshRNA cleavage site mutants The wild-type (wt) AgoshRNA is based on the AgoshRT5 hairpin.26 The AGO2 cleavage site is marked by an arrow. Mutants 1–5 have one or multiple mismatches introduced near the cleavage site. The mutated nt are marked in a gray box. The thermodynamic stability of the wt and mutant hairpins is indicated below (ΔG in kcal/mole). Deep sequencing of RNA mutant 1 products revealed an additional 5′-terminal C residue due to a-1 shift of the transcriptional start site in the H1 promoter.
We first analyzed the AgoshRNA processing products by northern blotting using probes that detect either the 3′ or 5′ side of the hairpin (Fig. 3, upper and lower panel, respectively). As expected, no regular Dicer products of ∼21 nt could be detected for the wt and mutant AgoshRNAs with these probes. As a control, we included a regular shRNA of 21 bp (shRT5) that produces such 21 nt siRNA products from both sides of the hairpin, but most prominently from the 3′ side (guide strand). The wt and mutant AgoshRNAs show only a vague signal of ∼40 nt with the 3′ side probe, which likely reflects the precursor transcript. The migration of these precursor RNAs on the denaturing gel seems to be affected by duplex disruption, with the slowest mobility for the transcripts with the largest internal loop.
Figure 3.
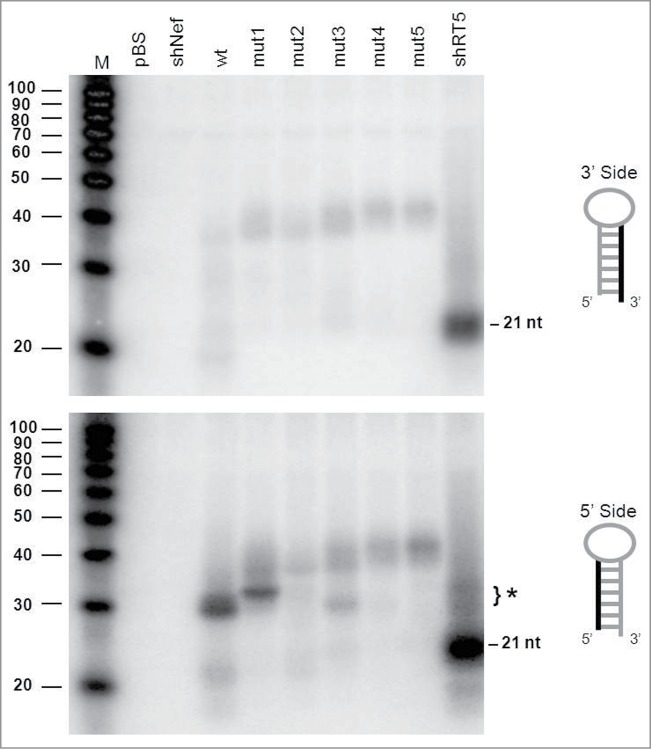
Northern blot analysis of the AgoshRNA cleavage site mutants An irrelevant shRNA (shNef) and Bluescript (pBS) were used as negative controls. The regular 21 nt shRNA products are indicated and the special AGO2-dependent ∼30 nt AgoshRNA products are marked (*). Lane M contains an RNA size ladder. The hairpin side that was probed is marked in the cartoon on the right.
The 5′ side probe detected the predicted AGO2-product of 33 nt for the wt AgoshRNA (marked with * in Fig. 3, bottom panel). Interestingly, mutant 1 with a mismatch at bp 11 produces a slightly longer RNA product. This may suggest that AGO2 cleavage was shifted toward the bottom of the hairpin stem between bp 9 and 10, thus creating an extended RNA product. However, AGO2 cleavage is known to be very precise and we therefore performed deep sequencing of the RNA products generated by mutant 1. Much to our surprise, the mutant 1, 2 and 3 transcripts were extended by a −1 shift in the transcription start site of the H1 promoter (Fig. 4), which explains the slightly longer RNA product. The shift in transcription start site is unexpected as the H1 promoter is known to have all important sequence motifs located upstream of the start site.32 Processing is reduced for mutant 1. The single mismatch at bp position 10 in mutant 2 completely abolished AGO2-mediated cleavage. Mutant 3 with a symmetric internal loop at bp positions 10 and 11 showed a minor band on the RNA gel blot corresponding to the original cleavage product of ∼30 nt. This was confirmed by deep sequencing of the transcripts. Mutants 4 and 5 with bigger internal loops produced no specific products. Loss of the specific signal for mutants 2–5 correlates with the appearance of a faint signal of the ∼40 nt precursor. Again, we observed the minor shift in migration behavior of these hairpin precursors with extended internal loops (Fig. 3).
