Abstract
Small RNAs are incorporated into Argonaute protein-containing complexes to guide the silencing of target RNAs in both animals and plants. The abundance of endogenous small RNAs is precisely controlled at multiple levels including transcription, processing and Argonaute loading. In addition to these processes, 3′ end modification of small RNAs, the topic of a research area that has rapidly evolved over the last several years, adds another layer of regulation of their abundance, diversity and function. Here, we review our recent understanding of small RNA 3′ end methylation and tailing.
Keywords: argonaute, methylation, small RNA, uridylation
Introduction
Small silencing RNAs (small RNAs) of 20–30 nucleotide (nt) in length are key regulators of gene expression in both animals and plants. Together with their effector protein called Argonaute (AGO), small RNAs repress gene expression at either the transcriptional or the post-transcriptional level and play important roles in various biological processes, such as cell differentiation and transgenerational inheritance. Small RNAs can be classified into microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi (a class of AGO protein)-interacting RNAs (piRNAs) based on their origin, biogenesis and/or the type of AGO they are associated with, although the boundaries of this classification become blurred and sometimes even difficult to discern.1,2 Dicer-like proteins, a class of RNAseIII-type enzymes, release miRNAs and siRNAs from their precursors as duplexes with 2 nt 3′ overhangs at each end.3 By contrast, piRNAs are processed via a Dicer-independent pathway.4 More details regarding the biogenesis and function of small RNAs have been reviewed elsewhere.3,4 After biogenesis, plant small RNAs as well as some animal small RNAs are 2′-O-methylated at their 3′ end by HUA ENHANCER1 (HEN1) in plants or its homologues in animals. 2′-O-methylation is critical for small RNA stability. Besides methylation, small RNAs are also subject to 3′ untemplated nucleotide addition (tailing), which in turn affects their function and stability. In this point of view, we summarize our recent understanding on small RNA methylation and tailing and discuss their biological relevance.
Methylation of small RNAs
Plant miRNAs and siRNAs contain a 2′-O-methyl group at their 3′ end. This modification is catalyzed by HEN1.5,6 HEN1 contains 2 double-stranded RNA binding domains (dsRBD1 and dsRBD2), a La-motif containing domain (LCD) and a methyltransferase domain (MTase).7 HEN1 specifically recognizes 21–24 base-pair (bp) dsRNAs with 2nt overhangs and deposits a methyl group to the 2′ OH position of the 3′ end in each strand in vitro.8 Structural analysis reveals that HEN1 recognizes dsRNAs via its dsRBD1 and dsRBD2 domains. The substrate length of HEN1 is determined by the distance between the LCD domain and the MTase domain.7 Loss of 2′-O-methylation in the hen1 mutants not only leads to the reduced small RNA abundance but also results in 3′ end untemplated uridine addition (uridylation) and trimming of small RNAs.9-11 These results demonstrate that methylation stabilizes small RNAs by preventing uridylation and degradation. A single uridine addition at miR171 in the hen1 mutant results in the production of secondary siRNAs from target RNAs that undergo miR171-mediated cleavage.11 This result indicates that methylation may also block miRNA-dependent secondary siRNA production, and, therefore, protects plants from undesired regulation by small RNAs.
HEN1 is an evolutionarily conserved protein in eukaryotes and bacteria.12 In bacteria and animals, HEN1 lacks dsRNA-binding domains and methylates single-stranded RNAs.13,14-21 In bacteria, Hen1 is involved in an RNA repair process.22,23 In animals, HEN1 methylates some small RNAs in a germ cell-specific manner.14-21 Intriguingly, among 297 animals homozygous for zebrafish hen1, only one was identified as female, suggesting a critical role of HEN1 in sex determination.18 In mice, HEN1 is specifically expressed in testes and methylates piRNAs.15,17 In addition, the zebrafish HEN1 is expressed both in ovary and testis, and is required for piRNA stability.18 Besides piRNAs, AGO2-associated siRNAs in Drosophila and ERGO-1 (a PIWI clade AGO protein)-associated 26G RNAs in C. elegans are also subject to 2′-O-methylation by DmHEN1/Pimet (piRNA methyltransferase) and HENN-1 (HEN1 of Nematodes 1), respectively, in a germline-specific manner. In all cases, loss-of-function mutations in HEN1 cause uridylation and 3′-to-5′ exonucleolytic trimming of small RNAs,16,18–21 reminiscent of the observations in Arabidopsis.9-11 Animal HEN1 homologs appear to act on AGO-bound small RNAs in vivo and fly HEN1 interacts with PIWI,14,16 suggesting that the physical interaction between HEN1 and AGOs may determine its substrate specificity in animals.
