Abstract
In order to gain insights on the nuclear organization in mammalian cells, we characterized ultrastructurally nuclear bodies (NBs) previously described as fluorescent foci. Using high resolution immunoelectron microscopy (I-EM), we provide evidence that CNoBs (CRM1-Nucleolar bodies) and INBs (Intranucleolar bodies) are distinct genuine nucleolar structures in untreated HeLa cells. INBs are fibrillar and concentrate the post-translational modifiers SUMO1 and SUMO-2/3 as strongly as PML bodies. In contrast, the smallest CRM1-labeled CNoBs are vitreous, preferentially located at the periphery of the nucleolus and, intricately linked to the chromatin network. Upon blockage of the CRM1-dependent nuclear export by leptomycin B (LMB), CNoBs disappear while p62/SQSTM1-containing fibrillar nuclear bodies are induced. These p62 bodies are enriched in ubiquitinated proteins. They progressively associate with PML bodies to form hybrid bodies of which PML decorates the periphery while p62/SQSTM1 is centrally-located. Our study is expanding the repertoire of nuclear bodies; revealing a previously unrecognized composite nucleolar landscape and a new mode of interactions between ubiquitous (PML) and stress-induced (p62) nuclear bodies, resulting in the formation of hybrid bodies.
Keywords: CNoB, INB, immunoelectron microscopy, nuclear organization, nucleolus, nuclear bodies, p62/SQSTM1, PML bodies, SUMO, ubiquitin
Abbreviations
- CB
Cajal(Coiled) body
- CNoB
CRM1-Nucleolar body
- CRM1
chromosome region maintenance 1 (or exportin1)
- DFC
dense fibrillar component
- EM
electron microscope
- FC
fibrillar center
- GC
granular component
- I-EM
immunogold electron microscopy
- INB
intranucleolar body
- LMB
leptomycin B
- NBs
nuclear bodies
- p62/SQSTM1
p62/sequestosome1 protein; p62 body p62/SQSTM1-containing nuclear body
- PML
promyelocytic leukemia protein.
Introduction
The cell nucleus is hosting complex biological functions such as the replication of the genome and the coordinated transcription, maturation and transport of many genes products such as mRNAs, tRNAs, rRNAs, miRNAs, lncRNA. The transient macromolecular complexes generated by these multiple nuclear processes are often small and hardly detectable within the cell nuclei. Even at the highest resolution of the electron microscope (EM), S-phase nuclear sections cannot be distinguished from G1 or G2-phase nuclei in absence of a specific labeling. In contrast, prominent nuclear bodies (NBs) have been recognized ultrastructurally long ago but establishing their functions has proved to be a lengthy process.1-3 Functional studies, initiated by identification of relevant antibodies and development of immuno-fluorescent (IF) and immuno-electron microscopic (I-EM) techniques, have revealed the dynamic behavior of such nuclear structures which like the Cajal bodies (CBs), the PML-bodies, or the paraspeckles, are ultrastructurally well-defined.4-11 They also showed that the inventory of the NBs is far from being complete. A recent, genome-wide microscopy-based screening indicated that 65 of the 325 proteins found in nuclear dots in HeLa cells reside in undefined nuclear foci.12 while nuclear foci like the Sam68 bodies,13 the “GLFG bodies,”14 the CNoBs,15 the INBs,16 the Pat1b PML-associated foci,17 the histone locus bodies18 were successively discovered by IF studies, but are not or poorly-characterized at the ultrastructural level.
Among the nuclear organelles, overlapping functional and ultrastructural compartments were first established for the nucleolus.19 Functions in rDNA transcription, accumulation and processing of primary rRNA transcripts, and in assembly of pre-ribosomal subunits were progressively assigned to the 3 nucleolar subcompartments identified under the EM, namely the fibrillar centers (FC), the dense fibrillar component (DFC) and granular component (GC), respectively. Further studies uncovered more diverse functions of the nucleolus,20,21 in line with proteomic analyses that revealed nucleolar proteins with no obvious relationship to ribosome biogenesis.22,23 For example, nucleolar sequestration of MDM2 (or HDM2 in humans) by p14ARF upon stress leads to p53 activation, and consequently to cell-cycle arrest or apoptosis.24 Independently, IF detection of specific proteins revealed novel intranucleolar foci like CNoBs and INBs.16
INBs were found in various human and hamster cell lines, and animal tissue like bovine lens or human skin.16 Although no specific marker for INB has been evidenced yet, a number of nuclear proteins accumulate within the INBs. Among these, some are involved in DNA replication and/or DNA repair, like PCNA, MCM3, MCM7, Ku70 and DNA-PKcs whereas some others, such as SF2/ASF, U2AF65, U1/U2 snRNP, PPM1G/PP2Cg and the Sm proteins, are pre-mRNA splicing factors. Interestingly, INBs are enriched in the post-translational modifiers SUMO1 and SUMO-2/3, suggesting that they concentrate or actively generate SUMOylated proteins. INBs are found in about 30–40% of HeLa cells, increasing to 71% in S-phase nuclei and they are promoted by DNA damaging agents like topoisomerase inhibitors, hydroxyurea or IR exposure. They disappear upon treatment with a low dose of Actinomycin D suggesting that they are dependent on rDNA transcription.16 Sensitivity to a low dose of Actinomycin D was also reported for CNoBs, intranucleolar foci initially characterized by IF-labeling of CRM1 (or exportin 1), the LMB-sensitive transporter of proteins that contain a Nuclear Export Signal (NES). CNoBs, as seen by video microscopy, are formed into the nucleolus and occasionally migrate, detached from the nucleolus, into the nucleoplasm. However, as CNoBs were not seen reaching the nuclear envelope, a function in transporting nucleolar components to nuclear pores for nuclear export seemed unlikely.15 The Lamond's group reported that INBs also contain CRM1, and concluded that CNoBs and INBs are a single nucleolar entity.16 However, despite being most frequent in HeLa cells these nucleolar bodies have no known ultrastructurally defined counterpart and a proof of their identity is lacking.
CNoBs were found to disappear after LMB-treatment.15 In this setting, a prominent nucleoplasmic p62/SQSTM1-containing nuclear body (hereafter p62 body) was described.25 P62/SQSTM1 is an abundant cytoplasmic protein which possesses an ubiquitin-binding domain and has a high affinity for LC3, a key factor of autophagosome formation. Mutated in the adult form of the Paget's bone syndrome, it is induced by a number of stresses such as proteasome inhibition, oxidative stress or expression of abnormal proteins generated by triplet-extension. P62/SQSTM1 plays a key role in mediating LC3 recruitment to cytoplasmic aggregates of poly-ubiquitinated proteins and autophagosome formation.26 As such, p62/SQSTM1 is a marker, a substrate and a key component of the autophagic control of proteinopathies induced by aggregation of misfolded proteins.27 p62/SQSTM1 is a shuttling protein containing 2 NLS and a NES, which has a capacity to homo-polymerise. In untreated HeLa cells, nuclear p62-foci are formed at low frequency (0.5% of the cells).25 Upon LMB treatment, cytoplasmic p62/SQSTM1 is extensively delocalized to numerous nuclear bodies in which poly-ubiquitinated proteins also accumulate,25 Intriguingly, these p62-bodies were shown either to be tightly-associated,25 or even to overlap,28 with PML bodies. PML bodies, like INBs are known to concentrate the post-transcriptional modifiers SUMO1 and SUMO-2/3.1 Using I-EM, we investigated the ultrastructure of these emerging nuclear territories in order to clarify their relationship. Our results indicate that CNoBs, INBs and p62 bodies are distinct nuclear structures, of different size and with different ultrastructural characteristics. Whereas INBs are strictly nucleolar and highly concentrating the post-translational modifiers SUMO1 and SUMO-2/3, CNoBs are partly nucleolar and partly nucleoplasmic and surrounded by dense chromatin-fibers. Upon LMB treatment, PML bodies and p62 bodies are found either as separate entities, often in close contact or else in the form of hybrid p62/PML bodies, suggesting fusion events between the 2 bodies.
Results
Ultrastructural characterization of INBs
Only endogenous proteins were used as NB markers in this study. Taking into account the list of proteins shown by IF to reside within the newly-described INBs,16 we choose anti-SUMO-2/3 and anti-SUMO1 antibodies as ligands to characterize these structures by I-EM. We previously showed that these 2 antibodies are highly efficient for detecting the nuclear aggregates of SUMOylated proteins that form upon proteasome-inhibition.29 The anti-SUMO-2/3 antibody specifically labeled intranucleolar fibrillar domains which were conspicuous, albeit at low frequency, on ultrathin-sections of HeLa cells (Fig. 1A and A′). These domains were obviously distinct from the classical nucleolar compartments FCs, DFC and GC which remained label-free. Compared to neighboring FCs (Fig. 1B), which are often numerous within a nucleolar section and surrounded by the irregularly-shaped DFC, SUMO-2/3 labeled structures showed a higher electron density, they were surrounded by a clear halo and were invariably found as a single unit within a nucleolar section. These SUMO-2/3 containing structures were always spherical and enclosed within a roundish nucleolar cavity (n = 41). The same ultrastructural features were observed using an anti-SUMO1 antibody (Fig. 1C), indicating that they correspond to genuine nucleolar bodies highly-enriched in SUMOylated proteins.