Figure 4.
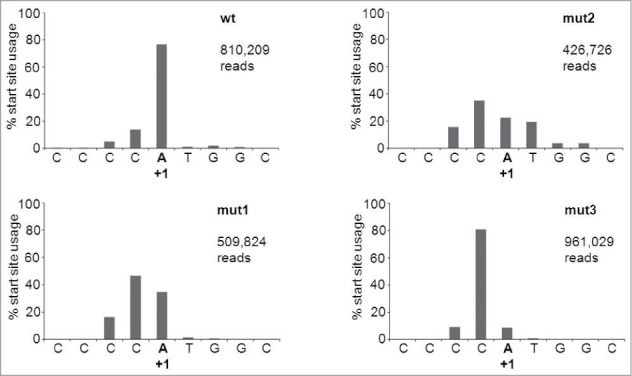
Transcription start site usage in AgoshRNA constructs. SOLiD deep sequencing was performed on the wt and mut 1, 2 or 3 AgoshRT5 constructs. The natural starting position is marked as +1.
We next tested the knockdown ability of these AgoshRNA hairpins on luciferase reporters with the sense or antisense target to probe the activity of the 3′ or 5′ strand, respectively (Fig. 5A, left and right panel). The 3′ strand of the AgoshRNA hairpin carries the mutations that result in incomplete pairing with the luciferase sense target, whereas the 5′ strand is not modified and thus perfectly complementary to the antisense luciferase reporter. We co-transfected an AgoshRNA and luciferase construct in HEK 293T cells. The pBluescript plasmid (pBS) mock-transfection served as negative control for which luciferase expression was set at 100% (Fig. 5A). The unrelated shNef served as another negative control. No active 3′ arm derived siRNA activity was scored for the wt and mutant AgoshRNAs on the luciferase-sense reporter, consistent with the absence of 3′ side processing products (Fig. 3). The shRT5 served as positive control for targeting the sense reporter via the dominant 5′ guide strand and the antisense reporter via the 3′ passenger strand.
Figure 5.

Knockdown activity of the AgoshRNA cleavage site mutants (A) The knockdown activity of the indicated AgoshRNAs was determined by co-transfection of 25 ng of hairpin construct with 100 ng of a luciferase reporter. This reporter encoded either the sense or antisense target sequence, which monitors the activity of the AgoshRNA 3′ or 5′ strand, respectively. Luciferase values measured for the pBS control were set at 100%. The mean values and standard deviations are based on 3 independent experiments. (B) The AgoshRNA knockdown activity (5′ strand) was carefully determined by transfecting different amounts of the indicated constructs (1.25, 5 and 20 ng) together with a fixed amount (100 ng) of the antisense reporter. For each condition, luciferase values obtained for pBS were set at 100%. The averages and standard deviations of 3 independent transfections are shown.
The wt AgoshRNA showed potent luciferase-antisense knockdown, at least comparable to that of the shRNA controls (Fig. 5A). Fairly good activity was scored for mutant 1, but most activity was lost for the other mutants, consistent with the gradual disappearance of the processed AgoshRNAs on northern blot (Fig. 3). To carefully determine the knockdown abilities of the different AgoshRNA variants, we performed another transfection series in which the AgoshRNA construct was titrated (Fig. 5B: 1.25, 5.0 and 20 ng). A clear dose-dependent inhibition was measured for wt and mutant 1, but none of the other hairpins showed significant reporter knockdown activity.
More precise ends for AgoshRNA than regular shRNAs
Recent literature indicates that there is imprecision in the 5′ and 3′ ends of processed shRNA molecules, which can have a serious impact on the silencing activity and target specificity.33-35 In particular the Dicer-mediated processing introduces quite some sequence variability due to imprecise cleavage events and we argued that more precise processing can be expected for AGO2-processed AgoshRNA molecules because the AGO2 enzyme is known to accurately cleave duplexes between bp 10 and 11.26,28,36 The earlier result with mutant 1 indicated that even a deliberate opening of the basepaired stem near the AGO2 processing site does not affect the actual site of cleavage, but only the processing efficiency. We decided to investigate this issue in more detail by deep sequencing.