Uridylation of small RNAs
In contrast to piRNAs, animal miRNAs are not methylated. Comprehensive small RNA deep sequencing analyses reveale that uridylation or adenylation (usually mono- or di-nucleotide additions) at the 3′ end of miRNAs is widespread and conserved across a variety of animal species.24–26 Uridylation of small RNAs is catalyzed by the terminal uridyltransferases (TUTase) (Fig. 1). While many TUTases show overlapping activities on some miRNAs, some of them act on miRNAs in a sequence-specific manner.24,26,27 Uridylation often serves as an RNA decay signal whereas adenylation appears to increase RNA stability.28 Besides stability, 3′ modification can also regulate small RNA activities. For example, uridylation of miR-26a prevents the miRNA from repressing its mRNA target, without affecting its abundance.25 PAPD4/GLD-2 mediated 3′ end adenylation has little effect on miRNA abundance but reduces their AGO loading efficiency.24 Interestingly, miRNAs derived from the 3′ arms of their pre-miRNAs are more frequently uridylated, suggesting that at least some of the uridylation occurs to the 3′ end of pre-miRNAs before they are channeled into Dicer processing.24 Indeed, mono-uridylation of group II pre-miRNAs (with a 1nt 3′ overhang instead of the canonical 2nt 3′ overhang after Drosha cleavage) is crucial for their further maturation.29 In C. elegans, 22G secondary siRNAs are not methylated and are extensively oligo-uridylated by CDE-1, a germline-specific ncPAP.30 CDE-1-mediated uridylation of CSR-1 bound 22G siRNAs prevents their over-accumulation through active degradation, which is important for both mitotic and meiotic chromosome segregation.30
Figure 1.
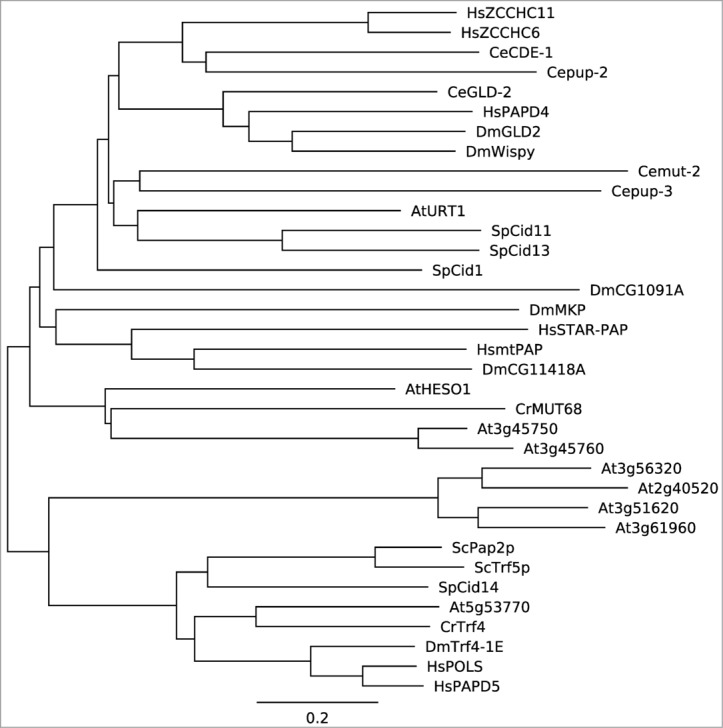
A phylogenetic tree of selected non-canonical poly(A) polymerases. Full-length proteins were downloaded from the Genbank database (http://www.ncbi.nlm.nih.gov/genbank). A neighbor joining phylogenetic tree was constructed using MEGA 5.1 52 and displayed using Geneious v7.1.5 (http://www.geneious.com/). Hs: Homo sapien; Ce: Caenorhabditis elegans; Dm: Drosophila melanogaster; At: Arabidopsis thaliana; Sp: Schizosaccharomyces pombe; Cr: Chlamydomonas reinhardtii; Sc: Saccharomyces cerevisiae.
