Abstract
Bordetella pertussis, the causative agent of human whooping cough (pertussis) produces a complex array of virulence factors in order to establish efficient infection in the host. The RNA chaperone Hfq and small regulatory RNAs are key players in posttranscriptional regulation in bacteria and have been shown to play an essential role in virulence of a broad spectrum of bacterial pathogens. This study represents the first attempt to characterize the Hfq regulon of the human pathogen B. pertussis under laboratory conditions as well as upon passage in the host and indicates that loss of Hfq has a profound effect on gene expression in B. pertussis. Comparative transcriptional profiling revealed that Hfq is required for expression of several virulence factors in B. pertussis cells including the Type III secretion system (T3SS). In striking contrast to the wt strain, T3SS did not become operational in the hfq mutant passaged either through mice or macrophages thereby proving that Hfq is required for the functionality of the B. pertussis T3SS. Likewise, expression of virulence factors vag8 and tcfA encoding autotransporter and tracheal colonization factor, respectively, was strongly reduced in the hfq mutant. Importantly, for the first time we demonstrate that B. pertussis T3SS can be activated upon contact with macrophage cells in vitro.
Keywords: Bsp22, Hfq, infection, T3SS, transcriptomics, virulence
Abbreviations
- ABC protein
ATP-binding cassette protein
- CFU
colony forming unit
- OMP
outer membrane protein
- P
P-value
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- SS medium
Steiner-Scholte medium
- T3SS
Type III secretion system
- wt
wild-type
Introduction
Bordetella pertussis is a Gram-negative bacterial pathogen of the human respiratory tract and causative agent of whooping cough (pertussis). Infection by B. pertussis is especially severe in infants and despite vaccination, B. pertussis remains a major cause of children mortality worldwide, predominantly in developing countries.1-3 However, pertussis is now on the rise also in developed countries and is reemerging in some highly vaccinated populations.3-5 The increase in pertussis cases has been mostly attributed to short-lived immunity induced by the acellular vaccine and escape from immunity due to pathogen adaptation and antigen variation.6-9 Therefore, there is an urgent need for a better understanding of molecular mechanisms underlying the pathogenesis of B. pertussis infection that would lead to identification of novel factors as potential components of new vaccine formulations.10,11
In order to successfully infect the host, B. pertussis produces a broad spectrum of virulence factors including filamentous hemagglutinin, pertactin, fimbriae, pertussis toxin and adenylate cyclase toxin.1,12 The type III secretion system (T3SS) allowing injection of proteins referred to as effectors directly into the cytosol of eukaryotic host cells has also been identified in Bordetella subspecies.13,14 The T3SS is a supramolecular structure consisting of an inner membrane export apparatus, a basal body that spans from inner to the outer membrane, an extracellular needle and the needle tip (translocon) complex.15,16 In the closely related B. bronchiseptica, this “injectisome” is composed of approximately 30 proteins including chaperones and effectors which are encoded by the bsc locus.13 Bordetella T3SS represents an important virulence factor as it subverts innate and adaptive immune responses of the host and is required for long term colonization of trachea of rats and mice.13,17,18 Although the T3SS of B. pertussis is highly homologous to that of B. bronchiseptica and bsc genes are actively transcribed,19,20 laboratory-adapted high-passage B. pertussis strains like Tohama I do not produce and secrete T3SS substrates when grown in vitro.20-22 However, the functionality of T3SS can be reverted upon contact with the host as evidenced by production of the T3SS tip complex protein Bsp22 in B. pertussis cells recovered from infected mice.22 Intriguingly, the T3SS activation is reversible as the production of Bsp22 protein is lost again after successive in vitro passages.22 Bsp22 is the most abundant T3SS substrate secreted by B. bronchiseptica and is considered as a hallmark of T3SS functionality.18 Deletion of bsp22 gene in B. bronchiseptica disrupts T3SS-mediated cytotoxicity and affects colonization levels and persistence in the lower respiratory tract of mice.18 Interestingly, production of Bsp22 was observed also in fresh clinical isolates of B. pertussis, thereby further confirming that contact with the host is crucial for switching on the expression of T3SS.21,22
The expression of the majority of B. pertussis virulence factors is controlled at transcriptional level by a two-component system encoded by the bvg locus (for a review see ref.23). This consists of the transmembrane sensor kinase BvgS and of the DNA-binding response regulator BvgA which in its phosphorylated form binds to promoter regions and activates transcription of dependent virulence genes.24,25 Expression of the bsc locus is regulated at both transcriptional and posttranscriptional levels by the BvgAS system and by factors encoded by the btr locus which is adjacent to bsc genes20 (Fig. 1A). The BtrS protein whose production is tightly activated by the BvgAS system bears homology to the family of extracytoplasmic function sigma factors and is required for transcription of bsc and btr loci. BtrU, BtrV and BtrW factors carry high similarity to a partner switching module known in Gram-positive bacteria and are required for secretion and stability of T3SS substrates.20
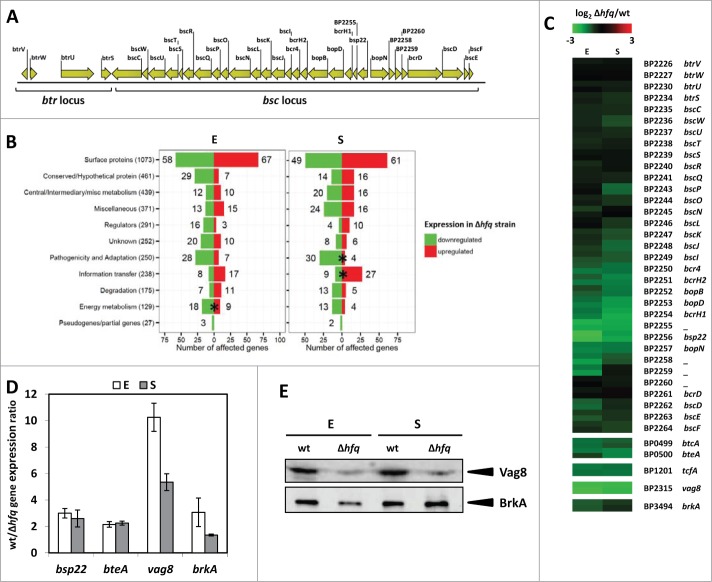
Figure 1.
Comparative transcriptomic analysis of laboratory-adapted wt and Δhfq strains of B. pertussis. (A) Genomic organization and open reading frames of the Type III secretion cluster in B. pertussis. The genes comprising both bsc and regulatory btr loci are presented as arrows. The drawing was created using CLC Sequence viewer version 6.6.2 (CLC bio A/S). (B) Functional categorization of genes significantly affected by loss of Hfq function. Data are expressed as numbers of genes within category which were significantly down- (green bars) or upregulated (red bars) in the hfq mutant cells grown to exponential (E) or stationary (S) phase of growth. Total numbers of B. pertussis genes associated with a specific category are shown in brackets. Significantly overrepresented functional categories are depicted with asterisks, P ˂ 0.05. (C) Changes in transcript abundance patterns of selected virulence factors in B. pertussis. The heat map presents comparison of gene expression profiles between Δhfq and wt cells (log2FC Δhfq/wt) grown to exponential (E) or stationary (S) phase as determined by microarray analysis. Data are averaged from 2 biological replicates, each gene is represented by 2 array elements. All expression profiles are in rows and are represented using the color scale on the top. (D) RT-qPCR analysis of bsp22, bteA, vag8 and brkA transcript levels in wt and Δhfq cells. Total RNA was isolated from cells grown to exponential (E; white columns) and stationary (S; gray columns) phases and RT-qPCR was performed using primers specific for bsp22, bteA, vag8 and brkA genes. Target expression levels in wt and Δhfq cells were normalized to the reference gene rpoB and the wt/Δhfq expression ratios were calculated. The values are means plus standard deviations from 3 experiments. (E) Western blot analysis of Vag8 and BrkA protein levels in Tohama I wt and Δhfq strain. Both strains were grown in SS medium and cells were harvested at exponential (E) and stationary (S) phase. Samples of cell lysates equivalent to 0.1 OD600 unit of initial culture were separated on 10% SDS-PAGE gel and analyzed by immunoblotting using anti-Vag8 and anti-BrkA antibodies. Relevant parts of the membranes are shown. The result is representative of 3 experiments.