We transfected 293T cells with the plasmids encoding either the 21 bp shRNA-RT5 or the 19 bp AgoshRNA-RT5 in combination with a flag-tagged AGO2 construct. The AGO2 complexes were pulled down after 2 days, size-selected for small RNA molecules and sequenced using SOLiD deep sequencing. We confirmed the inaccuracy of Dicer-mediated processing of the regular shRNA (Fig. 6A). Significant cleavage site variation is apparent on both the 5′ and 3′ arms of the molecule, consistent with previous reports.33-35 In fact, only few of the cleavage events are those expected based on the shRNA design (18% on the 5′ side and 0.01% on the 3′ side). Most 5′ and 3′ cleavages occur 2 bp lower in the duplex than expected, which may be related to breathing of the top U-G bp or imprecise 5′/3′ ends of the shRNA transcript as reported.37 In contrast, quite precise cleavage occurs between bp 10 and 11 for the new AgoshRNA design (Fig. 6B). The most abundant byproduct represents a shift to cleavage between bp 11 and 12 and accounts for just 5% of the reads. Similar results were apparent for mutants 1–3. Precise cleavage was also observed for mutants 1 and 3. The signal was lost for mutant 2, consistent with the RNA gel blot results (Fig. 3).
Figure 6.
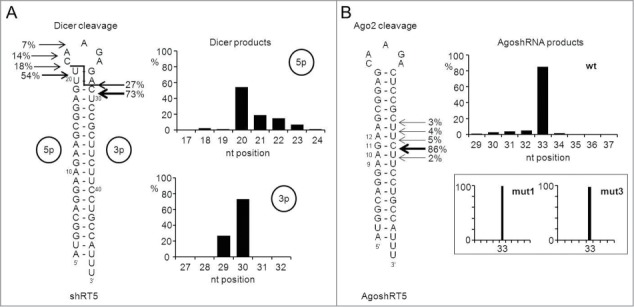
Probing the ends of a regular shRNA versus AgoshRNA molecule. SOLiD deep sequencing was performed on small RNAs isolated from shRT5 (21/5) or AgoshRT5 (19/5) expressing cells. (A) The predicted structure of the short hairpin shRT5. The predicted 2-nt staggered Dicer cleavage site is indicated with a line. Depicted are the percentages of the total reads that either end (5′ side) or start (3′ side) around the Dicer cleavage-site. The cut off was set at 5%. These percentages are also presented in a bar-graph. (B) Depicted on the predicted structure of AgoshRT5 are the percentages of the total reads that end around the AGO2 cleavage site on the 3′ side of the hairpin. The cut off was set at 1%. These percentages are also depicted in a bar-graph. The results for mutants 1 and 3 are shown as inserts.
Does trimming and tailing occur for AgoshRNAs?
The AGO2-processed miR-451 of ∼30 nt is further trimmed to a mature product of ∼22–26 nt.20,25 Because AgoshRNA processing closely resembles that of miR-451, we wondered whether tailing and trimming occurred. In Drosophila, trimming and tailing of a miRNA is triggered by extensive complementarity between the AGO1-bound miRNA and the target mRNA.38 Thus, we tested whether the inclusion of target RNA could induce this process. We compared 2 AgoshRNA constructs of 19 bp and a terminal loop of 3 nt (19/3) or 5 nt (19/5) with 2 regular shRNAs of 21 bp and 22 bp (21/5 and 22/5). These constructs were co-transfected with or without a luciferase reporter that encode the complementary target RNA for the AgoshRNAs. Processing of the AgoshRNAs yielded the typical AGO2-products of approximately ∼30 nt that are detected by the 5′ side probe (marked with * in Fig. 7, lower panel). As expected for AGO2-mediated processing on the 3 side of the stem, the larger loop in 19/5 results in a 2 nt extended product compared to that of construct 19/3. Most importantly, the products remain of the same discrete size in the presence of a complementary target RNA, providing no evidence that tailing and trimming occurs upon AGO2-mediated processing. Regular shRNA products of approximately 21 bp were detected for the 21/5 and 22/5 constructs with both the 5′ and 3′ side probes.
Figure 7.