Lack of HEN1 function causes extensive tailing and trimming of small RNAs in higher plants.9,11 An early study demonstrates that untemplated nucleotides added to the 3′ end of small RNAs in hen1 are predominantly uridines,9 which is subsequently shown to be the same in all other tested plant species.9–11 We recently showed that HESO1 acts in small RNAs uridylation.31,32 HESO1 is a TUTase (Fig. 1) and possesses robust RNA poly(U) polymerase activities in vitro.31,32 The activity of HESO1 is completely blocked by 2′-O-methylation.31,32 This maybe due to that HESO1 requires 2′ OH for its substrate recognition or catalysis. Alternatively, 2′-O-methylation may block substrate recognition or catalysis although HESO1 does not need 2′ OH for its activity. Loss-of-function mutations in HESO1 lead to a general reduction of U-tail length in hen1, accompanied by an increase of normal-sized, 3′ trimmed, and/or short-tailed small RNAs.31,32 In contrast, over-expression of HESO1 in the hen1 background accelerates miRNA turnover and causes more severe developmental defects.31 These results demonstrate that uridylation triggers small RNA destabilization. As in Arabidopsis, the Chlamydomonas TUTase MUT68 has also been implicated in both siRNA and miRNA uridylation.33 The increased extent of 3′ trimming in the hen1 heso1 mutants suggests that uridylation may antagonize the action of 3′-to-5′ trimming, of which the biological meaning is not yet clear.31,32 As 3′ truncated miRNA species are tailed by HESO1,32 it is likely that one function of uridylation is to remove 3′ truncated miRNAs. In Drosophila, the 3′-to-5′ exoribonuclease Nibbler trims a proportion of AGO1 associated miRNAs.34,35 There are 2 putative homologous proteins in Arabidopsis and it will be interesting to determine whether any of them is involved in 3′-to-5′ trimming of unmethylated small RNAs.
Uridylation of 5′ cleavage products (5′ CPs) generated by small RNA-mediated cleavage
The endonucleolytic cleavage of target RNAs is a mechanism used by miRNAs and siRNAs to silence gene expression. Slicing of target RNAs often happens at a position opposite to the middle of miRNAs and siRNAs and results in a 3′ cleavage product (3′ CP) and a 5′ CP.36 In C. elegans, 3′ CPs and 5′ CPs generated by siRNA-mediated cleavage are further removed by the 5′-to-3′ exonuclease XRN1 and the exosome, which degrade RNAs from 5′-to-3′ and 3′-to-5, respectively.37 In Arabidopsis, the XRN1-LIKE exonuclease XRN4 is responsible for the degradation of 3′ CPs generated by miRNA-mediated target cleavage.38 Intriguingly, the 3′ ends of 5′ CPs are often subject to oligo-uridylation or adenylation modifications in several evolutionarily distant plant species.39 The Chlamydomonas MUT68 adds adenosines to the 3′ end of 5′ CP of MAA7 (the endogenous target of siRNAs generated by a transgenic inverted-repeat seq-uence) and promotes its degradation from 3′-to-5′, likely involving the action of the exosome.40 In a recent study, we show that HESO1 is responsible for the uridylation of 5′ CPs in Arabidopsis.41 A reduction in uridylation in heso1–2 increases the accumulation of 5′ CP, demonstrating that uridylation triggers degradation of 5′ CP.41 The proportion of 3 ‘truncated 5′ CPs is increased by heso1 relative to wild-type plants (WT), suggesting that uridylation may cause 5′ CP degradation through a mechanism different from 3′-to-5′ trimming.41 This resembles the effect of heso1 on miRNAs in hen1.31,32 In humans, uridylation can trigger decapping of histone mRNAs followed by 5′-to-3′ degradation.42 This maybe not the case for 5′ CPs, as the status of 5′-to-3′ truncation of 5′ CPs is not altered by heso1. 5′ CPs can also be degraded by 5′-to-3′ and 3′-to-5′ trimming since both 5′ and 3′ truncated 5′ CPs exist.41 In xrn4, the amount of 5′ CPs is increased compared with that in WT, revealing that XRN4 is an enzyme responsible for the degradation of 5′ CPs although it is not clear whether it acts before or after uridylation.41 The enzymes that trim 5′ CPs from 3′-to-5′ remain to be identified.