Figure 1.
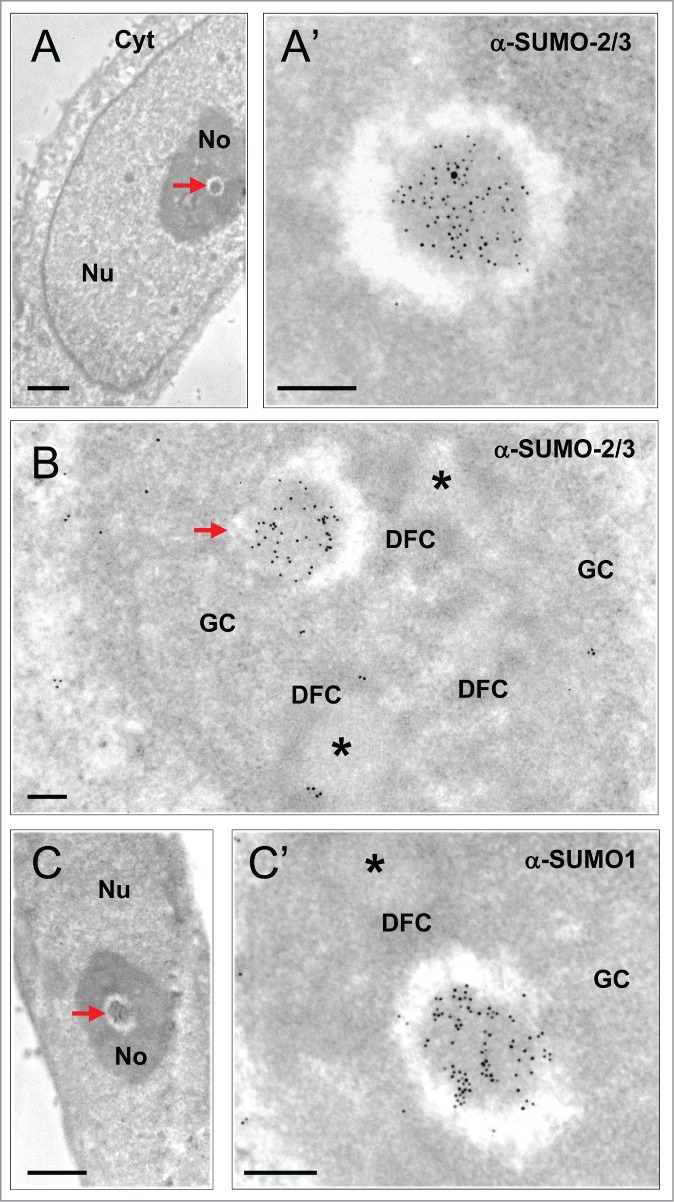
Characterization of INBs by I-EM localization of SUMO-2/3 and SUMO1 (ultra-thin sections of glutaraldehyde-fixed, Lowicryl embedded HeLa cells). (A) The INB (arrow) is electron-dense and centrally-located. (A’) Enlargement showing high SUMO-2/3 content as detected with primary anti-SUMO-2/3 antibody and a secondary antibody conjugated to 5 nm gold particles. Nu = Nucleus, No = Nucleolus, Cyt = cytoplasm. Scale bars: 1 and 0.2 µm in A and A’, respectively. (B) Compared to the classical nucleolar components, the INB (arrow) is surrounded by a white halo and highly labeled by the anti-SUMO-2/3 antibody (10 nm gold particles) whereas the FCs (asterisks) surrounded by the DFC and embedded within the GC are unlabelled. Scale bar = 0.2 µm (C–C’) As in A, except that the centrally-located INB (arrow) is labeled, as shown in C’, with an anti-SUMO1 antibody and secondary antibody conjugated to 10 nm gold particles. Scale bars: 1 and 0.2 µm, respectively.
Differentiating INBs from PML bodies
The anti-SUMO-2/3 and anti-SUMO1 antibodies also label nucleoplasmic PML bodies, which, being fibrillar, roundish and surrounded by a white halo, are ultrastructurally resembling the intra-nucleolar bodies.4,11 To investigate the relationship between INBs and PML-bodies, we first scrutinized PML bodies on the same cell samples using a rabbit anti-PML antibody. Among a large group of typical PML bodies (n = 45), none were located within the nucleolus, and 7 were adjacent to the nucleolus (Figure S1). Next, we performed a double-labeling experiment using a mouse anti-PML antibody and the rabbit anti-SUMO-2/3 antibody coupled to gold particles of 5 and 15 nm respectively (Fig. 2). The nucleoplasmic PML bodies (n = 10) were, as expected, decorated with both antibodies (Fig. 2A) and with similar level of labeling (a total of 90 and 101 gold particles for PML and SUMO-2/3 respectively). In contrast, intranucleolar SUMO-2/3 labeled bodies were either unlabelled with PML antibodies as in Figure 2B (n = 6) or very weakly labeled (n = 2) (4 and 78 gold particles for PML and, SUMO-2/3 respectively). These results are consistent with the reported absence of PML in the INBs.16 Moreover, none of the intranucleolar SUMO1 or SUMO-2/3-labeled bodies showed the finely fibrillar core surrounded by an electron dense fibrillar capsule that is typical of most PML-bodies in HeLa cells (Fig. 2A and Fig. S1). Finally, the size of the INBs was significantly higher than the one of PML bodies measured in thin-sections of the same sample (p < 0.05) (Fig. 3). We therefore concluded that the SUMO1 and SUMO-2/3 containing INBs described here are distinct from PML bodies and correspond to the INBs previously described in IF.16 The labeling of SUMO1 and SUMO-2/3 within the INBs was surprisingly high. SUMO-2/3 labeling density in INBs (n = 18) was 60 +/−11% of the one in nucleoplasmic PML bodies (n = 22), while SUMO1 labeling was similar (363 +/− 206 and 303 +/- 196 gold particles/µm2 in INBs (n = 13) and PML bodies (n = 18), respectively).Taking advantage of the ultrastructural definition of the INBs, we further analyzed their relationship with other NBs using antibodies directed against CRM1 and coilin.
Figure 2.
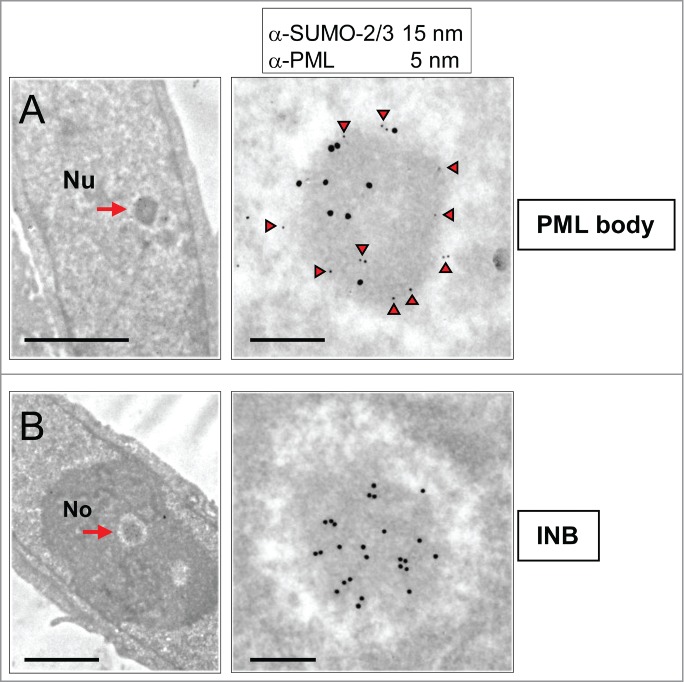
INBs are distinct from PML bodies as shown by differential PML and SUMO-2/3 content. (A) A nucleoplasmic NB (arrow, left frame), double-labeled (as shown in enlarged right frame) with mouse anti-PML (5 nm gold particles underlined by arrow-heads) and rabbit anti-SUMO-2/3 (15 nm gold particles) antibodies is identified as a PML body. Notice peripheral PML-labeling. Scale bars: 2 and 0.2 µm, respectively. (B) On the same thin-section, a centrally-located NB (arrow, left frame) labeled by anti-SUMO-2/3 (15 nm gold particles, right frame) but not anti-PML (lack of 5 nm gold particles) antibodies is defined as an INB. Nu: nucleoplasm, No: Nucleolus. Scale bars: 2 and 0.2 µm, respectively.
Figure 3.