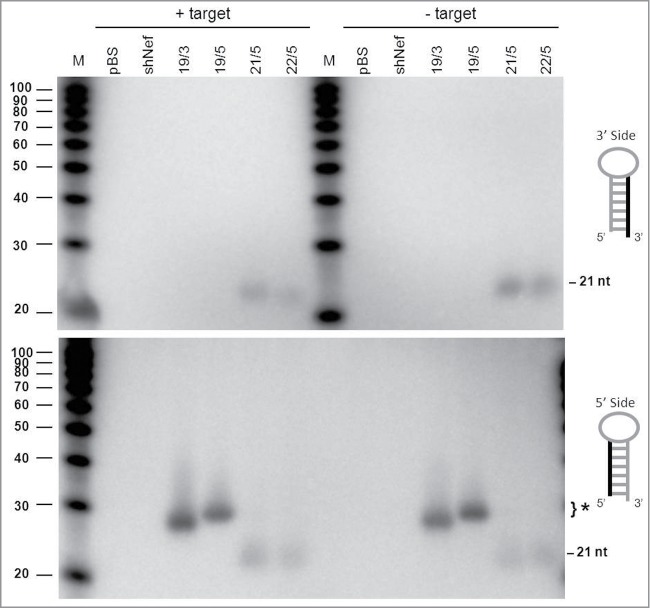
Processing of the 3′/5′ strands of AgoshRNAs in the presence of a complementary target RNA. HEK 293T cells were transfected with regular shRNA (21/5 and 22/5) or AgoshRNA (19/3 and 19/5) constructs and a reporter construct encoding a transcript with a complementary target sequence. As a control, a reporter encoding an irrelevant target sequence (shNef) was used. Total RNA was isolated from the transfected cells and subjected to RNA gel blot analysis to analyze the cleavage products and putative modification by tailing and/or trimming. Lane M contains an RNA size ladder. The hairpin side that was probed is marked in the cartoon on the right.
Full AgoshRNA activity in Dicer-minus cells
The Dicer-independence of AgoshRNAs predicts that these research tools or therapeutic agents can be used in Dicer-deficient (Dcr-) cell types. To critically test this idea, we compared the efficacy of AgoshRNA and regular shRNA constructs in the HCT-116 cell line in which Dicer is affected by disruption of the helicase domain.39 We again used related constructs that differ in size of the hairpin stem: the regular shRNA 21/5 and the AgoshRNA 19/5 of which the activity can be scored on the same luciferase reporter. The knockdown ability of the regular shRNA (21/5) was reduced in these Dcr- cells (Fig. 8, left panel). In contrast, AgoshRNA (19/5) knockdown activity was not affected (Fig. 8, right panel). Notably, the AgoshRNA 19/5 activity was even enhanced in Dcr- cells, which may cautiously suggest that AGO2 loading occurs more efficiently in the absence of competitive Dicer loading.
Figure 8.

AgoshRNA activity in Dicer-deficient cells HCT-116 cells that express the wt Dicer protein and a non-functional Dicer protein (Dcr-) were transfected with the regularly Dicer-processed shRNA construct (21/5) or the AGO2-processed AgoshRNA construct (19/5) together with a luciferase reporter encoding the sense or antisense target sequence, respectively. Activity scored in transfection of the luciferase reporter with the control plasmid pBS was set at 100%. The averages and standard deviations of 3 independent transfections are shown.
Discussion
In this study we probed several mechanistic aspects of the recently described class of Dicer-independent shRNAs that are processed by AGO2. These AgoshRNA molecules are characterized by a relatively short basepaired stem that likely prevents recognition by Dicer, such that the alternative AGO2-mediated processing route becomes operational. For AGO2 recognition of the hairpin one would expect the requirement of perfect basepairing around the actual cleavage site between bp position 10 and 11. This was tested by introduction of a single or multiple mismatches around the cleavage site in an AgoshRNA, followed by northern blot analysis and activity testing on a matching reporter construct. Indeed, AgoshRNA mutants with 2–4 mismatches in the center of the duplex could not be processed by AGO2 and lost all luciferase knockdown activity. This mimics the exquisite dependence on perfect basepairing for AGO2 cleavage of regular siRNA-mRNA duplexes.29-31 A mixed pattern was observed for the mutants with a single mismatch. Mutant 1 with a mismatch at bp position 11 directly above the cleavage site remained fairly active, whereas mutant 2 with a mismatch at bp position 10 directly below the cleavage site abolished AGO2 processing and lost all reporter silencing activity. Deep sequencing indicated that the introduction of the mismatches near the AGO2 cleavage site did not affect the cleavage site specificity.