In many pathogenic bacteria the RNA chaperone Hfq and small non-coding regulatory RNAs (sRNAs) emerged as key players in posttranscriptional regulation of virulence and physiological fitness.26,27 Posttranscriptional regulation of B. pertussis virulence has not yet been studied extensively, however, several sRNA were identified in B. pertussis28 and recently, we have shown that the RNA chaperone Hfq is required for virulence of B. pertussis.29 The Tohama I Δhfq strain produced decreased amounts of adenylate cyclase toxin and secreted reduced amounts of pertussis toxin. Consequently, the hfq mutant was clearly attenuated in the mouse respiratory model of infection as its lethality as well as its capacity to colonize mouse lungs was strongly reduced when compared to the wt strain. These data encouraged us to apply microarray analysis in order to evaluate the global impact of hfq deletion on gene expression with specific focus on virulence. Through differential analysis of expression profiles of laboratory-adapted and mouse-passaged wt and Δhfq strains we aimed at the identification of Hfq regulon in B. pertussis. In this work we show that loss of Hfq function has a deep impact on expression of several virulence factors including T3SS. Our data clearly indicate that Hfq is strongly required for functionality of T3SS in B. pertussis.
Results
Hfq affects global gene expression in B. pertussis
To evaluate the effect of hfq deletion on global gene expression profiles, DNA microarrays covering 3554 B. pertussis protein-coding genes were hybridized with total RNA purified from wt and Δhfq cells grown to exponential and stationary phase of growth. A cutoff of at least twofold variation in expression (log2FC >1; <−1; P < 0.05) was applied to identify genes with significant changes in transcript abundance between wt strain and hfq mutant. In the exponential phase, 368 genes (≈10.6%) fulfilled these criteria and displayed differences in transcript abundance (Fig. 1B and Table S1). Among them, 212 genes were downregulated and 156 upregulated in the Δhfq strain. In the stationary phase, the transcriptional profiles exhibited similar pattern with 351 (10.1%) significantly affected genes (Table S2). Among them, 186 genes were downregulated and 165 upregulated. Functional class categorization of filtered Hfq-dependent genes showed significant enrichment (P < 0.05) in some gene categories (Fig. 1B). In the exponential phase, “Energy metabolism” was the only functional category significantly enriched for Hfq-dependent genes (20.9%; P < 0.002) since the “Pathogenicity and Adaptation” category did not reach the statistical significance (14.0%; P = 0.054). In the stationary phase, genes related to “Pathogenicity and Adaptation” (13.6%; P < 0.05) and “Information transfer” (15.1%; P < 0.015) categories were significantly overrepresented. Notably, in the exponential and stationary phase 80% (28/35) and 88% (30/34) of genes related to “Pathogenicity and adaptation” category were downregulated in the hfq mutant, respectively, being fully in accordance with reduced virulence of the mutant. Interestingly, a large portion of affected genes (17/25 and 20/36, respectively) within “Information transfer” category belonged to genes encoding 30S and 50S ribosomal proteins. While not significantly enriched, the highest numbers (125 and 110, respectively) of affected genes were related to largest functional category of “Surface proteins.” Besides large number of genes predicted to encode putative membrane proteins this group also included several genes encoding ATP binding cassette (ABC) transport proteins which expression in the exponential phase was largely upregulated in the mutant (Tables S1 and S2). Interestingly, 16 of 19 genes (84.2%) associated with regulatory function were downregulated in the Δhfq strain in the exponential phase.
Lack of Hfq results in reduced expression of several virulence genes including T3SS
Within the set of genes related to pathogenicity and displaying significantly lower transcript levels in the mutant regardless of the growth phase, we identified the T3SS bsc gene cluster (Fig. 1C). Among the significantly downregulated genes were bsp22, bopD and bopN encoding the T3SS tip complex protein, the pore-forming translocator protein and the T3SS effector, respectively. In contrast, the expression of the btr locus which comprises T3SS regulatory genes was not significantly affected. Notably, expression of bteA and btcA genes which are not part of the bsc locus and encode another T3SS effector protein BteA and its chaperone BtcA, respectively, was also significantly downregulated in the hfq mutant. Similarly, expression of other virulence genes including vag8 autotransporter, serum resistance gene brkA (only in the exponential phase) and tracheal colonization factor tcfA was also significantly decreased in the mutant strain, suggesting that Hfq has an impact on a broad spectrum of virulence genes (Fig. 1C). In contrast to the majority of virulence genes and in agreement with our previous data,29 the ptx/ptl locus was significantly upregulated in the hfq mutant (Tables S1 and S2).
In order to verify our microarray data, we have analyzed transcript levels of several virulence genes including the T3SS locus. We have quantified the levels of the bsp22, bteA, vag8 and brkA transcripts in both wt and Δhfq strains of B. pertussis by RT-qPCR. As shown in Figure 1D, the relative expression levels of bsp22, bteA, vag8 and brkA genes were decreased in the hfq mutant thereby confirming our microarray data. Furthermore, Western blot analysis of wt and Δhfq cell lysates showed that levels of Vag8 and BrkA proteins mirrored the relative transcript levels and thereby provided another layer of evidence for credibility of our microarray data (Fig. 1E).
Hfq is required for regained production of Bsp22 protein upon passage in mice
Transcriptional profiling of wt and Δhfq strains adapted to laboratory culture conditions suggested that Hfq is required for expression of several virulence factors including T3SS. Therefore, we asked whether the differential expression of the T3SS locus would have an impact on production of T3SS components. Laboratory-adapted B. pertussis strains like Tohama I do not produce T3SS substrates under standard in vitro conditions, however, Gaillard and coworkers have shown that after passage through mice the recovered Tohama I cells produced Bsp22 into SS medium and kept the production for 3 additional passages in vitro.22 Therefore, we decided to passage both strains in mice and monitor their ability to produce and secrete the T3SS tip complex protein Bsp22 (marker of T3SS functionality).Toward this aim, CD-1 mice were infected with sublethal doses of both wt and Δhfq strain (1 × 105 CFU/mouse) and colonization profiles of both strains were examined over 23 days. As illustrated by Figure 2A and in line with our previous observations,29 the wt strain was able to proliferate in lungs and its CFU counts increased by more than one order of magnitude within 8 days of infection whereas the hfq mutant was unable to multiply and was cleared from mouse lungs with a significantly higher efficacy. In the course of the infection, the viable wt and Δhfq bacteria were recovered from mouse lungs at each experimental time point, subcultivated in SS medium and assayed for Bsp22 accumulation in whole-cell lysates and in culture supernatants. As shown in Figure 2B, Bsp22 protein accumulated in whole cell lysates as well as in culture supernatants of mouse-passaged wt bacteria recovered at days 8, 12 and 23 post infection. Strikingly, Bsp22 protein was absent in cell lysates and culture supernatants of passaged Δhfq bacteria. These results indicated that Hfq is required for both expression and production of Bsp22 and consequently, for T3SS function in B. pertussis.