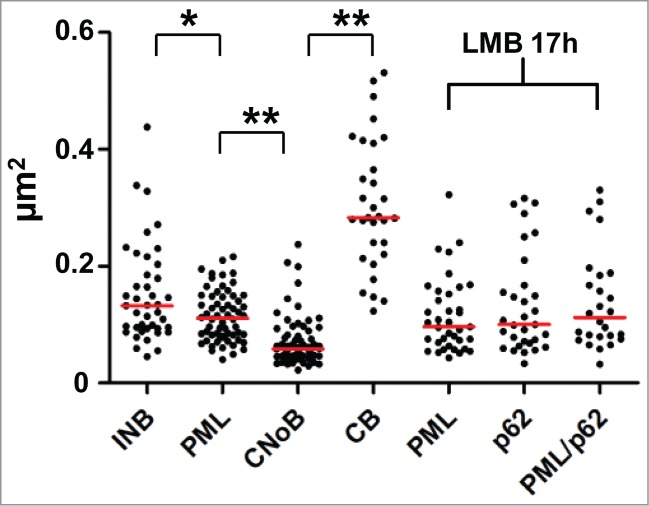
Comparative size distribution of NBs in HeLa cell ultrathin-sections. Surface of NB sections were measured after I-EM and plotted in µm2. Statistical significance was determined by a 2-tailed Mann-Whitney U test. (*) and (**) represent p-values <0.05 and <0.0001, respectively.
CNoBs ultrastructure
CNoBs were defined in IF by the presence of CRM1. Observation in live cells showed that CNoBs form within the nucleolus and migrate occasionally into the nucleoplasm.15 As a result, CNoBs, in contrast to INBs, are not exclusively intranucleolar (Fig. 4A). On HeLa cells thin-sections (Fig. 4B–E), the CRM1 antibody labeled heavily and specifically NBs which were distinct from the INBs for the reasons explained below and therefore maintained hereafter in their original designation of CNoBs. At the EM level, CNoBs were frequently associated with the nucleolus and, most frequently at the periphery of this organelle. Analyzing 70 CNoBs sections in HeLa cells, we found 9 that were enclosed and 31 juxtaposed to the nucleolus (as in Fig. 4 B and C) whereas 30 were nucleoplasmic and had no visible nucleolar contacts (Fig. 4D and E). Among the latter, it is likely that some were nucleolus-associated but that the orientation of the thin-sectioning was such that only the CRM1-positive body was sectioned. Independently of their nuclear localization, CNoBs have a uniform ultrastructural aspect, so finely fibrillar that they appear to be vitreous, an aspect that is not found in previously-described NBs in human cells. Considering their common nucleolar origin and their similar vitreous ultrastructure, the nucleoplasmic CRM1 bodies were hereafter referred to as “nucleoplasmic CNoBs.”
Figure 4.
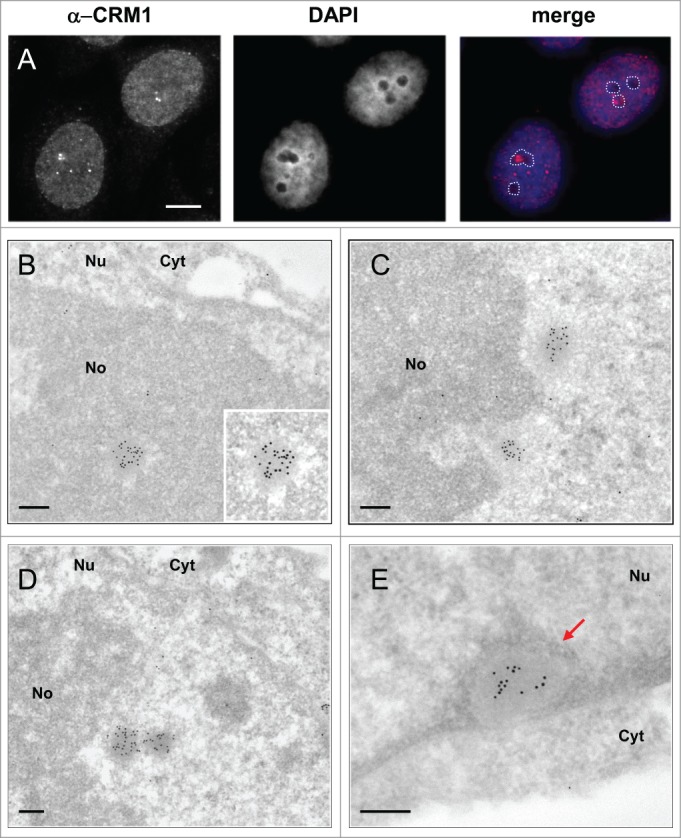
Ultrastructural characterization and nuclear distribution of CNoBs. (A) IF localization of CNoBs in HeLa cells. Nucleolar association is detected by merging CRM1 foci with nucleoli (unstained with DAPI, dotted lines). CNoBs associated with - and distant from - nucleoli are evidenced. Scale bar: 5 µm. (B–D) I-EM detection of CRM1 (glutaraldehyde-fixed, lowicryl-embedded HeLa cells, 10 nm gold particles) reveals uniform, vitreous aspect of CNoBs within the nucleolus in B, at the nucleolar periphery in C and D or within the nucleoplasm as in D, (right object) and E. CNoB in (E), in close proximity to the nuclear envelope, is surrounded by peripheral nuclear heterochromatin (arrow). Nu: nucleus, No: nucleolus, Cyt: cytoplasm. Scale bars: 0.2 µm.
Further, the intranucleolar CNoBs were devoid of the marked electron-lucent halo observed around the INBs (Fig. 4B). Also consistent with IF observation (Fig. 4A), and further contrasting with INBs, several nucleoplasmic CNoBs could be found within the same nuclear ultra-thin section (Fig. 4C and D). We found one occurrence of 5, one occurrence of 3 and 6 occurrences of 2 nucleoplasmic CNoBs which represented as many as 20 of the 70 CNoBs sections analyzed. CNoBs, independently of their nuclear localization, were smaller than INBs (p < 0.0001, Fig. 3). We finally noticed that nucleoplasmic CNoBs close to the nuclear envelope (NE) were embedded into peripheral heterochromatin (Fig. 4E). Upon closer examination, such close contacts with dense chromatin-like fibers were also noticeable for CNoBs present at the periphery of the nucleolus (Fig. 5A and Supplemental Figure S2B). To substantiate this observation, we used an anti-histone H3 antibody to detect chromatin at the proximity of CNoBs. Indeed, there was a ring of chromatin fibers surrounding NBs with all ultrastructural characteristics of CNoBs (Fig. 5B). This was confirmed with a double-labeling of histone H3 and CRM1. As both antibodies were raised in rabbit, the anti-histone antibody was tagged with biotin in vitro and detected with a goat anti-biotin antibody (see Material and Methods). This experiment confirmed that CNoBs are surrounded by dense chromatin fibers in HeLa cells (Fig. 5C and C′). Noteworthily, nucleoplasmic CNoBs, although intricately linked to the chromatin network, do not contain appreciable amount of histone H3-labeled chromatin. Finally, because of their intranucleolar assembly and their sensitivity to low dose of Actinomycin D, which selectively inhibits RNA polymerase I, it was postulated that CNoBs may play a role in pre-ribosome biogenesis.15 By electron-microscopic in situ hybridization (EM-ISH), we analyzed CNoB content in 18S ribosomal rRNA (or precursors thereof) with a complementary biotinylated DNA probe. This experiment revealed that CNoBs are devoid of significant amount of rRNA (Fig. 5D).
Figure 5.
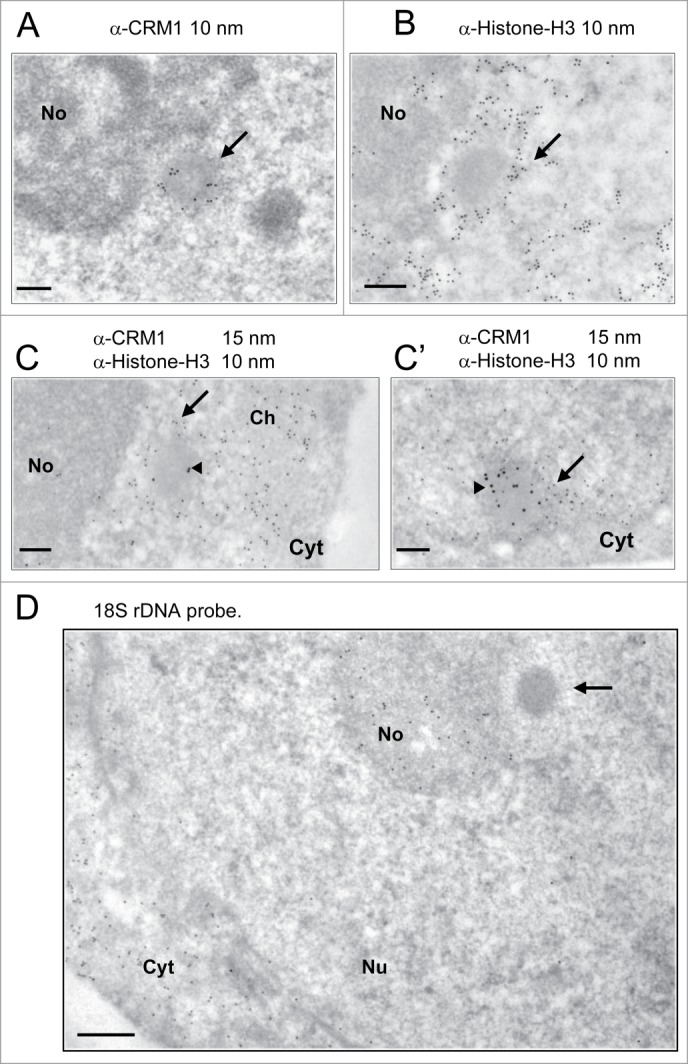
CNoBs do not contain rRNA and are associated with dense chromatin fibers. (A) A nucleolar peripheral CNoB, identified by CRM1 staining (10 nm gold particles), is surrounded by a crown of chromatin–like dense fibers (arrow). (B) Histone H3 labeling (10 nm gold particles) identifies chromatin fibers (arrow) surrounding a nucleolus-associated NB with a characteristic CNoB vitreous aspect. (C–C’) CNoBs and chromatin fibers were labeled with anti-CRM1 (15 nm gold particles, arrow-head) and a biotinylated anti-Histone-H3 antibody (10 nm particles), respectively. CNoBs, being nucleolus-associated as in C or closely-apposed to the nuclear envelope as in C’ are surrounded by a ring of dense, histone-H3 labeled chromatin fibers (arrow). No: nucleolus, Cyt: cytoplasm, Ch: chromatin. Scale bars: 0.2 µm. (D) EM-ISH with a biotinylated 18S rDNA probe and detection of hybrids with an anti-biotin antibody (10 nm gold particles) reveals presence of complementary rRNA sequences within the nucleolus (No) and within ribosome-rich cytoplasmic areas (Cyt) but not in CNoBs (arrow). Bar 0.2 µm.