We did observe an unexplained effect of the single nt variation in mutant 1 on the start site of transcription from the H1 promoter. Deep sequencing revealed a distinct shift from the regular A at position +1 to the neighboring C at position −1, which explains the notable shift in the RNA band observed on the RNA gel blot of Figure 3. Heterogeneity of transcription start site usage has been reported for H1 transcription35, but we observed an unexpected shift in initiation site that is induced by a single mutation some 33 nt further downstream in the double-stranded DNA of the H1 promoter. It remains to be seen whether this shift is a coincidence, e.g. by inactivation or activation of a binding site for a transcription factor, or that such effects can be expected more generally. This phenomenon adds further potential sequence variation to the already heterogeneous small RNAi inducers.
The non-canonical processing pathways of man-made AgoshRNAs and the natural miR-451 are very similar in the sense that both precursors are processed by AGO2 instead of Dicer.36 The AGO2-cleaved miR-451 is subject to 3′-5′ exonucleolytic trimming by the poly(A)-specific ribonuclease (PARN) to its mature length of 23–26 nt.25 We tested whether trimming also occurs for an AgoshRNA, but did not find evidence for this based on northern blotting and a previous sequence analysis of small RNAs that were in complex with wild-type and catalytically inactive AGO2.26 As trimming could depend on the presence of a complementary target RNA38, we also studied AgoshRNA processing in the presence of target RNA, but no evidence for trimming was obtained. For miR-451, it was suggested that the actual sequence of the 3′ arm of the hairpin stem affects trimming, in particular the presence of uridines at specific stem positions.25,40 It can therefore not be excluded that other AgoshRNA molecules will be subjected to trimming. Recently, Yoda et al showed that 3′ trimming of AGO2-cleaved pre-miRNAs is not essential for target silencing.25 It remains to be determined whether trimming does modulate the activity of miR-451.
We previously discussed the putative advantages of AgoshRNAs over regular shRNAs.26,36 The major advantage is the fact that only a single RNAi-active guide strand is produced by an AgoshRNA, which is an important safety feature to restrict off target effects caused by the passenger strand. It was proposed that AgoshRNAs would be the silencing method of choice for cell types that express a limited amount or no Dicer at all, such as monocytes.41 In this study, we confirm that AgoshRNAs, unlike regular shRNAs, remain fully active in such Dicer-deficient cells. In fact, superior AgoshRNA activity was scored in Dcr- cells, likely because of the absence of competitive loading in Dicer. We suggested that toxicity due to saturation of Dicer as critical component of the cellular RNAi pathway will not likely occur with AgoshRNAs and this potential advantage was recently confirmed experimentally.42 We reasoned that the smaller AgoshRNAs may exhibit an improved safety profile because innate immunity mechanisms will not be triggered that easily. On the other hand, it cannot be excluded that the typical AgoshRNA product, consisting of a duplex of 9 bp with 10 nt overhang, is recognized by innate sensors. Yet another potential advantage that was proposed is that AGO2-mediated processing of shRNAs may yield more precise ends compared to Dicer processing36, which shows considerable variation.33,34 It is notoriously challenging to design RNAi reagents that are processed at a precise position, e.g., recent evidence indicates that the exact composition of the Dicer complex influences the actual Dicer cleavage site.43,44 In this study, we demonstrate that AgoshRNA processing yields much more precise RNA products. Overall, our combined results and those of other laboratories42 suggest that there is a future for potent and specific AgoshRNA reagents in basic biology research and therapeutic applications.
Materials and Methods
DNA constructs
The wt and mutant AgoshRNA constructs were made by annealing of complementary oligonucleotides (containing the BamHI and HindIII restriction enzyme sites) and inserting them into the BglII and HindIII sites of the pSUPER vector.12 All DNA constructs were verified by sequence analysis using the BigDye Terminator Cycle Sequencing kit (ABI, Foster City, CA, USA). Hairpin RNA constructs were sequenced using a sample denaturation temperature of 98˚C and upon addition of 1 M Betaine. The RNA secondary structure and the thermodynamic stability of the shRNA-AgoshRNA transcripts were predicted with the Mfold program.45
Firefly luciferase reporters (based on pGL3; Promega, Madison, WI, USA) were constructed by insertion of a 50- to 70-bp fragment with the actual 19-nt target sequence in the center into the 3′ untranslated region between the EcoRI and PstI sites of pGL3.46 The Luc-RT5 sense and antisense reporters were described previously.47
Cell culture and DNA transfections
Human embryonic kidney (HEK) 293T, HCT-116 wt and Dcr-48 adherent cells were grown as monolayer in Dulbecco's modified Eagle's medium (Life Technologies, Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), streptomycin (100 μg/ml) and minimal essential medium non-essential amino acids (DMEM/10% FCS) in a humidified chamber at 37°C and 5% CO2.