Figure 2.
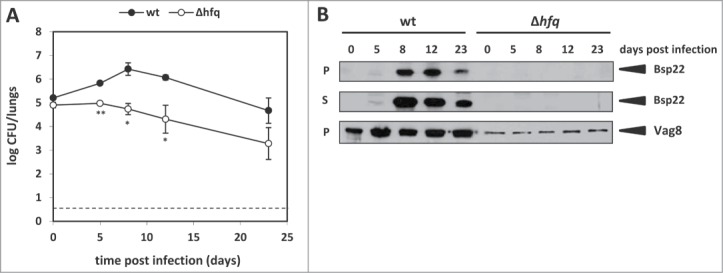
Hfq is required for Bsp22 and Vag8 production in B. pertussis. (A) Colonization of mouse lungs after intranasal challenge with the wt or Δhfq strain. 15 mice/strain were infected intranasally with 1 × 105 CFU/mouse and groups of 3 animals were sacrificed 2 h (day 0) and 5, 8, 12 and 23 days after infection. Lung homogenates were plated in duplicates on BGA plates and CFU of the wt (•) and Δhfq (○) strains were counted. The values are means plus standard deviations. *, P˂ 0.05; **, P˂ 0.01 (Δhfq versus wt). The dashed line represents the limit of detection. (B) Westernblot analysis of Bsp22 and Vag8 protein levels in the wt or Δhfq strains recovered from lung homogenates in the course of colonization experiment (see panel A). Samples equivalent to 0.1 OD600 unit (pellets, P) or 1 OD600 unit (supernatants, S) of initial culture were analyzed by immunoblotting using anti-Bsp22 and anti-Vag8 antibodies. Relevant parts of the membranes are shown.
Transcriptional profiling of passaged strains confirms requirement of Hfq for virulence
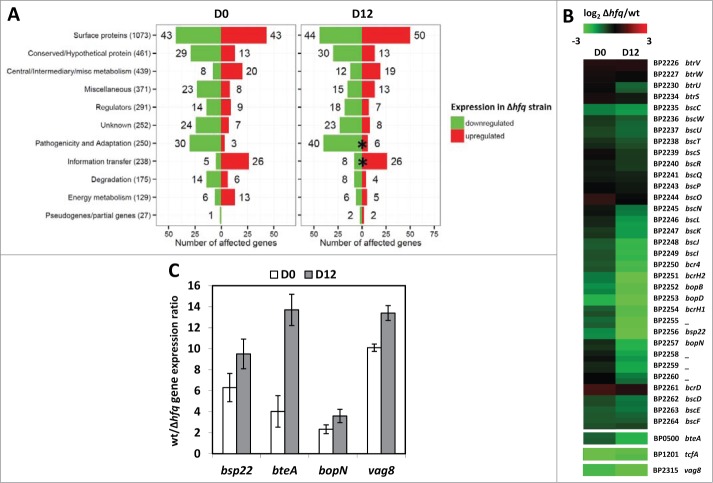
Next, we assumed that transcriptional profiling of wt and Δhfq bacteria passaged in mice would not only allow us to further corroborate the effect of hfq deletion on production of T3SS components but could also enlighten the global contribution of Hfq to B. pertussis adaptation and survival during infection. Therefore, in order to elaborate this assumption, total RNA was purified from wt and Δhfq bacteria recovered at days 0 and 12 post infection and used for microarray analysis to determine changes in gene expression occurring upon passage in mice. A cutoff of at least twofold variation in expression (log2FC >1; <−1; P < 0.05) between wt and hfq mutant strains (Δhfq versus wt at day 0 and at day 12) or in the course of infection (day 12 vs. day 0 for both strains) was applied to identify genes with significant changes in transcript abundance. Comparison of expression profiles of both strains at day 0 revealed 346 (10.1%) significantly affected genes (Fig. 3A, left histogram) showing a distribution into functional categories similar to our initial experiment (Fig. 1B). In the hfq mutant, 198 genes were downregulated and 148 genes were upregulated when compared to the wt strain (Table S3). None of the functional categories was enriched significantly, however, categories “Energy metabolism” (14.7%; P = 0.062), “Information transfer” (13%; P = 0.074) and “Pathogenicity and Adaptation” (13.2%; P = 0.058) were highly represented. Within the latter category, expression of majority of the bsc locus and bteA and vag8 genes was significantly diminished in the hfq mutant. The difference in expression of these virulence factors became even more apparent when we compared the transcriptomic profiles of wt and Δhfq bacteria recovered 12 days post infection (Table S4). As indicated in Figure 3A (right histogram), the total number of affected genes (359, 10.4%) did not surprisingly differ from day 0, however, the “Pathogenicity and Adaptation” category became the only significantly overrepresented functional category (18.4%; P < 1 × 10−4) with 40 out of 46 genes downregulated in the hfq mutant. Likewise, 12 days post infection the difference in expression of vag8 and T3SS-specific genes between wt and Δhfq strains further increased (Fig. 3B). Interestingly, expression of the btrU gene which plays a role in secretion of T3SS substrates was also significantly downregulated in the hfq mutant upon infection. In addition, genes involved in iron metabolism such as bhuU and BP0546 encoding hemin permease and bacterioferritin, respectively, also exhibited lower transcript levels in the Δhfq strain at day 12 (Table S4). Within the set of genes observed to have significantly higher transcript levels in the hfq mutant 12 days post infection were bvgR and fim2 encoding BvgR regulatory factor and serotype 2 fimbrial subunit, respectively.
Figure 3.
Comparative transcriptomic analysis of mouse-passaged wt and Δhfq strains. (A) Functional categorization of genes significantly affected by loss of Hfq function in B. pertussis cells recovered at day 0 (D0) and at day 12 (D12) post infection. Data are expressed as numbers of genes within category which were significantly down- (green bars) or upregulated (red bars) in the hfq mutant. Total numbers of B. pertussis genes associated with a specific category are shown in brackets. Significantly overrepresented functional categories are depicted with asterisks, P ˂ 0.05. (B) Changes in transcript abundance patterns of selected virulence factors in B. pertussis. The heat map presents comparison of gene expression profiles between Δhfq and wt cells recovered (log2FC Δhfq/wt) at day 0 and day 12 post infection as determined by microarray analysis. All expression profiles are in rows and are represented using the color scale on the top, each gene is represented by 2 array elements. (C) RT-qPCR analysis of bsp22, bteA, bopN and vag8 transcript levels in wt and Δhfq bacteria upon infection. Differential expression analysis was performed using total RNA isolated from wt and Δhfq bacteria recovered from infected mice at day 0 (white columns) and day 12 (gray columns). The values are means plus standard deviations from 3 replicates.