Comparing CNoBs, INBs and CBs
Previous IF studies have reported the presence of CRM1 in CBs.30,31 However, none of the vitreous CRM1-positive bodies that we identified by I-EM resembled CBs. In addition, visual inspection of the CBs, ultrastructurally characterized by 40–60 nm thick coiled threads on the same thin-sections, revealed that they were not labeled (Fig. S2). Since this discrepancy could result from the different antibodies used in these studies, we labeled thin-sections with a distinct anti-CRM1 antibody that again decorated the same vitreous bodies, but not the CBs (Fig. S2B). On the other hand, the CB marker coilin,32 was detected in INBs,16 and in intranucleolar CBs in breast cancer cell lines,33,34 or upon okadaic acid treatment.35 Using an anti-coilin antibody on HeLa cell thin-sections, heavily labeled CBs (297 +/− 87 gold particles/µm2, n = 16) were found within the nucleoplasm whereas the nucleolar-associated and nucleoplasmic vitreous CNoBs remained unlabelled (Fig. S3A and A′), in agreement with previous IF observations.15 In the same cell sample, the INBs recognized by their ultrastructural features (n = 5) were also unlabeled (Fig. S3B). Overall, as illustrated in Figure 3 and Figures S2 and S3, CBs, INBs and CNoBs are so dissimilar in size and so distinct ultrastructurally in HeLa cells that they cannot be confounded; confirming that the CRM1-positive nucleoplasmic bodies seen under the EM are CNoBs and the SUMO-labeled intranucleolar bodies are INBs, with no contribution of the CBs.
Ultrastructure of LMB-induced p62 bodies
While CNoBs disappear following inhibition of CRM1-dependent nuclear export by LMB, prominent PML-associated p62 bodies are formed.15,25,28 To investigate the relationship between CNoBs, PML and p62 bodies, Hela cells were treated with LMB and analyzed after staining with anti-CRM1, PML, p62/SQSTM1 and ubiquitin antibodies. Despite extensive observations, no residual CNoBs were seen on CRM1-labeled ultrathin sections, in agreement with previous IF observations.15 This indicated that not only the CRM1-labeling is dispersed upon LMB treatment but the CNoB structure altogether. Meanwhile, pale and larger fibrillar NBs became conspicuous under the EM (Fig. 6A). In IF, p62/SQSTM1 which is essentially cytoplasmic in untreated cells was drastically relocated within brilliant nuclear foci which were distinct and smaller than the CBs (Figures 6B and 3, p < 0.0001 ). To correlate the p62 foci seen by IF and the pale NBs seen by EM, we carried out an I-EM localization of p62/SQSTM1 in LMB-treated Hela cells. As illustrated in Figure 6C, p62/SQSTM1 was highly enriched within the LMB-induced fibrillar NBs. So was also ubiquitin (Fig. 6D), and finally the same LMB-induced NBs were labeled with anti-ubiquitin (5 nm gold particles) and p62/SQSTM1 (15 nm gold particles) antibodies (Fig. 6E). We conclude that we have ultrastructurally characterized a fibrillar LMB-inducible p62 body that also concentrates ubiquitinated proteins.25,28
Figure 6.
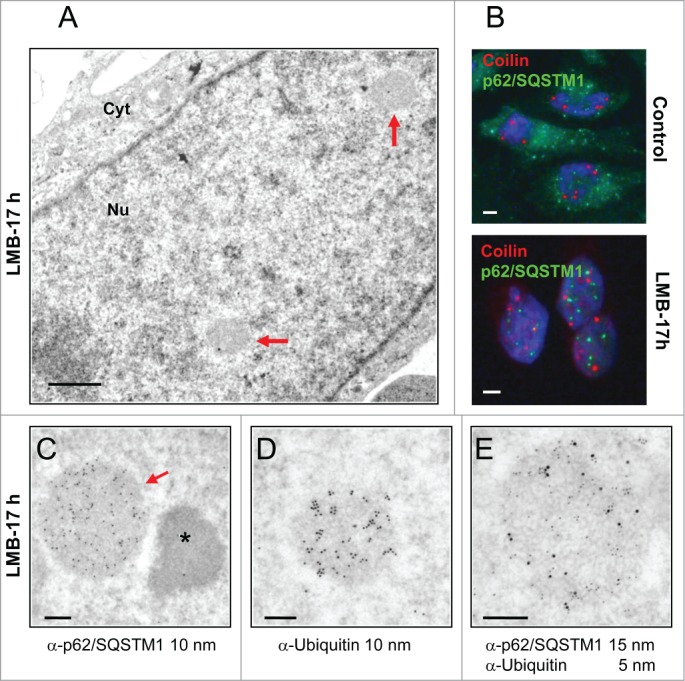
Ultrastructure of p62/SQSTM1-containing nuclear bodies induced by LMB-treatment. (A) Upon LMB-treatment of HeLa cells (10 nM, 17 h), inducible roundish nuclear bodies of low electron density (arrows) are revealed by conventional EM (glutaraldehyde-fixation, Epon embedding). Cyt: cytoplasm. Scale bar: 1 µm. (B) IF double-labeling of control and LMB-treated HeLa cells (10 nM, 17 h) with anti-p62/SQSTM1 (green) and anti-coilin (red) show relocation of cytoplasmic p62/SQSTM1 into nuclear foci upon nuclear export inhibition. These LMB-induced p62 bodies are distinct from the coilin-stained CBs. DNA stained by DAPI (blue). Scale bars: 2 µm. (C) I-EM characterization of the p62 bodies with an anti-p62/SQSTM1 antibody. The labeled p62 body (arrow, 10 nm gold particles) is structurally equivalent to the LMB-induced bodies shown in A by conventional EM. The asterisk denotes an adjacent characteristic electron-dense paraspeckle. (Glutaraldehyde-fixation, Lowicryl-embedding). (D) I-EM detection of ubiquitin within a LMB-induced NB (formaldehyde-fixation, 10 nm gold particles). (E) Double-labeling of a p62 body with an anti-p62/SQSTM1 and an anti-ubiquitin antibody (5 nm and 15 nm gold particles, respectively) (formaldehyde fixation). The ratio of 5 to 15 nm gold particles does not reflect the relative amount of the 2 proteins because antibodies conjugated to 15 nm gold particles are less efficient in detecting primary antibodies than the ones conjugated to 5 nm gold particles. Scale bars in C, D and E: 200 nm.
High resolution analysis of p62 body association with PML body
Although they appear ultrastructurally distinct from classical ring-shaped PML bodies, p62 bodies were previously shown either to overlap or to be tightly associated with PML bodies.25,28 To investigate the relationship between the 2 bodies, we carried out double-labeling experiments at IF and EM levels. After 3h of LMB treatment, assembly of p62 bodies was conspicuous in IF, but not uniform. Only a fraction of the nuclei (26%, n = 377) showed multiple p62 bodies (Fig. 7A), independently of their content in PML bodies. In contrast, multiple p62 bodies were present within every nucleus after long-term treatment (17 h) (Fig. 7B),. Within a single nucleus, p62 bodies were well-separated, partly-overlapping or totally overlapping with PML bodies (Fig. 7B, insets). Consistently, at both time-points in I-EM (Fig. 7C), PML and p62 bodies were found as separated entities (upper frames) or as adjacent, docking bodies (middle frames) or finally as double-labeled hybrid bodies. In the latter case, invariably, p62/SQSTM1 was present within the central region surrounded by a peripheral layer of PML. From these I-EM experiments we determined that the percentage of hybrid bodies increased progressively after 3 and 17 h of leptomycin-treatment at the detriment of the PML bodies (Fig. 7D). This suggests a gradual amalgamation of the newly-formed p62 bodies with pre-existing PML bodies, or the diversion of PML proteins from the nucleoplasm. In several instances, EM observations as in Figure 8 and Figure S3 depicted intermediate figures of partial, asymmetrical PML coverage, encircling a fully-formed p62 body. These recurrent observations clearly show that PML recruitment is a polarized event, mobilizing numerous PML molecules at once rather than gradually. From this we propose that PML and p62 bodies are formed independently, with distinct ultrastructural features, that they come into contact frequently and that they fuse forming hybrid PML/p62 bodies (Fig. 8.)