For siRNA analysis by RNA gel blotting, HEK 293T cells were plated one day before transfection in T25 flasks at a density of 1.5 × 106 cells in 4 ml DMEM/10% FCS without antibiotics. The next day, cells were transfected with 5 μg of the hairpin RNA constructs with Lipofectamine 2000 reagent (Invitrogen) as suggested by the manufacturer.
For luciferase assays, HEK 293T, HCT-116 wt or Dcr- cells were seeded one day prior transfection in 24-wells plates at a density of 1.4 × 105 cells per well in 500 μl DMEM/10%FCS without antibiotics. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
siRNA detection by northern blot analysis
RNA gel blotting was performed as previously described.49 Briefly, 1.5 × 106 HEK 293T cells were transfected with 5 μg of wt and mutant AgoshRNA construct using Lipofectamine 2000. Total cellular RNA was extracted 2 d post-transfection with the mirVana miRNA isolation kit (Life Technologies, Ambion and Austin, TX) according to the manufacturer's instructions. The RNA concentration was measured by Nanodrop 1000 (Thermo Fisher Scientific). For northern blot analysis, 15 μg total RNA was electrophoresed in a 15% denaturing polyacrylamide gel (precast Novex TBU gel, Life Technologies). The Decade RNA molecular weight marker (Life Technologies) was prepared according to the manufacturer's protocol and run alongside the cellular RNA. rRNA was stained with 2 μg/ml ethidium bromide and visualised under UV light to ensure equal sample loading. The RNA in the gel was electro-transferred to a positively charged nylon membrane (Boehringer Mannheim, GmbH and Mannheim, Germany) and crosslinked to the membrane using UV light at a wavelength of 254 nm (1200 μJ × 100). Overnight hybridization was performed at 42°C with radiolabeled locked nucleic acid (LNA) oligonucleotides in 10 ml ULTRAhyb hybridization buffer (Life Technologies, Austin, TX) according to the manufacturer's instructions. LNA oligonucleotide probes were 5′-end labeled with the kinaseMax kit (Life Technologies) in the presence of 1 μl [γ-32P] ATP (0.37 MBq/μl, Perkin Elmer). To remove unincorporated nucleotides, the probes were purified on Sephadex G-25 spin columns (Amersham Biosciences) according to the manufacturer's protocol. We used the following oligonucleotides to detect the antisense and sense strand of the RT5 siRNA (LNA positions underlined): 5′-ATGGCAGGAAGAAGCGGAG-3′ and 5′-CTCCGCTTCTTCCTGCCAT-3′. After overnight hybridization, the membranes were washed twice for 5 min at 42°C in 2 × SSC/0.1% SDS and twice for 15 min at 42°C in 0.1 × SSC/0.1% SDS. Signals were detected by autoradiography using a phosphorimager (Amersham Biosciences).
Small RNA library preparation and SOLiD deep sequencing
293T cells were co-transfected with 5 μg AGO2-FLAG plasmid and several shRNA- or AgoshRNA-expressing plasmids. At 48 h post-transfection, cytoplasmic cell extracts were prepared by the treatment of cells on ice for 20 min with IsoB-NP-40 [10 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1.5 mM MgCl2, 1% NP-40] followed by a centrifugation at 12000 g for 10 min at 4°C. The supernatant was incubated with 75 μl of anti-FLAG M2 agarose beads (Sigma) with constant rotation overnight at 4°C. The beads were washed 3 times in NET-1 buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5% Tween 20]. Small RNAs associated with AGO2 were isolated by phenol chloroform extraction followed by DNAse treatment using the TURBO DNA-free kit (Life Technologies). 5 μg RNA was loaded on a denaturing 15% PAGE gel for size fractionation. The 15–55 nt RNA fragments were isolated using a Spin Column (Ambion). The quality of the RNA was assayed on a Bioanalyzer 2100 (Agilent) using a small RNA chip and served as template to create an RNA library that is compatible with the SOLiD sequencing platform. We used the SOLiD Small RNA Library Preparation protocol according to manufacturer's instructions (Applied biosystems; 4452437 Rev. B; page 51–66). Samples were run on a SOLiD Wildfire system (Applied biosystems).