When compared to wt strain, relative abundance of bsp22, bteA, bopN and vag8 transcripts as determined by RT-qPCR (Fig. 3C) was highly decreased in the Δhfq strain and this trend was further intensified in samples collected at day 12 post infection, thereby confirming the output of microarray data. In addition, Vag8 levels were determined in cell lysates of both passaged strains by Westernblot analysis. As seen in Figure 2B and in agreement with microarray and RT-qPCR data, the hfq mutant produced significantly reduced amounts of Vag8 protein while the Vag8 production in wt strain was further increased upon infection.
In order to characterize the infection-associated changes in gene expression in individual strains we have also compared transcriptional profiles in RNA samples collected at day 12 and day 0 in both wt and Δhfq strains by microarray analysis using the same threshold parameters (Fig. S1A). Comparison of transcript levels determined at day 12 with those obtained at day 0 revealed 115 and 78 genes to be affected in the wt and Δhfq strain, respectively (Fig. S1A, Tables S5 and S6). This analysis showed that upon contact with the host the T3SS-specific genes as well as the vag8 gene are significantly upregulated in the wt strain (Fig. S1B). This pattern of expression was observed also in the hfq mutant although to a much lower extent. Notably, expression of the btrS gene encoding T3SS regulatory factor was upregulated in both strains while transcript levels of btrU were increased only in the wt strain. Relative transcript levels of selected genes as determined by RT-qPCR were in agreement with microarray data. In contrast to Δhfq strain, the expression levels of bsp22, bteA, bopN and vag8 genes assessed in wt bacteria recovered at day 12 were increased when compared to those recovered at day 0 (Fig. S1C).
Hfq is required for expression of T3SS in B. pertussis cells recovered from infected macrophages
Our data indicated that Hfq is required for T3SS activation in mouse-passaged bacteria. During infection, resident and infiltrating macrophages are one of the first cells of the innate immune system to sense B. pertussis cells and thus play a very important role in the clearance of the pathogen.30,31 Therefore, we asked if the contact with macrophages in vitro would be sufficient for activation of T3SS in B. pertussis cells. Thus, in order to corroborate this hypothesis, we infected RAW 264.7 macrophages with wt and Δhfq bacteria and viable bacteria were recovered 24 and 48 hours post infection. Control bacteria which were not in contact with macrophages (time 0) together with recovered bacteria were further subcultivated in SS medium and culture supernatants were tested for presence of the Bsp22 protein. As shown in Figure 4A, wt cells recovered at both time points produced Bsp22 while the Δhfq cells did not. This result indicated that upon contact with macrophage cells the functionality of T3SS (assessed as production of Bsp22 protein) can be regained only in wt bacteria and that similarly to mouse-passaged bacteria, the production of Bsp22 can be observed in subsequent in vitro cultures of recovered bacteria. In order to further refine our observation, we have repeated macrophage infection experiment only with the wt strain and this time the viable cells were recovered already 3, 6 and 12 hours post infection. Control bacteria (time 0) and recovered bacteria were cultivated in SS medium and samples for RNA as well as for Westernblot analysis were harvested at each time point. As presented in Figure 4B, wt cells which were recovered from macrophages produced and secreted Bsp22 protein already 3 hours upon infection indicating that relatively short-time interaction with immune cells was sufficient for activation of T3SS. In addition, quantitative PCR analysis of RNA samples (Fig. 4C) revealed that upon infection of macrophages, expression of bopN, bsp22 and bteA genes was increased and thus copied the expression patterns observed in mouse-passaged strains.
Figure 4.
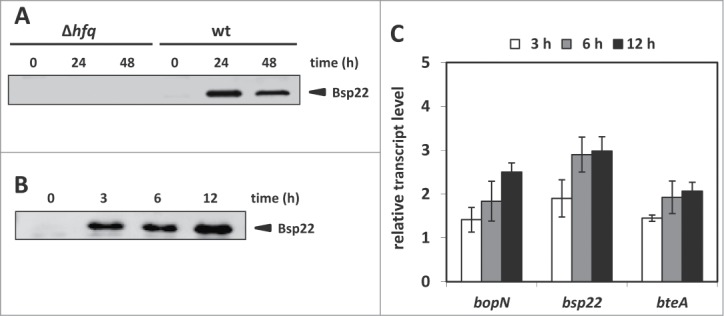
Expression of T3SS is activated only in wt B. pertussis cells upon infection of macrophages. (A) Westernblot analysis of Bsp22 protein levels in culture supernatants of wt or Δhfq bacteria recovered from RAW 264.7 macrophages. Bacteria recovered from macrophages 24 and 48 hours post infection were used to inoculate liquid cultures. Samples of supernatants equivalent to 1 OD600 unit of initial culture were analyzed by immunoblotting using anti-Bsp22 antibody. Relevant part of the membrane is shown. The result is representative of 3 experiments. (B) Westernblot analysis of Bsp22 protein levels in culture supernatants of wt bacteria recovered from RAW 264.7 macrophages. Viable bacteria, recovered from macrophages 3, 6 and 12 hours post infection, were used to inoculate liquid cultures. Samples of supernatants equivalent to 1 OD600 unit of initial culture were analyzed by immunoblotting using anti-Bsp22 antibodies. Relevant part of the membrane is shown. The result is representative of 3 experiments. (C) RT-qPCR analysis of bopN, bsp22 and bteA transcripts levels in wt bacteria upon infection of macrophages. Differential expression analysis was performed using total RNA isolated from viable wt bacteria recovered from macrophages 3 (white columns), 6 (gray columns) and 12 hours (black columns) post infection. The values are means plus standard deviations from 3 replicates.
