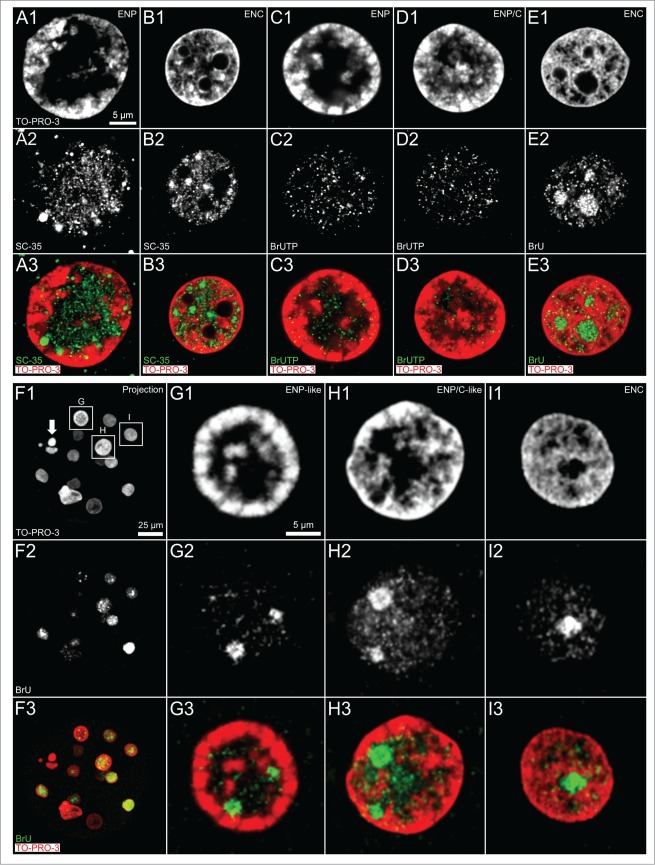
Figure 4.
(See previous page). Arrangements of splicing speckles and de novo synthesized RNA in nuclei of in vitro fertilized and cloned embryos. Panels A and B. Midplane sections recorded by 3D-CLSM in an ENP (panel A) and an ENC (panel B) from IVF embryos stained with TO-PRO-3 DNA (red) show the enrichment of the splicing factor SC-35 (green) in splicing speckles both in the major lacuna of the ENP and distributed throughout the interchromatin compartment of the ENC. Panels C–E. Immunocytochemical detection of bromine-labeled RNA after incubation for 45 minutes with BrUTP or BrU precursors. Panel F. Projection of a confocal image stack from a 15-cell cloned embryo stained with TO-PRO-3 (red) following 45 minutes incubation with BrU shows nuclei with strikingly different phenotypes, including a pyknotic nucleus (arrow). Note that the visibility of all nuclei in this projection is precluded by nuclear overlays. Panels G–I. Enlarged views of nuclei, framed in F1 by boxes G, H and I, include an ENP-like nucleus (G), an ENP/C-like nucleus (H) and an ENC (I) (for definition of these nuclear phenotypes see Results and compare Figs. 1 and 2). These Panels also present evidence for de novo RNA synthesis in nuclei of this cloned embryo independent of differences between nuclear phenotypes. Note strong RNA synthesis in nucleoli of ENCs of both fertilized (panel E) and cloned embryos (panel I). Bars: 5 μm in A1 representative for Panels A–E, 25 μm in Panel F; 5 μm in G1 for Panels G–I.