Bioinformatics
Analysis of the SOLiD colorspace reads was performed with LifeScope Genomic Analysis Software version 2.5 (Applied biosystems) using the small RNA pipeline. First the libraries were mapped against filter-sequences to eliminate reads generated from irrelevant sources (like tRNA, adaptors sequences etc). The remaining reads of the first steps are subsequently filtered against known miRNA sequences from miRBase (http://www.mirbase.org/). Finally, the unmapped reads from step 2 are aligned to reference sequences of AgoshRNA or shRNA expressing plasmids, allowing no mismatches during alignment.
Luciferase assays
HEK 293T cells were co-transfected with 100 ng of the firefly luciferase expression plasmid, 1 ng of renilla luciferase expression plasmid (pRL-CMV) and 1.25, 5, 20 or 25 ng of AgoshRNA expression construct. HCT wt or Dcr- cells were transfected with 100 ng of firefly luciferase expression plasmid, 1 ng of pRL-CMV and 12.5 ng of AgoshRNA or shRNA construct. The pRL plasmid served as an internal control for cell viability and transfection efficiency. We added pBluescript SK- (pBS) (Promega) to the transfection mixtures to obtain equal DNA concentrations. Two days post-transfection, firefly and renilla luciferase activities were assessed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Relative luciferase activities were calculated from the ratio between firefly and renilla luciferase activities. Transfection experiments were corrected for between-session variation as described previously.50
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Ted Bradley and Frank Baas for advice concerning the deep sequencing strategy.
Funding
This research was supported by grants from NWO-Chemical Sciences (TOP grant to BB) and ZonMw (Translational gene therapy grant to BB).
Reference
- 1. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 2. Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990; 2:279-89; PMID:12354959; http://dx.doi.org/ 10.1105/tpc.2.4.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004; 432:231-5; PMID:15531879; http://dx.doi.org/ 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 5. Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004; 432:235-40; PMID:15531877 [DOI] [PubMed] [Google Scholar]
- 6. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005; 123:607-20; PMID:16271386; http://dx.doi.org/ 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- 7. Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005; 123:621-9; PMID:16271385; http://dx.doi.org/ 10.1016/j.cell.2005.10.020 [DOI] [PubMed] [Google Scholar]
- 8. Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003; 115:209-16; PMID:14567918; http://dx.doi.org/ 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- 9. Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003; 115:199-208; PMID:14567917; http://dx.doi.org/ 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- 10. Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000; 404:293-6; PMID:10749213; http://dx.doi.org/ 10.1038/35005107 [DOI] [PubMed] [Google Scholar]
- 11. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411:494-8; PMID:11373684; http://dx.doi.org/ 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- 12. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296:550-3; PMID:11910072; http://dx.doi.org/ 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 13. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007; 130:89-100; PMID:17599402; http://dx.doi.org/ 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 2007; 448:83-6; PMID:17589500; http://dx.doi.org/ 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell 2010; 37:135-42; PMID:20129062; http://dx.doi.org/ 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science 2004; 304:734-6; PMID:15118162; http://dx.doi.org/ 10.1126/science.1096781 [DOI] [PubMed] [Google Scholar]
- 17. Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 2008; 22:2773-85; PMID:18923076; http://dx.doi.org/ 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519-28; PMID:19026782; http://dx.doi.org/ 10.1016/j.molcel.2008.10.017 [DOI] [PubMed] [Google Scholar]
- 19. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010; 465:584-9; PMID:20424607; http://dx.doi.org/ 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010; 328:1694-8; PMID:20448148; http://dx.doi.org/ 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 2012; 40:4626-40; PMID:22270084; http://dx.doi.org/ 10.1093/nar/gks026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics 2010; 284:95-103; PMID:20596726; http://dx.doi.org/ 10.1007/s00438-010-0556-1 [DOI] [PubMed] [Google Scholar]