Discussion
Recently we demonstrated that the hfq mutant of B. pertussis is significantly attenuated in virulence and explained this observation by reduced production and/or secretion of the key virulence factors adenylate cyclase and pertussis toxins by the mutant strain. Nevertheless, we realized that additional factors may be responsible for the reduced virulence of the mutant. In the present study, we therefore aimed at full characterization of the Hfq regulon in B. pertussis with focus on factors involved in pathogenicity. Transcriptional profiling has shown that Hfq is a global regulator in B. pertussis as the deletion of hfq gene had a large effect on gene expression with more than 10% of significantly affected genes in all experiments. Among the most overrepresented functional categories were “Information transfer,” “Energy metabolism” and “Pathogenicity and Adaptation” categories. Within the last category, comparative transcriptomic analysis identified additional virulence factors that play important roles in pathogenicity of B. pertussis and whose expression was significantly downregulated in the Δhfq strain. Besides tracheal colonization factor tcfA, serum resistance factor brkA and autotransporter vag8, a major part of the T3SS locus and T3SS effector gene bteA were the most prominent virulence factors downregulated in the hfq mutant. Quantitative PCR analysis confirmed our transcriptomic data and provided further evidence for Hfq involvement in T3SS expression. T3SS is one of the most remarkable results of long-standing association of bacteria with eukaryotic hosts and Hfq was reported to play specific roles in T3SS activity in several pathogens.32-35 Therefore, in our next experiments we concentrated on elucidation of the Hfq requirement for T3SS expression. Since the expression and production of T3SS components such as Bsp22 in laboratory-adapted strains is switched on only when the cells come into contact with the host,22 both wt and Δhfq strains were passaged through mice. Comparative analysis of transcriptomic profiles of Tohama I wt and Δhfq cells recovered from mouse lung revealed that expression of several T3SS-specific genes including the bteA effector was significantly induced in the wt strain, while this effect was much less obvious in the mutant strain. Hfq is known to act at the posttranscriptional level and therefore, we speculated that Hfq may exert its effect rather indirectly via T3SS-specific regulator BtrS encoded within the btr locus. However, we did not observe any substantial difference in btrS gene expression between wt and Δhfq strains. In fact, when we compared gene expression profiles in bacteria recovered at day 12 and day 0, the btrS expression was similarly increased in both wt (log2FC = 1.3; P < 1 × 10−6) and Δhfq strains (log2FC = 0.9, P < 2× 10−5, data not shown) suggesting that Hfq does not affect btrS expression and may employ another mechanism for T3SS activation. Remarkably, our microarray data indicated that several putative transcriptional factors are expressed at significantly reduced levels in the hfq mutant. Although none of them belongs to the AraC family of transcriptional activators which were shown to activate T3SS in Yersinia,36 it would be of interest to determine their possible role in regulation of T3SS activity in B. pertussis. In this study we confirmed the observation that a) in the laboratory-adapted Tohama I strain, T3SS genes are actively transcribed in vitro19,20 and b) while T3SS substrates such as Bsp22 protein are not produced and secreted under in vitro growing conditions,21,22,37 this ability can be regained upon passage in the host.22,38 Furthermore, infection experiment showed that differential expression of the bsp22 gene translated into strikingly altered levels of Bsp22 protein, a hallmark of T3SS functionality in B. pertussis. Indeed, Westernblot analysis of bacteria recovered from infected mice showed that in remarkable contrast to the wt strain, Bsp22 protein did not accumulate in both cell lysates and culture supernatants of the hfq mutant. The absence of Bsp22 protein in spite of reduced but detectable bsp22 transcription indicated that Hfq affects T3SS functionality also at posttranscriptional level and consequently that Hfq is required for synthesis of T3SS substrates. Recently, Hfq was reported to activate synthesis of T3SS proteins via a small RNA in Y. pestis39 and it would be of high importance to search for an analogous sRNA in B. pertussis. It should be mentioned that in contrast to the Δhfq strain, the btrU gene was found upregulated in the wt strain upon infection, however, this factor was shown to be required for secretion but not production of T3SS substrates.20 Therefore, its reduced expression cannot explain undetectable accumulation of Bsp22 protein inside the Δhfq cells. The precise mechanism of B. pertussis T3SS activation upon contact with the host remains unclear, however, the rapid activation of Bsp22 production upon infection and persistence of this effect for several in vitro passages suggests an epigenetic regulation.40
Among other virulence factors which were significantly affected upon passage in mice, expression of tracheal colonization factor tcfA was significantly reduced in the Δhfq strain when compared to the wt strain. Recently, we reported that the hfq mutant of B. pertussis exhibits a striking colonization defect when used in mixed infection experiment with the wt strain.29 We understood this result primarily as a consequence of reduced production of adenylate cyclase toxin by the hfq mutant, however, herein presented data indicate that impaired expression of T3SS gene cluster and tracheal colonization factor tcfA may as well significantly contribute to reduced capacity of the hfq mutant to colonize mouse lungs. In support, T3SS-deficient strains of B. bronchiseptica13,17,41 and B. pertussis21 were cleared much faster from the respiratory tract of infected animals. Remarkably, we could recapitulate our data obtained in in vivo infection experiments also in in vitro infection model of murine macrophages. In contrast to Δhfq cells, we observed increased expression of T3SS genes and regained ability to produce Bsp22 protein in wt cells recovered from RAW 264.7 macrophages. These data represent the first evidence that T3SS can be activated also upon contact with immune cells in vitro and therefore, we consider macrophage cells as a very versatile and cost-effective infection model for gene expression studies in B. pertussis.
Notably, the expression of autotransporter vag8 gene as determined by microarray and RT-qPCR analyses was also strongly decreased in the hfq mutant and almost mirrored T3SS expression patterns under all tested conditions. Likewise, as evidenced by immunoblotting, the production of Vag8 protein was significantly reduced in the hfq mutant under standard laboratory conditions. Furthermore, in contrast to Δhfq cells, the expression and production of Vag8 was further increased in wt cells upon infection and thereby again copied the T3SS expression pattern. These results suggested that besides being regulated by the BvgAS system the expression of non-adjacent bsc and vag8 genes is coordinated by some yet unknown trans-encoded factor. Vag8 plays an important role during infection as it interacts with C1 esterase inhibitor and confers to serum resistance.42 In addition, Vag8 is presumed but not shown to be an accessory factor of T3SS20 and our data would support this hypothesis. In contrast to other virulence factors, the transcription of the ptx/ptl locus, encoding pertussis toxin and its secretion apparatus, was significantly upregulated in the mutant. This observation is fully in line with our previous study where we have shown that in spite of increased transcription of ptx/ptl locus the amount of toxin secreted by the hfq mutant was lower compared to the wt.29
While focused on virulence genes, microarray analysis revealed a great number of Hfq-dependent genes which classified into several categories. Within the set of genes with known function and which were discovered to have higher transcript abundance in the hfq mutant emerged genes encoding the components of ABC transporter systems and amino acid transporters (“Surface proteins” category) and 30S and 50S ribosomal proteins (“Information transfer” category). Overexpression of both ABC transporter and ribosomal genes seems to be a typical phenomenon observed in hfq mutants of other bacteria.43-46 Among genes displaying deregulated transcript levels in the hfq mutant were also genes encoding several periplasmic and outer membrane proteins. This was not surprising as we have previously observed some differences in protein composition of the membrane fractions between the wt and Δhfq strain.29 Several studies indicated that lack of Hfq results in irregular production of OMPs which affects the membrane integrity and cell envelope stability.33,47,48 Collectively, misregulation of ABC transporter genes, unbalanced expression of ribosomal genes and aberrant expression/production of OMPs might result in pleiotropic defects of hfq mutants such as slower growth and therefore, may as well explain the observed growth deficiency of the Tohama I hfq mutant.
Materials and Methods
Bacterial strains and growth conditions
The Bordetella pertussis Tohama I strain and its isogenic hfq deletion mutant29 were grown on Bordet-Gengou agar (BGA) plates supplemented with 15% defibrinated sheep blood for 3 to 4 days at 37°C. For liquid cultures, bacteria were grown in modified Stainer-Scholte (SS) medium49 supplemented with 0.1% cyclodextrin (Sigma) and 0.5% casamino acids (Difco) at 37°C. Samples for RNA isolation and protein analyses were taken at exponential (OD600 ≈1.3 and 0.7 for wt and Δhfq strains, respectively) and early stationary (OD600 ≈3.0 and 1.3 resp.) phases of growth.
Immunoblotting
B. pertussis wt and Δhfq strains were cultivated in SS medium at 37°C and cell pellets from 1-ml aliquots were lysed and boiled in sample buffer. For analysis of supernatant fractions, 20-ml aliquots of cell-free supernatants were filtered through 0.22 μm filters, precipitates obtained with 10% (w/v) trichloracetic acid were washed with 80% acetone, dissolved in sample buffer and boiled. Samples equivalent to 1 OD600 unit (supernatants) or 0.1 OD600 unit (whole-cell lysates) were separated on SDS-PAGE gels and transferred on nitrocellulose membrane. Membranes were probed with polyclonal antibodies raised against Bsp22 (laboratory stock), Vag8 (kindly provided by Dimitri Diavatopoulos) and BrkA (kindly provided by Rachel Fernandez) followed by incubation with corresponding IgG antibodies conjugated with horse radish peroxidase. The antibody-antigen complexes were visualized using SuperSignal West Femto chemiluminescent substrate (Thermo) according to standard protocol.