Figure 7.
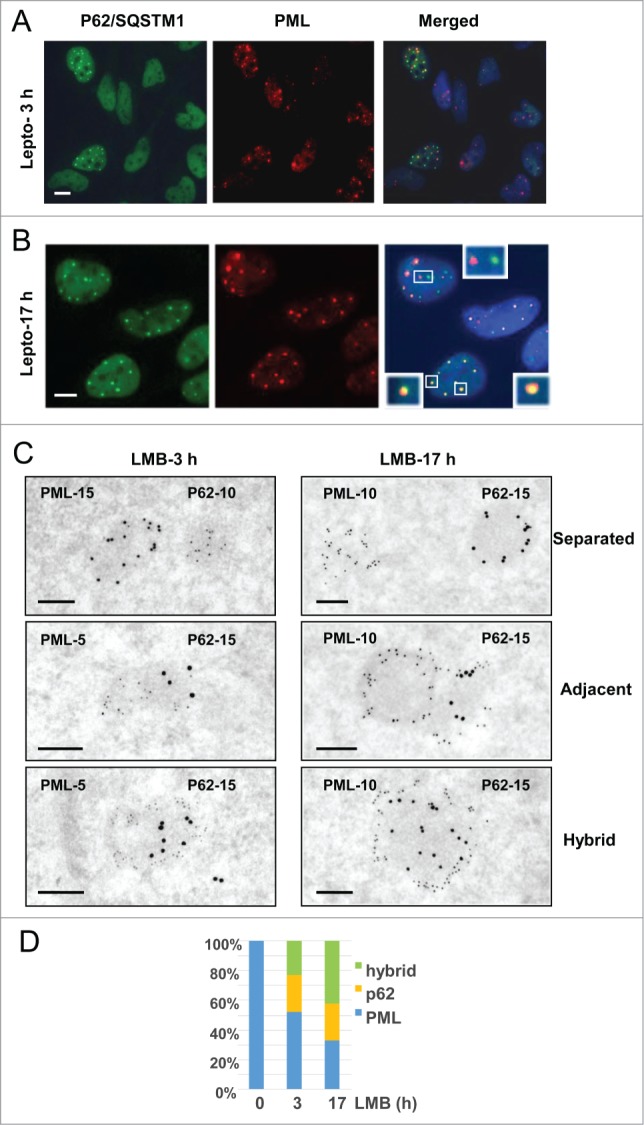
Interactions between PML-bodies and LMB-induced p62 bodies. (A) IF double-labeling of p62/SQSTM1 (green) and PML (red) in LMB-treated HeLa cells (10 nM, 3 h). Merging of signals with DAPI-stained DNA (blue) show p62 bodies in a subset of the nuclei while PML bodies are present in all cells. Scale bar: 5 µm. (B) As in A, except that after a17h long LMB treatment p62 bodies are found in most nuclei. Merging demonstrates either physically distinct PML bodies (red) and p62 bodies (green) (top inset) or adjacent and partially overlapping bodies (left inset) or overlapping bodies with p62/SQSTM1 surrounded by PML (yellow in red as in lower right inset). Scale bar: 5 µm. (C) High resolution analysis, by double-labeling I-EM, of the interactions between PML-containing and p62/SQSTM1-containing NBs in HeLa cells treated with LMB for 3 and 17h. Experiments have been repeated using various sizes of gold particles, as indicated. Images of physically separated bodies (upper frames), adjacent bodies (middle frames) and hybrid double-labeled bodies (lower frames) were taken from thin-sections after 3 or 17 h long LMB treatment (left and right). Notice that PML (middle frames) is apparently invading the periphery of the adjacent p62 body, leading to the gradual absorption of the latter. Scale bars: 200 nm. (D) Percentage of PML, p62 and hybrid bodies was determined from I-EM observations of control cells and cells treated with LMB for 3h or 17 h (number of bodies = 51, 171 and 158, respectively).
Figure 8.
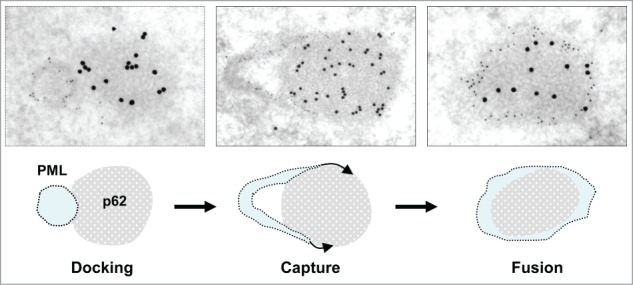
Proposed mechanism for PML/p62 hybrid body formation. PML and p62 bodies, as detected by I-EM (top row), are often found as distinct bodies in close contact (docking). The docking phase is followed by progressive invasion of the p62 body by the PML body (capture) This leads to the formation of an hybrid body with a p62-rich fibrillar core surrounded by a shell of PML (fusion). Further images evoking progressive capture of p62 bodies by PML bodies are displayed in Figure S3.
Discussion
Four recently described NBs are characterized ultrastructurally in this paper, the INBs, the CNoBs, the p62 bodies and the hybrid PML/p62 bodies. Furthermore, INBs and the CNoBs are evidenced as separate intranucleolar bodies, illustrating the structurally composite nature of the nucleolus. To facilitate comparison between NBs analyzed here and elsewhere,9,29 their ultrastructural features and their nuclear distribution are summarized in Figure 9, along with their demonstrated or candidate functions.
Figure 9.
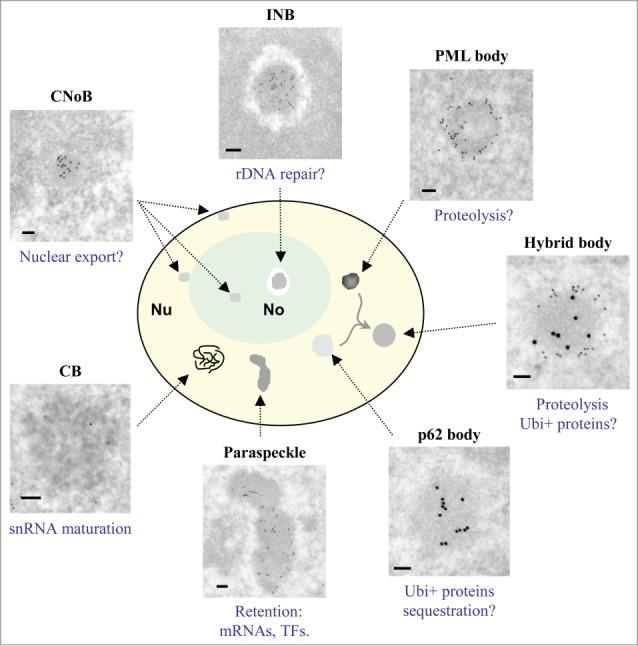
Comparative ultrastructure and localization of NBs. NBs in untreated HeLa cells, except for p62 bodies and fused PML/p62 bodies which formed readily only after LMB treatment, are depicted. Schematic representation illustrates that only INBs and CNoBs are found within the nucleolus and that hybrid bodies are found only between p62 and PML bodies. The frames exemplify the variable sizes and structures of NBs labeled with CRM1 (CNoB), SUMO-2/3 (INB), PML (PML body), PML (15 nm)/p62 (10 nm) (Hybrid body), p62/SQSTM1 (p62 body) and PSPC1 (Paraspeckle). The CB is unlabelled to highlight its characteristic large coiled threads. Their presumed (?) or reported functions are mentioned below each frame. TFs = transcription factors; Ubi+ proteins = ubiquitinated proteins. Scale bars: 100 nm.
INBs and CNoBs are genuine nucleolar structures
The nucleolus, in addition to its role in pre-ribosome synthesis, monitors stress sensitivity and regulates the cell cycle .20,24,36 From that, the nucleolus was progressively recognized as being functionally heterogeneous. Here we show that the nucleolus is also structurally heterogeneous, with various intranucleolar bodies. INBs and CNoBs were ignored by classical EM studies,19 probably because of their paucity. INBs were shown to be present in no more than 10–20% of the nuclei in a number of different cell lines;16 with the noticeable exception of HeLa cells peaking at 40%. Similarly, CNoB occurrence depends on the cell line, with a higher frequency in HeLa cells (DW, personal communication). This low frequency combined to a small size renders random ultrathin sectioning rarely fruitful. IF studies15,16 were instrumental for spotting these new nucleolar domains which, as shown in this paper, obviously differ from the classical nucleolar compartments.