- 23. Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A 2010; 107:15163-8; PMID:20699384; http://dx.doi.org/ 10.1073/pnas.1006432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43:892-903; PMID:21925378; http://dx.doi.org/ 10.1016/j.molcel.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3'-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep 2013; 5:715-26; PMID:24209750; http://dx.doi.org/ 10.1016/j.celrep.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res 2013; 41:3723-33; PMID:23376931; http://dx.doi.org/ 10.1093/nar/gkt036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 2012; 40:9850-62; PMID:22844086; http://dx.doi.org/ 10.1093/nar/gks705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herrera-Carrillo E, Harwig A, Liu YP, Berkhout B. Probing the shRNA characteristics that hinder Dicer recognition and consequently allow Ago-mediated processing and AgoshRNA activity. RNA 2014; 20:1410-18; PMID:25035295; http://dx.doi.org/16271386 10.1261/rna.043950.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005; 123:607-20; PMID:16271386; http://dx.doi.org/ 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- 30. Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002; 110:563-74; PMID:12230974; http://dx.doi.org/ 10.1016/S0092-8674(02)00908-X [DOI] [PubMed] [Google Scholar]
- 31. Czech B, Zhou R, Erlich Y, Brennecke J,Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 2009; 36:445-56; PMID:19917252; http://dx.doi.org/ 10.1016/j.molcel.2009.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res 2001; 29:2502-9; PMID:11410657; http://dx.doi.org/ 10.1093/nar/29.12.2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denise H, Moschos SA, Sidders B, Burden F, Perkins H, Carter N, Stroud T, Kennedy M, Fancy SA, Lapthorn C, et al. Deep sequencing insights in therapeutic shRNA processing and siRNA target cleavage precision. Mol Ther Nucleic Acids 2014; 3:e145; PMID:24496437; http://dx.doi.org/ 10.1038/mtna.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gu S, Jin L, Zhang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of Dicer processing in vivo. Cell 2012; 151:900-11; PMID:23141545; http://dx.doi.org/ 10.1016/j.cell.2012.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma H, Wu Y, Dang Y, Choi JG, Zhang J, Wu H. Pol III promoters to express small RNAs: delineation of transcription initiation. Mol Ther Nucleic Acids 2014; 3:e161; PMID:24803291; http://dx.doi.org/ 10.1038/mtna.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berkhout B, Liu YP. Towards improved shRNA and miRNA reagents as inhibitors of HIV-1 replication. Future Microbiology 2014; 9: in press; PMID:24810353; http://dx.doi.org/ 10.2217/fmb.14.5 [DOI] [PubMed] [Google Scholar]
- 37. Ma H, Wu Y, Dang Y, Choi JG, Zhang J, Wu H. Pol III promoters to express small RNAs: delineation of transcription initiation. Mol Ther Nucleic Acids 2014; 3:e161; PMID:24803291; http://dx.doi.org/ 10.1038/mtna.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010; 328:1534-9; PMID:20558712; http://dx.doi.org/ 10.1126/science.1187058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A 2006; 103:3687-92; PMID:16505370; http://dx.doi.org/ 10.1073/pnas.0511155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA 2012; 18:945-57; PMID:22461413; http://dx.doi.org/ 10.1261/rna.032938.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Coley W, Van DR, Carpio L, Guendel I, Kehn-Hall K, Chevalier S, Narayanan A, Luu T, Lee N, Klase Z, et al. Absence of DICER in monocytes and its regulation by HIV-1. J Biol Chem 2010; 285:31930-43; PMID:20584909; http://dx.doi.org/ 10.1074/jbc.M110.101709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma H, Zhang J, Wu H. Designing Ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol Ther Nucleic Acids 2014; 3:e176; PMID:25025466; http://dx.doi.org/ 10.1038/mtna.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 2012; 151:533-46; PMID:23063653; http://dx.doi.org/ 10.1016/j.cell.2012.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res 2013; 41:6568-76; PMID:23661684; http://dx.doi.org/ 10.1093/nar/gkt361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res 2005; 33:796-804; PMID:15687388; http://dx.doi.org/ 10.1093/nar/gki220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 2006; 14:883-92; PMID:16959541; http://dx.doi.org/ 10.1016/j.ymthe.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 48. Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A 2006; 103:3687-92; PMID:16505370; http://dx.doi.org/ 10.1073/pnas.0511155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu YP, Haasnoot J, Ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res 2008; 36:2811-24; PMID:18346971; http://dx.doi.org/ 10.1093/nar/gkn109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 2006; 3:1-8; PMID:16398928; http://dx.doi.org/ 10.1186/1742-4690-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]