Preparation of the microarrays
Oligonucleotide probes were designed for 3554 annotated coding sequences of Bordetella pertussis Tohama I strain (BX470248.1, GI:33591069) using the OligoArray v2.1 software.50 One oligonucleotide for each of the annotated genes was designed except for the non-coding genes (rRNA, tRNA and smRNA(ssrA)) and for the transposase genes of IS elements. Out of the total 3554 probes 3462 were 60-nucleotides long with a Tm between 80 and 99°C, 60 probes were 50-nucleotides long with a Tm between 70 and 99°C and 32 probes were 45-nucleotides long with a Tm between 70 and 99°C. Aminated oligonucleotides were obtained from Sigma Aldrich and were resuspended in 20 μL of spotting solution (sciSPOT-cDNA, Scenion) at a final concentration of 20 μM. 96 oligonucleotide probes (randomly selected among the 3554 probes to be produced in duplicate) were mixed together, spotted as one probe and used as positive control of hybridization (intrinsic control providing an average intensity value for each study). All designed probes including the control mixture of 96 probes and human gene oligonucleotide-probes (negative controls) were spotted in duplicate (non-adjacent) on aldehydsilane coated glass slides (Nexterion Slide AL, Schott) using a QArray II spotter (Genetix) equipped with 12 ArrayHit 946MP3 pins. Spotting was done at 20-22°C and in 50–55% relative humidity. After spotting, glass slides were incubated at 120°C for 1 h and then treated just before usage with 132 mM NaBH4 in PBS containing 25% of ethanol for 30 minutes. The slides were then washed twice in 0.2% SDS solution and twice in distilled water before being dried by centrifugation.
RNA preparation and microarray analysis
B. pertussis wt and Δhfq cells were grown in 2 independent biological replicates in SS medium and harvested by centrifugation (6000 rpm, 10 min, 4°C) at exponential and stationary phases of growth. Total RNA was extracted from pellets using Trizol (Sigma) as recommended by the manufacturer and further treated with DNase I (Promega) to remove any contaminating genomic DNA. Purified RNA was checked for concentration and integrity using the Bioanalyzer 2100 device (Agilent Technologies). Labeling with Cyanine-5 (Cy5) and Cyanine-3 (Cy3) was performed by random primed reverse transcription starting from 10 μg of purified total RNA and using SuperscriptIII (Invitrogen) reverse transcriptase according to manufacturer protocol and amplified cDNA was purified on Qiaquick PCR purification kit (Qiagen). A mix of 1 μg of Cy5-cDNA + 1 μg of Cy3-cDNA was then prepared in 490 μL of hybridization buffer (5× SSC, 40% deionised formamide, 5× Denhart's buffer, 0.1% SDS, 1 mM sodium pyrophosphate). The labeled mixture was used for hybridization using a gasket slide (Agilent Technologies) for 14 to 16 h at 52°C under agitation. For each sample a Cy5/Cy3 dye-swap technical replicate starting from the same RNA preparation was prepared and hybridized. After hybridization, slides were subsequently washed at room temperature in 2× SSC, 0.2%SDS for 5 minutes, in 0.5× SSC for 10 minutes, in 0.05× SSC for 5 minutes, and 0.01× SSC for 2 minutes before being dried and scanned using InnopScan700 (Innopsys). Background correction as well as within- and between-array normalization51 was performed from raw data using LIMMA (Linear Models for Microarray Data) package.52 After normalization, identification of genes with significantly modulated expression was performed using moderated Student's t-test with empirical Bayes shrinkage of standard errors.53 Statistics were corrected for multiple testing using false-discovery rate approach. In order to select genes significantly affected by hfq deletion, absolute log fold change values (log2FC) of transcript abundance was set to at least 1 (2-fold variation) and threshold for adjusted P-value (P) of less than 0.05 was considered as indicative of significance. Significantly affected genes were classified into functional categories as assigned by Park and coworkers54 with following modifications. The categories “Degradation of small molecules” and “Degradation of large molecules” were combined into category “Degradation,” category “Pathogenicity, Adaptation and Chaperones” was abbreviated to “Pathogenicity and Adaptation.” Genes comprising T3SS bsc and btr loci were moved from “Surface protein” category into “Pathogenicity and Adaptation” category. Statistical significance of overrepresentation of functional categories within the sets of genes with modified transcript abundance was assessed using the Fisher exact test and corrected for multiple testing using Benjamini and Hochberg method.55 Clustering of microarray data was performed using MeV software from TIGR.56
Quantitative PCR (RT-qPCR)
To study the quantitative effects on gene expression, 1 μg of total isolated RNA was reverse transcribed in duplicates into cDNA following the manufacturer's instructions in a 25-μl reaction using the Reverse Transcription System (Promega). All primers (Table S7) were designed to anneal at 60°C and were analyzed for secondary structures and for cross-dimers in primer pairs. RT-qPCR was performed on Bio-Rad CFX96 instrument using SYBR® Green JumpStart™ Taq ReadyMix™ (Sigma). Briefly, 200 nmol/l of each primer together with 40 ng of reverse transcribed RNA were used in a 20-μl qPCR reaction volume, with an initial step at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, 72°C for 30 s, followed by recording of the melting curve. The rpoB gene was used as reference gene and relative gene expression was quantified using amplification efficiency values.57
Infection of macrophages
B. pertussis cells were suspended in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% bovine serum albumin and the RAW 264.7 murine macrophages were infected at a multiplicity of infection of 10 bacteria per cell (MOI 10). After addition of bacteria, tissue culture plates were centrifuged for 5 min at 300 g to facilitate bacterial interaction with macrophage cells. After 60 min of incubation at 37°C with 5% CO2, non-adherent bacteria were removed by 3 washing steps in DMEM. Then, medium containing 100 μg/ml of polymyxin B sulfate was added for 60 min at 37°C with 5% CO2 to kill extracellular bacteria. Next, infected macrophages were washed with fresh medium containing 30 μg/ml of polymyxin B sulfate and further incubated at 37°C with 5% CO2. In order to recover viable intracellular bacteria, infected macrophages were lysed with sterile water at different times post infection as specified in legends of Figure 4A, B. Lysates as well as bacteria which were not in contact with macrophages (time point 0; control sample) were plated onto BGA plates and obtained bacteria were subsequently cultivated in SS medium for RNA isolation and Westernblot analysis.
In vivo infection of mice
Four-week-old CD-1 Swiss outbred mice were obtained from Charles River Laboratories and maintained under specific pathogen free conditions. Overnight cultures of B. pertussis strains were diluted in sterile phosphate buffered saline (PBS, pH 7.4) to adjust appropriate cell concentrations. Sublethal doses (1 × 105 CFU/mouse) of both wt and Δhfq bacteria were administered intranasally in suspensions of 50 μl and in parallel, the number of delivered bacteria was determined by plating serial dilutions of challenge suspensions on BGA plates. Cohorts of 15 animals were challenged per strain and 3 mice per group were sacrificed 2 hours and 5, 8, 12 and 23 days post infection. The lungs were removed aseptically and homogenized in 2 ml of PBS using a cell grinder (Heidolph RZR2020). The resulting homogenates were serially diluted with PBS and plated in duplicates on BGA plates and colonies were counted to calculate the CFU values per mouse. Student's t test (SigmaPlot 1, Systat Software Inc., California, USA) was used to compare the mean CFU values between wt and Δhfq strains at each time point. Differences were considered significant at P values of ≤ 0.05. Recovered bacteria were subcultured in SS medium and samples for RNA isolation and westernblot analysis were taken at exponential phase of growth.