Ultrastructural characterization of the INBs
INBs analyzed at high resolution following SUMO1 or SUMO-2/3 labeling were embedded within the GC of the nucleolus with little contact either with the FCs or the DFC. Roundish and surrounded by an electron-lucent halo, the INBs were finely fibrillar, homogeneous, with no apparent granules. The size of their twisted fibers as in Figure 9 was measured at about 8 nm. Over 50 INBs, we found no single evidence for a connecting canal to the nucleoplasm strongly suggesting that their intranucleolar location is not resulting from a nucleoplasmic invagination. As a transient invagination permitting the entry of a NB would not be easily detected under the EM, a definitive confirmation would require to visualize INB formation in vivo and to determine whether it takes place during or after the post-mitotic reformation of the nucleolus.
Importantly, although both INBs and PML-bodies contained SUMO1 and SUMO-2/3, INBs contained little or no PML. They were also significantly smaller and strictly intranucleolar while PML bodies were nucleoplasmic. It has been shown that only conjugated SUMO1 is detected within INBs,37 indicative of the presence of SUMOylated proteins, and with a very low turn-over, as demonstrated by FRAP experiments.16 Some proteins that reside in INBs (e.g. PCNA,38 Ku70,39 PA28γ/PSME3,40 TRIM28/KAP-1,41) are known to be SUMOylated, when other (e.g., DNA-PKcs, Cdc5L, Prp19, PNUTS/p99) are predicted to be SUMOylated (using both JASSA, (www.jassa.fr), and GPS-SUMO softwares).42 In addition, TRIM28/Kap-1,41 is a presumptive E3 SUMO ligase when SF2/ASF is a positive regulator of SUMOylation.43 These data are consistent with our quantitation of SUMO in INBs and PML bodies, which reveals that INBs are another major nuclear site where SUMOylated proteins accumulate and/or are post-transcriptionally modified.
The function of the INBs is still largely undefined. Because of their components playing a role in genome repair and their reported proximity with silent rDNA fibers, it was suggested that INBs play a role in rDNA genome maintenance.16 As revealed in this study, their location within the GC, which is devoid of rDNA genes,44 does not support this hypothesis. In fact, our efforts to show an association between INBs and histone H3-labeled chromatin were unsuccessful, indicating no or very transient links with intranucleolar chromatin.
Characterization and nuclear distribution of CNoBs
CNoBs under the EM are mainly characterized by their smooth aspect, their small size and their bimodal distribution with about half being nucleolus-associated and half being nucleoplasmic. While CNoBs were originally defined as CRM1-containing bodies,15 CRM1 was also reported to accumulate within CBs.30,31 In this study, all the CRM1-labeled nucleoplasmic bodies detected by I-EM in HeLa cells were CNoBs with no contribution of the CBs, as supported by their distinct ultrastructure, size, molecular composition and nuclear distribution; e.g. in HeLa cells, CBs are never in close contact to the NE. Conversely, all the coilin-labeled nucleoplasmic bodies were CBs with no contribution of the CNoBs. The divergence between IF and I-EM data on CRM1 localization in HeLa cells is surprising and unexplained but it helped to discriminate ultrastructurally the nucleoplasmic CNoBs from the CBs.
By their size, their occasional clustering and their ultrastructure, CNoBs resemble the Nup98 containing GFLG-bodies which have been described in a Xenopus cell line.14 Moreover, CRM1 interacts with nuclear pore proteins which like Nup98 are GFLG-repeats-containing proteins.45 However, GFLG bodies were not found specifically associated with the nucleolus or with the NE and are present in no more than 5% of HeLa cells,14,46 a frequency far below the one reported for CNoBs.15 Further studies will be needed to determine if - or to what extent - CNoBs and GFLG-bodies overlap.
CNoB formation, recorded in vivo, was shown to take place within the nucleolus.15 Our observations of intranucleolar CNoBs, tightly-embedded within the GC of the nucleolus, with no evidence for a cavity or a nucleoplasmic invagination are consistent with this assumption. We further show that CNoBs are most often located at the periphery of the nucleolus where they become tightly associated with dense chromatin fibers. Chromatin association is also observed for CNoBs abutting the inner face of the NE, raising the possibility of nuclear trafficking tightly bound to the chromatin network. Because of their sensitivity to Actinomycin D, it was postulated that CNoBs may play a role in pre-ribosome biogenesis. However, as they were not detected at the NE by IF studies,15 they would not participate to their transport to the nuclear pores. Despite our contrasting observation of a subset (∼10%) of CNoBs at the NE, our results do not either support a role for CNoBs in transporting pre-ribosomal particles: first, their constant vitreous aspect is not compatible with the presence of nucleolar GC-like granules and second, 18 S rRNA (or precursors thereof) is absent in CNoBs.
We confirmed that upon LMB-treatment CNoBs disappear, and not just its CRM1 content, linking CNoB stability to active CRM1 activity. Our observations upon LMB-treatment of HeLa cells significantly differ from a previous report showing that upon a 30 nM 3 h long treatment, U2-snRNA and Coilin were depleted from CBs and Coilin relocalized to the nucleolus.47 By IF as in Figure 6B or by I-EM as in Figure S5, following a 10 nM 17 h long treatment, Coilin was found in CBs with a seemingly normal content of SMN and of U2 snRNA. The different concentration of LMB used in these studies is likely the source of this discrepancy.
Originally, one CRM1 substrate, CPEB1, was shown to accumulate in CNoBs.15 Here, by I-EM and EM-ISH we found that this observation is not extendable to all CRM1 substrates (Fig. 7B), (neither p62/SQSTM1, the polyA binding protein PABP1, β-actin, nor U2 snRNA, data not shown). Finally, CNoBs did not contain PML, SUMO-1, SUMO-2/3, ubiquitin, Sam68, SMN, coilin, NONO, PSPC1 (not shown). Altogether, this is substantiating their specificity with respect to other NBs, including the INBs but also underlines how enigmatic they are with CRM1 and the translational inhibitor CPEB1 being their only components identified so far.15
LMB-induced p62 bodies form hybrid bodies with PML
Numerous pale and fibrous NBs are induced upon LMB treatment of HeLa cells. Made of 8 nm fibers like the INBs, their electron-density is however comparatively reduced which reflects a loose packing of their fibers. They contain a high amount of p62/SQSTM1, a shuttling protein containing 2 NES whose nuclear export is LMB-sensitive,25,48 and are therefore the p62 bodies previously described in IF studies.25,28 In the cytoplasm, p62/SQSTM1 through dual binding to LC3 and ubiquitin plays a key role in the clearing of pathological poly-ubiquitinated protein aggregates by recruiting the autophagic machinery. In this setting, p62/SQSTM1 interacts and co-localizes with a ubiquitously expressed large phosphoinositide-binding protein Alfy (autophagy-linked FYVE).28 Upon LMB treatment, both proteins, are relocated within, and are essential for the assembly of nuclear p62 bodies which contain a high amount of poly-ubiquitinated proteins. Noteworthily, although p62 bodies are generated by blockage of CRM1-dependent nuclear export, they do not accumulate LMB-bound CRM1 or, notwithstanding P62/SQSTM1, its substrates like PABP1, β-actin or U2-snRNA (not shown). Intriguingly, p62 bodies belong to a long list of fluorescent foci such as those containing E2F3,49 p53-HDM2,50 PLZF,51 Bach2,52 BCL6,53 Pat1b,17 KRAB and KAP1,54 which were found associated with PML bodies. It has been suggested that PML bodies may act as nuclear sensors that detect highly localized, abnormal concentration of proteins.55
Our observations confirm that PML bodies and p62 bodies interact specifically. Under the EM, neither PML bodies nor p62 bodies were found docking to other nuclear bodies like the paraspeckles or CBs. Furthermore, hybrid bodies consistently composed of a shell of PML surrounding a characteristic p62 body progressively accumulate during leptomycin treatment. Several scenarios might account for the mixture of PML, p62 and hybrid bodies juxtaposed within nuclei of LMB-treated HeLa cells. First, p62 bodies could form independently of - and progressively fuse with - PML bodies. Alternatively, hybrid bodies could form first and progressively segregate PML and p62/SQSTM1 so as to generate p62-only and PML-only bodies. We cannot either exclude more complex patterns with repeated fusion/separation or fusion/degradation cycles. Nevertheless, the respective increasing and decreasing frequency of hybrid and PML bodies over time favors the scenario of a fusion process, which is likely caught in the act in the EM images displayed in Fig. 8 and Figure S3.