How does uridylation trigger the degradation of small RNAs and 5′ CPs?
Both small RNAs and 5′ CPs at least in transient associate with the AGO proteins. This suggests that a common mechanism maybe used to degrade both uridylated small RNAs and 5′ CPs. Agreeing with this notion, the exosome is responsible for the degradation of both uridylated small RNAs and adenylated 5′ CPs in Chlamydomonas.33,40 Thus, it is possible that a nuclease may simultaneously target both uridylated 5′ CPs and small RNAs in higher plants. Recently, Dis3l2, which is a 3′-to-5′ RNAse II nuclease encoded by a paralog of RRP44, a core component of the exosome, was shown to degrade uridylated pre-let-7 (precursor of let-7 miRNA).43,44 In yeast, the Dis3l2 homolog can trigger rapid degradation of uridylated RNAs in a tail-length dependent manner.45 Arabidopsis SOV (SUPPRESSOR OF VARICOSE) is the closest homologous protein of Dis3l2.46 It is worth testing if SOV is the enzyme responsible for the degradation of uridylated small RNAs and 5′ CPs in the near future. HESO1 can add long U-tails (Up to hundreds of nt long) to its substrates in vitro.31,32,41 However both miRNAs and 5′ CPs only contain short U-tails (less than 15 nt) in vivo.32,41 This provides an alternative explanation for the degradation of uridylated miRNAs and 5′ CPs: long U-tail may disassociate miRNAs and 5′ CPs from the AGO1 complex, resulting in their rapid degradation such that no long-tailed products can be detected.
HEN1 protects small RNAs from AGO-associated tailing and trimming activities that normally acts on 5′ CPs
The dual activities of HESO1 and MUT68 on both small RNAs and 5′ CPs and the fact that both small RNAs and 5′ CPs associate with AGO proteins suggest that the terminal uridyl transferase may recognize their substrates in the AGO complex (Fig. 2). Several evidences demonstrate that HESO1 indeed uridylates AGO1-associated miRNAs. First, HESO1 associates with AGO1, as revealed by colocalization and co-immunoprecipitation assays.41 Second, HESO1 is able to uridylate AGO1-bound miR165/166 in vitro.41 Third, in hen1, AGO1-bound miRNAs are tailed.11 Fourth, suppression of AGO1 function in hen1 diminishes miRNA uridylation.11,41 We envision that HESO1 also recognizes siRNAs in vivo through its physical interaction with respective AGO proteins. Further analyses show that the PAP domain rather than the unconserved C-terminal domain of HESO1 interacts with the PAZ and PIWI domain of AGO1.41 These interactions may facilitate HESO1 to recognize its substrates, as PAZ binds the 3′ end of small RNAs, which can be released by base pairing between small RNAs and their targets, and PIWI cleaves the targets. In addition, the interaction of terminal uridyl transferases with AGO may be common among different species since PAP (Fig. 1), PAZ and PIWI are evolutionarily conserved.47
Figure 2.