All animal experiments were approved by the Animal Welfare Committee of the Institute of Microbiology of the ASCR, v.v.i., in Prague, Czech Republic. Handling of animals was performed according to the Guidelines for the Care and Use of Laboratory Animals, the Act of the Czech National Assembly, Collection of Laws No. 149/2004, inclusive of the amendments on the Protection of Animals against Cruelty and Public Notice of the Ministry of Agriculture of the Czech Republic, Collection of Laws No. 207/2004, on care and use of experimental animals.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Rachel Fernandez (University of British Columbia, Vancouver) and Dimitri Diavatopoulos (Radboud University Medical Centre, Nijmegen) for provision of antibodies. IB is a doctoral student of the Institute of Chemical Technology in Prague and OC is a doctoral student of the Faculty of Science of the Charles University in Prague.
Funding
This work was supported by the Czech Science Foundation (www.gacr.cz) (grant P302/11/1940 to BV) and by funding from RVO61388971. This work was also supported by the Institut Pasteur de Lille and the Institut National de la Santé et de la Recherche Médicale (Inserm) (to DH and SS) and by German ministry of science in frame of the e:Bio initiative (to FA). This publication was supported by the Ministry of Education, Youth and Sports of the Czech Republic projects (CZ.1.07/2.3.00/20.0055 to BV and IB, CZ.1.07/2.3.00/30.0003 to BV and KK, and 7AMB14AR028 to BV and OC) and by the project “BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University” (CZ.1.05/1.1.00/02.0109 to BV and IB) from the European Regional Development Fund. We also acknowledge European Science Foundation for short visit grant (FFG 4135 to B.V.) within the activity entitled “Frontiers of Functional Genomics.”
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 2005; 18:326-82; PMID:15831828; http://dx.doi.org/ 10.1128/CMR.18.2.326-382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crowcroft NS, Stein C, Duclos P, Birmingham M. How best to estimate the global burden of pertussis? Lancet Infect Dis 2003; 3:413-8; PMID:12837346; http://dx.doi.org/ 10.1016/S1473-3099(03)00669-8 [DOI] [PubMed] [Google Scholar]
- 3. Cherry JD. The present and future control of pertussis. Clin Infect Dis 2010; 51:663-7; PMID:20704492; http://dx.doi.org/ 10.1086/655826 [DOI] [PubMed] [Google Scholar]
- 4. Raguckas SE, VandenBussche HL, Jacobs C, Klepser ME. Pertussis resurgence: diagnosis, treatment, prevention, and beyond. Pharmacotherapy 2007; 27:41-52; PMID:17192161; http://dx.doi.org/ 10.1592/phco.27.1.41 [DOI] [PubMed] [Google Scholar]
- 5. de Melker HE, Schellekens JF, Neppelenbroek SE, Mooi FR, Rumke HC, Conyn-van Spaendonck MA. Reemergence of pertussis in the highly vaccinated population of the Netherlands: observations on surveillance data. Emerg Infect Dis 2000; 6:348-57; PMID:10905967; http://dx.doi.org/ 10.3201/eid0604.000404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bart MJ, van Gent M, van der Heide HG, Boekhorst J, Hermans P, Parkhill J, Mooi FR. Comparative genomics of prevaccination and modern Bordetella pertussis strains. BMC Genomics 2010; 11:627; PMID:21070624; http://dx.doi.org/ 10.1186/1471-2164-11-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mooi FR, Van Der Maas NA, De Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation – two sides of the same coin. Epidemiol Infect 2014; 142:685-94; PMID:23406868; http://dx.doi.org/ 10.1017/S0950268813000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bouchez V, Brun D, Cantinelli T, Dore G, Njamkepo E, Guiso N. First report and detailed characterization of B. pertussis isolates not expressing Pertussis Toxin or Pertactin. Vaccine 2009; 27:6034-41; PMID:19666155; http://dx.doi.org/ 10.1016/j.vaccine.2009.07.074 [DOI] [PubMed] [Google Scholar]
- 9. Octavia S, Maharjan RP, Sintchenko V, Stevenson G, Reeves PR, Gilbert GL, Lan R. Insight into evolution of Bordetella pertussis from comparative genomic analysis: evidence of vaccine-driven selection. Mol Biol Evolut 2011; 28:707-15; PMID:20833694; http://dx.doi.org/ 10.1093/molbev/msq245 [DOI] [PubMed] [Google Scholar]
- 10. Hewlett EL, Burns DL, Cotter PA, Harvill ET, Merkel TJ, Quinn CP, Stibitz ES. Pertussis pathogenesis–what we know and what we don't know. J Infect Dis 2014; 209:982-5; PMID:24626533; http://dx.doi.org/ 10.1093/infdis/jit639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robbins JB, Schneerson R, Kubler-Kielb J, Keith JM, Trollfors B, Vinogradov E, Shiloach J. Toward a new vaccine for pertussis. Proc Natl Acad Sci U S A 2014; 111:3213-6; PMID:24556987; http://dx.doi.org/ 10.1073/pnas.1324149111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Locht C. Molecular aspects of Bordetella pertussis pathogenesis. Int Microbiol 1999; 2:137-44; PMID:10943406 [PubMed] [Google Scholar]
- 13. Yuk MH, Harvill ET, Miller JF. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol Microbiol 1998; 28:945-59; PMID:9663681; http://dx.doi.org/ 10.1046/j.1365-2958.1998.00850.x [DOI] [PubMed] [Google Scholar]
- 14. Fauconnier A, Veithen A, Gueirard P, Antoine R, Wacheul L, Locht C, Bollen A, Godfroid E. Characterization of the type III secretion locus of Bordetella pertussis. Int J Med Microbiol 2001; 290:693-705; PMID:11310448; http://dx.doi.org/ 10.1016/S1438-4221(01)80009-6 [DOI] [PubMed] [Google Scholar]
- 15. Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006; 444:567-73; PMID:17136086; http://dx.doi.org/ 10.1038/nature05272 [DOI] [PubMed] [Google Scholar]
- 16. Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol 2006; 4:811-25; PMID:17041629; http://dx.doi.org/ 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- 17. Skinner JA, Pilione MR, Shen H, Harvill ET, Yuk MH. Bordetella type III secretion modulates dendritic cell migration resulting in immunosuppression and bacterial persistence. J Immunol 2005; 175:4647-52; PMID:16177111; http://dx.doi.org/ 10.4049/jimmunol.175.7.4647 [DOI] [PubMed] [Google Scholar]
- 18. Yuk MH, Harvill ET, Cotter PA, Miller JF. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol Microbiol 2000; 35:991-1004; PMID:10712682; http://dx.doi.org/ 10.1046/j.1365-2958.2000.01785.x [DOI] [PubMed] [Google Scholar]
- 19. Hot D, Antoine R, Renauld-Mongenie G, Caro V, Hennuy B, Levillain E, Huot L, Wittmann G, Poncet D, Jacob-Dubuisson F, et al. Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol Genet Genomics 2003; 269:475-86; PMID:12768411; http://dx.doi.org/ 10.1007/s00438-003-0851-1 [DOI] [PubMed] [Google Scholar]
- 20. Mattoo S, Yuk MH, Huang LL, Miller JF. Regulation of type III secretion in Bordetella. Mol Microbiol 2004; 52:1201-14; PMID:15130135; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04053.x [DOI] [PubMed] [Google Scholar]