Nuclear body assembly
Various models of nuclear body assembly have been proposed.2,3 A “stochastic-assembly” model was inferred from chromatin tethering experiments showing that many CB components initiate CBs assembly.56 In contrast, an “ordered assembly” model was deduced from the characterization of seeding elements, which like SUMOylated PML and the lncRNA NEAT1 promote assembly of PML bodies and paraspeckles respectively.2,57 The formation of the hybrid p62/PML bodies as reported in this study would add a new layer to a hierarchically “ordered assembly” model with 2 different pre-assembled bodies, one being dependent on p62 and Alfy, the other dependent on PML, merging to form a new NB in a response to stress. The recent finding that protein and RNP granules can result from phase transitions that lead to the local aggregation of otherwise diffuse components sheds new light on these observations.58 Indeed, membrane-less organelles such as nucleoli or cytoplasmic germ granules have all physical characteristics of viscous droplets. In this context, the occasional fusions previously observed for nucleoli in life cells is a manifestation of their inherent liquid nature.59 In the current study, the distribution of PML at the periphery of the hybrid p62/PML bodies, as observed in Figures 7C and 8, is reminiscent of the behavior of a surfactant, and suggests that it actively interfaces with both components of the fused granule and the surrounding nucleoplasm. Finally, phase transitions are tightly controlled events, as illustrated by the chronology of nucleolus reassembly after mitosis or by the formation of grP-bodies in quiescent C. elegans oocytes.60 Thus, the fusion observed specifically between PML and p62 bodies is likely to fulfil a function in cells. This is consistent with the scenario proposing that p62/SQSTM1 transfers ubiquitinated proteins to PML bodies for proteasomal degradation.25 In line with the formation of specialized PML bodies enriched in components of the ubiquitin-proteasome proteolytic pathway, the clastosomes,61 which prevent accumulation of aggregate-prone polyglutamine and/or midfolded proteins,62,63 it is tempting to speculate that the “capture” of stress-induced nuclear bodies by PML bodies results in their degradation.
Material and Methods
Cells and reagents
HeLa cells were grown in DMEM supplemented with 10% fetal calf serum (FCS) at 37°C with 5% CO2. LMB was added at a 10 nM final concentration from a 10 µm stock solution in 70% methanol (Sigma-Aldrich). An equivalent amount of methanol was added to control cells. The following rabbit polyclonal antibodies were used in this study: anti-SUMO-2/3 (Abcam 3742), anti-SUMO1 (Santa Cruz sc FL-101), anti-CRM1 (Santa Cruz sc-5595), anti-Histone H3 (Abcam1791), anti-p62/SQSTM1 (Santa Cruz sc-25575), anti-Ubiquitin (DAKO Z 0458), anti-Coilin (Santa Cruz sc-32860), rabbit anti-PML.64 Monoclonal mouse anti-CRM1 (BD 611 832), anti-PML (Santa Cruz sc PG-M3) and anti-p62/SQSTM1 (BD transduction Laboratories 610832) were used single or in conjunction with rabbit antibodies for double-labeling experiments.
Immuno-fluorescence Microscopy (IF)
For immunofluorescence (IF), HeLa cells were fixed in 4% formalin/H2O for 20 min at RT, permeabilized in 0.2% Triton-X-100/PBS for 20 min, and incubated with primary (e.g anti-coilin + anti-p62/SQSTM1 or anti-PML + anti-p62/SQSTM1, all diluted at 1:200) and secondary Alexa Fluor® 594 anti-rabbit and/or Alexa Fluor® 488 anti-mouse antibodies, diluted at 1:500 (Life technologies, Carlsbad, CA). All antibodies were diluted in PBST/BSA (PBS/Tween20 0.5%/BSA 3%). For Crm1, cells were fixed in 4% PAF for 10 min before permeabilization. Cells were 4,6-diamidino-2-phenylindole (DAPI) stained, mounted in Vectashield (Vector Laboratories, Burlingame, CA), and imaged with an inverted Olympus microscope (Nikon, Tokyo, Japan) using a 40/0.75 objective, except for CRM1 microscopy performed on a Leica DMR microscope (Leica, Heidelberg, Germany) using a 63X1.32 oil immersion objective. Photographs were taken using a Micromax CCD camera (Princeton Instruments) driven by Metamorph software.
Ultrastructural observations
Conventional ultrastructural microscopy after Epon embedding was as in.65 Thin sections were analyzed with a Tecnai Spirit (FEI, Hillsboro, OR) and digital images were taken with a SIS MegaviewIII charge-coupled device camera (Olympus, Tokyo, Japan).
Immuno-electron microscopy (I-EM)
Protein and nucleic acid localization were performed on thin-sections of cells embedded at low temperature in Lowicryl K4M (Polysciences Inc., PA, USA) as in.66 Duplicated cell samples were fixed in situ for 1h at 4°C either with 1.6% glutaraldehyde or with 4% formaldehyde freshly prepared from paraformaldehyde). Both fixatives were in 0.1M Sörensen phosphate buffer pH 7.3. Cells were scrapped-off plastic containers and centrifuged. After rinsing in phosphate buffer, cell pellets were equilibrated in 30% methanol and deposited in a Leica EM AFS2/FSP automatic reagent handling apparatus (Leica Microsystems). Lowicryl polymerization under UV was for 40 h at – 20°C and 40 h at + 20°C. Ultra-thin sections were incubated at room temperature for 1 h with the primary antibody and for 30 min with the secondary anti-mouse or anti-rabbit antibody coupled to 10-nm gold particles (BBInternational, Cardiff, UK). All antibodies, diluted 1:10 to 1:30 in PBS were tested on thin sections obtained with both fixatives. The DAKO anti-ubiquitin antibody was usable only on PF-fixed cell samples as it produced very high background on GLUT-fixed cell samples. Anti-PML antibodies gave cleaner labeling on GLUT-fixed cells.
For double-labeling, thin-sections were incubated on primary antibodies mixed in PBS, then with a mix of secondary antibodies conjugated to different gold particles sizes (5, 10 or 15 nm).
To label 2 proteins with antibodies derived from the same species e.g rabbit as in Fig. 5, one of the antibody was biotinylated in vitro by incubating 10 µl of the antibody for 3–6 h at 4°C with 0.3 µl of a solution of 2 mg of the EZ-link® Sulfo-NHS-LC-LC-Biotin reagent (Thermoscientific) freshly dissolved in 300 µl H2O. The double-labeling was carried out by incubating the thin section sequentially with: the unmodified primary antibody, the corresponding secondary antibody coupled to 5 or 15 nm gold particles and, after washing, the biotinylated primary antibody and a goat anti-biotin antibody conjugated to 10 nm gold particles (BBInternational).
Surface areas of nuclear body sections were determined with AnalySIS (Olympus Soft Imaging Solutions, Munster, Germany). The scatterplot and the Mann-Whitney U test were performed using GraphPad Prism (GraphPad, San Diego, CA). Gold particles were counted by eye. Calculations and standard deviations for labeling densities were obtained with Excel (Microsoft, Redmond, WA).
Electron microscopic in situ hybridization (EM-ISH)
Biotinylated 18S rDNA probe was as described in.65 Hybridization conditions and detection of RNA/DNA hybrids with goat anti-biotin antibody conjugated to 10 nm gold particles (BBInternational) was as described in.9,66
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank H. de Thé for kind gift of polyclonal anti-PML antibody, Guillaume Beauclair for analysis of INB proteins with SUMO-SIM predictors software's.