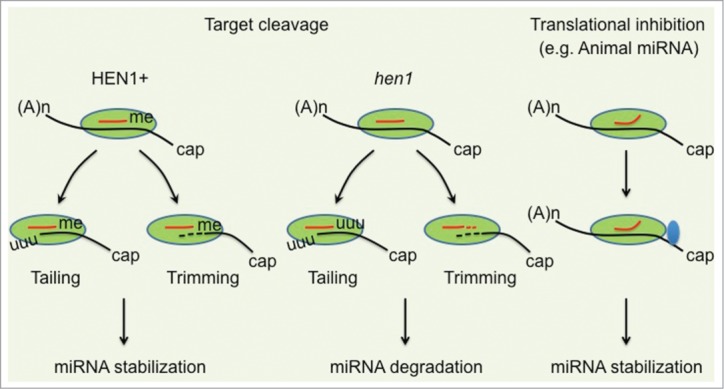
A proposed model for target cleavage mediated small RNA destabilization. In this model, small RNA mediated target cleavage (A and B) but not translational inhibition (C) may provide a signal in the recruitment both tailing and trimming enzymes to AGO for the modification and further degradation of 5′ cleavage products. (A) HEN1 mediated 2′-O-methylation protects small RNAs from both tailing and trimming activities. (B) In the absence of HEN1, small RNAs become frequently tailed and trimmed. (C) In the case of partially complementary target RNA (e.g. animal miRNAs), small RNAs predominantly suppresses gene expression via translational inhibition. In this scenario, tailing and trimming enzymes will be barely recruited to AGO and small RNAs are less tailed and trimmed, regardless of methylated or not. Green oval, AGO protein; Blue oval, translational machinery.
Both small RNAs and 5′ CPs are also subject to 3′-to-5′ trimming activities. By analogy, it is possible that the enzymes catalyzing 3′-to-5′ trimming may also associate with AGO proteins. In fact, Nibbler has been shown to interact with AGO in Drosophila.34 The tailing and trimming activities are likely recruited to AGO to eliminate the 5′ CPs. The presence of these AGO1-associated activities confers the necessity of small RNA methylation by HEN1 to ensure their stability in the AGO complex in plants (Fig. 2). In flies, AGO2-assoicated siRNAs are protected by methylation.16 In contrast, AGO1-associated miRNAs are not methylated16 and display very limited tailing and trimming. This may be due to the fact that animal miRNAs are less complementary to their targets.48 Consistent with this, introduction of artificial target RNAs with high complementarity to some endogenous miRNAs triggers their tailing and trimming.49,50 Moreover, siRNAs, which often have highly complementary targets, become tailed and trimmed when misloaded into AGO1.49,50 A possible explanation for these observations is that less complementarity between miRNAs and targets inhibits target cleavage, which may signal tailing and trimming. Alternatively, the extensive complementarity between small RNA and its target may help to release its 3′ end from PAZ protection.51
Conclusions and Perspectives
In addition to transcription and processing, 3′ end modification also contributes to the accumulation of small RNAs. We are just beginning to understand the underlying biochemical pathway of this process. While it is clear that methylation plays a crucial role in stabilizing small RNAs through antagonizing uridylation and trimming activities, future studies will be required to determine whether the degree of complementarity to targets or target cleavage serves as a signal for the recruitment of tailing and trimming enzymes. There are more challenges in this field. An immediate one is the identification of all the enzymes involved in the small RNAs modification and catabolic pathway, with those responsible for trimming and degradation in particular. Other outstanding questions include how these enzymes are recruited, and more importantly, whether small RNA stability control can be regulated in response to any developmental or environmental signals. We believe that addressing these questions will greatly advance our understanding of the regulation of small RNAs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Yongchao Dou for the preparation of Figure 1.
Funding
This work was supported by a start-up grant from Fudan University (to G.R.), a National Science Foundation Grant MCB-1121193 (to B.Y.), and grants from National Institutes of Health (GM061146) and Gordon and Betty Moore Foundation (GBMF3046) (to X.C.)