- 21. Fennelly NK, Sisti F, Higgins SC, Ross PJ, van der Heide H, Mooi FR, Boyd A, Mills KH. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect Immun 2008; 76:1257-66; PMID:18195025; http://dx.doi.org/ 10.1128/IAI.00836-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaillard ME, Bottero D, Castuma CE, Basile LA, Hozbor D. Laboratory adaptation of Bordetella pertussis is associated with the loss of type three secretion system functionality. Infect Immun 2011; 79:3677-82; PMID:21730086; http://dx.doi.org/ 10.1128/IAI.00136-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cotter PA, Jones AM. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol 2003; 11:367-73; PMID:12915094; http://dx.doi.org/ 10.1016/S0966-842X(03)00156-2 [DOI] [PubMed] [Google Scholar]
- 24. Uhl MA, Miller JF. BvgAS is sufficient for activation of the Bordetella pertussis ptx locus in Escherichia coli. J Bacteriol 1995; 177:6477-85; PMID:7592423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uhl MA, Miller JF. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J 1996; 15:1028-36; PMID:8605872 [PMC free article] [PubMed] [Google Scholar]
- 26. Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 2010; 13:24-33; PMID:20080057; http://dx.doi.org/ 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 27. Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe 2010; 8:116-27; PMID:20638647; http://dx.doi.org/ 10.1016/j.chom.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 28. Hot D, Slupek S, Wulbrecht B, D'Hondt A, Hubans C, Antoine R, Locht C, Lemoine Y. Detection of small RNAs in Bordetella pertussis and identification of a novel repeated genetic element. BMC Genomics 2011; 12:207; PMID:21524285; http://dx.doi.org/ 10.1186/1471-2164-12-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bibova I, Skopova K, Masin J, Cerny O, Hot D, Sebo P, Vecerek B. The RNA chaperone Hfq is required for virulence of Bordetella pertussis. Infect Immun 2013; 81:4081-90; PMID:23980112; http://dx.doi.org/ 10.1128/IAI.00345-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Higgs R, Higgins SC, Ross PJ, Mills KH. Immunity to the respiratory pathogen Bordetella pertussis. Mucosal Immunol 2012; 5:485-500; PMID:22718262 [DOI] [PubMed] [Google Scholar]
- 31. Lamberti YA, Hayes JA, Perez Vidakovics ML, Harvill ET, Rodriguez ME. Intracellular trafficking of Bordetella pertussis in human macrophages. Infect Immun 2010; 78:907-13; PMID:20065021; http://dx.doi.org/ 10.1128/IAI.01031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schiano CA, Bellows LE, Lathem WW. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect Immun 2010; 78:2034-44; PMID:20231416; http://dx.doi.org/ 10.1128/IAI.01046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 2007; 63:193-217; PMID:17163975; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shakhnovich EA, Davis BM, Waldor MK. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol 2009; 74:347-63; PMID:19703108; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06856.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol 2013; 195:1706-17; PMID:23378513; http://dx.doi.org/ 10.1128/JB.02056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cornelis G, Sluiters C, de Rouvroit CL, Michiels T. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol 1989; 171:254-62; PMID:2644192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brickman TJ, Cummings CA, Liew SY, Relman DA, Armstrong SK. Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J Bacteriol 2011; 193:4798-812; PMID:21742863; http://dx.doi.org/ 10.1128/JB.05136-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Villarino Romero R, Bibova I, Cerny O, Vecerek B, Wald T, Benada O, Zavadilova J, Osicka R, Sebo P. The Bordetella pertussis type III secretion system tip complex protein Bsp22 is not a protective antigen and fails to elicit serum antibody responses during infection of humans and mice. Infect Immun 2013; 81:2761-7; PMID:23690400; http://dx.doi.org/ 10.1128/IAI.00353-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schiano CA, Koo JT, Schipma MJ, Caulfield AJ, Jafari N, Lathem WW. Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J Bacteriol 2014; 196(9):1659-70; PMID:24532772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 2006; 70:830-56; PMID:16959970; http://dx.doi.org/ 10.1128/MMBR.00016-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pilione MR, Harvill ET. The Bordetella bronchiseptica type III secretion system inhibits gamma interferon production that is required for efficient antibody-mediated bacterial clearance. Infect Immun 2006; 74:1043-9; PMID:16428751; http://dx.doi.org/ 10.1128/IAI.74.2.1043-1049.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marr N, Shah NR, Lee R, Kim EJ, Fernandez RC. Bordetella pertussis autotransporter vag8 binds human C1 esterase inhibitor and confers serum resistance. Plos One 2011; 6:e20585; PMID:21695123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao M, Barnett MJ, Long SR, Teplitski M. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol Plant Microbe Interact 2010; 23:355-65; PMID:20192823; http://dx.doi.org/ 10.1094/MPMI-23-4-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Torres-Quesada O, Oruezabal RI, Peregrina A, Jofre E, Lloret J, Rivilla R, Toro N, Jimenez-Zurdo JI. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol 2010; 10:71; PMID:20205931; http://dx.doi.org/ 10.1186/1471-2180-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dietrich M, Munke R, Gottschald M, Ziska E, Boettcher JP, Mollenkopf H, Friedrich A. The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS J 2009; 276:5507-20; PMID:19691497; http://dx.doi.org/ 10.1111/j.1742-4658.2009.07234.x [DOI] [PubMed] [Google Scholar]
- 46. Wilms I, Moller P, Stock AM, Gurski R, Lai EM, Narberhaus F. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J Bacteriol 2012; 194:5209-17; PMID:22821981; http://dx.doi.org/ 10.1128/JB.00510-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun 2008; 76:3019-26; PMID:18458066; http://dx.doi.org/ 10.1128/IAI.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol Microbiol 2006; 62:838-52; PMID:16999834; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05413.x [DOI] [PubMed] [Google Scholar]
- 49. Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 1970; 63:211-20; PMID:4324651; http://dx.doi.org/ 10.1099/00221287-63-2-211 [DOI] [PubMed] [Google Scholar]
- 50. Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 2003; 31:3057-62; PMID:12799432; http://dx.doi.org/ 10.1093/nar/gkg426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 2002; 30:e15; PMID:11842121; http://dx.doi.org/ 10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol 2003; 224:111-36; PMID:12710670 [DOI] [PubMed] [Google Scholar]
- 53. Lonnstedt I, Britton T. Hierarchical Bayes models for cDNA microarray gene expression. Biostatistics 2005; 6:279-91; PMID:15772106; http://dx.doi.org/ 10.1093/biostatistics/kxi009 [DOI] [PubMed] [Google Scholar]
- 54. Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, Ahuja U, Liu M, Miller JF, Sebaihia M, Bentley SD, et al. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 2012; 13:545; PMID:23051057; http://dx.doi.org/ 10.1186/1471-2164-13-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B 1995; 57:289-300 [Google Scholar]
- 56. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34:374-8; PMID:12613259 [DOI] [PubMed] [Google Scholar]
- 57. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/ 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.