Funding
This work was supported by the Centre National de la Recherche Scientifique, Fondation ARC pour la Recherche, and the Agence Nationale pour la Recherche contract ANR-14-CE09-0013-01.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000661; PMID:20452955; http://dx.doi.org/ 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet 2011; 27:295-306; PMID:21680045; http://dx.doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 2009; 17:639-47; PMID:19922869; http://dx.doi.org/ 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al.. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. Embo J 1994; 13:1073-83; PMID:8131741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res 1969; 27:266-88; PMID:5813971; http://dx.doi.org/ 10.1016/S0022-5320(69)80017-1 [DOI] [PubMed] [Google Scholar]
- 6.Navascues J, Berciano MT, Tucker KE, Lafarga M, Matera AG. Targeting SMN to Cajal bodies and nuclear gems during neuritogenesis. Chromosoma 2004; 112:398-409; PMID:15164213; http://dx.doi.org/ 10.1007/s00412-004-0285-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 1991; 195:27-37; PMID:2055273; http://dx.doi.org/ 10.1016/0014-4827(91)90496-H [DOI] [PubMed] [Google Scholar]
- 8.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol 1990; 104:120-7; PMID:2088441; http://dx.doi.org/ 10.1016/1047-8477(90)90066-L [DOI] [PubMed] [Google Scholar]
- 9.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 2010; 21:4020-7; PMID:20881053; http://dx.doi.org/ 10.1091/mbc.E10-08-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visa N, Puvion-Dutilleul F, Bachellerie JP, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur J Cell Biol 1993; 60:308-21; PMID:8330629 [PubMed] [Google Scholar]
- 11.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 1994; 76:345-56; PMID:8293468; http://dx.doi.org/ 10.1016/0092-8674(94)90341-7 [DOI] [PubMed] [Google Scholar]
- 12.Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol 2013; 203:149-64; PMID:24127217; http://dx.doi.org/ 10.1083/jcb.201303145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell 1999; 10:3015-33; PMID:10473643; http://dx.doi.org/ 10.1091/mbc.10.9.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 2002; 13:1282-97; PMID:11950939; http://dx.doi.org/ 10.1091/mbc.01-11-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell 2009; 20:176-87; PMID:18923137; http://dx.doi.org/ 10.1091/mbc.E08-09-0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutten S, Prescott A, James J, Riesenberg S, Boulon S, Lam YW, Lamond AI. An intranucleolar body associated with rDNA. Chromosoma 2011; 120:481-99; PMID:21698343; http://dx.doi.org/ 10.1007/s00412-011-0327-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marnef A, Weil D, Standart N. RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol Biol Cell 2012; 23:213-24; PMID:22090346; http://dx.doi.org/ 10.1091/mbc.E11-05-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci USA 2008; 105:16964-9; PMID:18957539; http://dx.doi.org/ 10.1073/pnas.0809273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjolov AA. The nucleolus and Ribosome biogenesis. 1985 [Google Scholar]
- 20.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8:574-85; PMID:17519961; http://dx.doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 21.Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell 2005; 16:3401-10; PMID:15857956; http://dx.doi.org/ 10.1091/mbc.E05-02-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol 2002; 12:1-11; PMID:11790298; http://dx.doi.org/ 10.1016/S0960-9822(01)00650-9 [DOI] [PubMed] [Google Scholar]
- 23.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol Biol Cell 2002; 13:4100-9; PMID:12429849; http://dx.doi.org/ 10.1091/mbc.E02-05-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40:216-27; PMID:20965417; http://dx.doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem 2010; 285:5941-53; PMID:20018885; http://dx.doi.org/ 10.1074/jbc.M109.039925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 27.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603-14; PMID:16286508; http://dx.doi.org/ 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, et al.. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010; 6:330-44; PMID:20168092; http://dx.doi.org/ 10.4161/auto.6.3.11226 [DOI] [PubMed] [Google Scholar]
- 29.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al.. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014; 25(1):169-83; PMID:24173718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al.. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777-87; PMID:15574332; http://dx.doi.org/ 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 31.Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 2007; 120:1540-50; PMID:17405816; http://dx.doi.org/ 10.1242/jcs.001529 [DOI] [PubMed] [Google Scholar]
- 32.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med 1991; 173:1407-19; PMID:2033369; http://dx.doi.org/ 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochs RL, Stein TW Jr., Tan EM. Coiled bodies in the nucleolus of breast cancer cells. J Cell Sci 1994; 107(Pt 2):385-99; PMID:8207070 [DOI] [PubMed] [Google Scholar]
- 34.Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 2010; 119:527-40; PMID:20449600; http://dx.doi.org/ 10.1007/s00412-010-0276-7 [DOI] [PubMed] [Google Scholar]
- 35.Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res 1998; 243:290-304; PMID:9743589; http://dx.doi.org/ 10.1006/excr.1998.4135 [DOI] [PubMed] [Google Scholar]
- 36.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol 2009; 184:771-6; PMID:19289796; http://dx.doi.org/ 10.1083/jcb.200812014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desterro JM, Keegan LP, Jaffray E, Hay RT, O'Connell MA, Carmo-Fonseca M. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell 2005; 16:5115-26; PMID:16120648; http://dx.doi.org/ 10.1091/mbc.E05-06-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005; 436:428-33; PMID:15931174 [DOI] [PubMed] [Google Scholar]
- 39.Hang LE, Lopez CR, Liu X, Williams JM, Chung I, Wei L, Bertuch AA, Zhao X. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J Biol Chem 2014; 289:10308-17; PMID:24567323; http://dx.doi.org/ 10.1074/jbc.M113.526178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Wang L, Zhou P, Wang G, Zeng Y, Wang Y, Liu J, Zhang B, Liu S, Luo H, et al.. Regulation of REGgamma cellular distribution and function by SUMO modification. Cell Res 2011; 21:807-16; PMID:21445096; http://dx.doi.org/ 10.1038/cr.2011.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 2011; 18:831-9; PMID:21642969; http://dx.doi.org/ 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 42.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 2014; 42:W325-30; PMID:24880689; http://dx.doi.org/ 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelisch F, Gerez J, Druker J, Schor IE, Munoz MJ, Risso G, Petrillo E, Westman BJ, Lamond AI, Arzt E, et al.. The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc Natl Acad Sci U S A 2010; 107:16119-24; PMID:20805487; http://dx.doi.org/ 10.1073/pnas.1004653107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol 1994; 6:354-9; PMID:7917325; http://dx.doi.org/ 10.1016/0955-0674(94)90026-4 [DOI] [PubMed] [Google Scholar]
- 45.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol 1997; 7:767-75; PMID:9368759; http://dx.doi.org/ 10.1016/S0960-9822(06)00335-6 [DOI] [PubMed] [Google Scholar]
- 46.Xu S, Powers MA. Nup98-homeodomain fusions interact with endogenous Nup98 during interphase and localize to kinetochores and chromosome arms during mitosis. Mol Biol Cell 2010; 21:1585-96; PMID:20237156; http://dx.doi.org/ 10.1091/mbc.E09-07-0561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 1999; 147:715-28; PMID:10562276; http://dx.doi.org/ 10.1083/jcb.147.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. Identification of CRM1-dependent Nuclear Export Cargos Using Quantitative Mass Spectrometry. Mol Cell Proteomics 2013; 12:664-78; PMID:23242554; http://dx.doi.org/ 10.1074/mcp.M112.024877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernier M, Bourdeau V, Gaumont-Leclerc MF, Moiseeva O, Begin V, Saad F, Mes-Masson AM, Ferbeyre G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev 2013; 25:41-50; http://dx.doi.org/ 10.1101/gad.1975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lain S, Xirodimas D, Lane DP. Accumulating active p53 in the nucleus by inhibition of nuclear export: a novel strategy to promote the p53 tumor suppressor function. Exp Cell Res 1999; 253:315-24; PMID:10585254; http://dx.doi.org/ 10.1006/excr.1999.4672 [DOI] [PubMed] [Google Scholar]
- 51.Ruthardt M, Orleth A, Tomassoni L, Puccetti E, Riganelli D, Alcalay M, Mannucci R, Nicoletti I, Grignani F, Fagioli M, et al.. The acute promyelocytic leukaemia specific PML and PLZF proteins localize to adjacent and functionally distinct nuclear bodies. Oncogene 1998; 16:1945-53; PMID:9591778; http://dx.doi.org/ 10.1038/sj.onc.1201722 [DOI] [PubMed] [Google Scholar]
- 52.Tashiro S, Muto A, Tanimoto K, Tsuchiya H, Suzuki H, Hoshino H, Yoshida M, Walter J, Igarashi K. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol Cell Biol 2004; 24:3473-84; PMID:15060166; http://dx.doi.org/ 10.1128/MCB.24.8.3473-3484.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchberger E, El Harchi M, Payrhuber D, Zommer A, Schauer D, Simonitsch-Klupp I, Bilban M, Brostjan C. Overexpression of the transcriptional repressor complex BCL−6/BCoR leads to nuclear aggregates distinct from classical aggresomes. PLoS One 2013; 8:e76845; PMID:24146931; http://dx.doi.org/ 10.1371/journal.pone.0076845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Briers S, Crawford C, Bickmore WA, Sutherland HG. KRAB zinc-finger proteins localise to novel KAP1-containing foci that are adjacent to PML nuclear bodies. J Cell Sci 2009; 122:937-46; PMID:19258395; http://dx.doi.org/ 10.1242/jcs.034793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol 2000; 2:871-8; PMID:11146650; http://dx.doi.org/ 10.1038/35046510 [DOI] [PubMed] [Google Scholar]
- 56.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; http://dx.doi.org/ 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- 57.Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de The H, et al.. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol 2014; 204:931-45; PMID:24637324; http://dx.doi.org/ 10.1083/jcb.201305148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol 2013; 203:875-81; PMID:24368804; http://dx.doi.org/ 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 2011; 108:4334-9; PMID:21368180; http://dx.doi.org/ 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell 2013; 27:161-73; PMID:24176641; http://dx.doi.org/ 10.1016/j.devcel.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lafarga M, Berciano MT, Pena E, Mayo I, Castano JG, Bohmann D, Rodrigues JP, Tavanez JP, Carmo-Fonseca M. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biol Cell 2002; 13:2771-82; PMID:12181345; http://dx.doi.org/ 10.1091/mbc.E02-03-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo L, Giasson BI, Glavis-Bloom A, Brewer MD, Shorter J, Gitler AD, Yang X. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol Cell 2014; 55:15-30; PMID:24882209; http://dx.doi.org/ 10.1016/j.molcel.2014.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, Brice A, Trottier Y, Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol 2006; 174:65-76; PMID:16818720; http://dx.doi.org/ 10.1083/jcb.200511045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daniel MT, Koken M, Romagne O, Barbey S, Bazarbachi A, Stadler M, Guillemin MC, Degos L, Chomienne C, de The H. PML protein expression in hematopoietic and acute promyelocytic leukemia cells. Blood 1993; 82:1858-67; PMID:8400236 [PubMed] [Google Scholar]
- 65.Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci 2009; 122:3619-26; PMID:19812307; http://dx.doi.org/ 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- 66.Souquere S, Pierron G. Ultrastructural analysis of nuclear bodies using electron microscopy. Methods Mol Biol 2015; 1262:105-18; PMID:25555578; http://dx.doi.org/ 10.1007/978-1-4939-2253-6_7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