References
- 1. Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 2013; 64:137-59; PMID: 23330790; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- 2. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10:126-39; PMID:19165215; http://dx.doi.org/ 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 3. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006; 313:320-4; PMID:16809489; http://dx.doi.org/ 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- 4. Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet 2013; 14:523-34; PMID:23797853; http://dx.doi.org/ 10.1038/nrg3495 [DOI] [PubMed] [Google Scholar]
- 5. Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005; 307:932-5; PMID:15705854; http://dx.doi.org/ 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 2002; 129:1085-94; PMID:11874905; http://dx.doi.org/ 10.1242/dev.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang Y, Ji LJ, Huang QC, Vassylyev DG, Chen XM, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 2009; 461:823-U86; PMID:19812675; http://dx.doi.org/ 10.1038/nature08433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2‘ OH of the 3’ terminal nucleotide. Nucleic Acids Res 2006; 34:667-75; PMID:16449203; http://dx.doi.org/ 10.1093/nar/gkj474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol 2005; 15:1501-7; PMID:16111943; http://dx.doi.org/ 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, Itoh J. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol 2010; 154:1335-46; PMID:20805329; http://dx.doi.org/ 10.1104/pp.110.160234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhai J, Zhao Y, Simon SA, Huang S, Petsch K, Arikit S, Pillay M, Ji L, Xie M, Cao X, Yu B, Timmermans M, Yang B, Chen X, Meyers BC. Plant microRNAs display differential 3'- truncation and tailing, modifications which are ARGONAUTE1-dependent and conserved across species. Plant Cell 2013; 25:2417-28; PMID:23839787; http://dx.doi.org/ 10.1105/tpc.113.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tkaczuk KL, Obarska A, Bujnicki JM. Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol 2006; 6:6; PMID:16433904; http://dx.doi.org/ 10.1186/1471-2148-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan CM, Zhou C, Brunzelle JS, Huang RH. Structural and biochemical insights into 2‘-O-methylation at the 3’-terminal nucleotide of RNA by Hen1. Proc Natl Acad Sci U S A 2009; 106:17699-704; PMID:19822745; http://dx.doi.org/ 10.1073/pnas.0907540106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2‘-O-methylation of Piwi- interacting RNAs at their 3’ ends. Genes Dev 2007; 21:1603-8; PMID:17606638; http://dx.doi.org/ 10.1101/gad.1563607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3‘ termini of mouse Piwi-interacting RNAs are 2’-O-methylated. Nat Struct Mol Biol 2007; 14:349-50; PMID:17384646; http://dx.doi.org/ 10.1038/nsmb1220 [DOI] [PubMed] [Google Scholar]
- 16. Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 2007; 17:1265-72; PMID:17604629; http://dx.doi.org/ 10.1016/j.cub.2007.06.030 [DOI] [PubMed] [Google Scholar]
- 17. Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 2007; 13:1397-401; PMID:17652135; http://dx.doi.org/ 10.1261/rna.659307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. Embo J 2010; 29:3688-700; PMID:20859253; http://dx.doi.org/ 10.1038/emboj.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 2012; 8:e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet 2012; 8:e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 2012; 8:e1002702; PMID:22829772; http://dx.doi.org/ 10.1371/journal.pgen.1002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan CM, Zhou C, Huang RH. Reconstituting bacterial RNA repair and modification in vitro. Science 2009; 326:247; PMID:19815768; http://dx.doi.org/ 10.1126/science.1179480 [DOI] [PubMed] [Google Scholar]
- 23. Jain R, Shuman S. Bacterial Hen1 is a 3‘ terminal RNA ribose 2’-O-methyltransferase component of a bacterial RNA repair cassette. Rna 2010; 16:316-23; PMID:20007328; http://dx.doi.org/ 10.1261/rna.1926510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, Daub CO. A comprehensive survey of 3‘ animal miRNA modification events and a possible role for 3’ adenylation in modulating miRNA targeting effectiveness. Genome Res 2010; 20:1398-410; PMID:20719920; http://dx.doi.org/ 10.1101/gr.106054.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu SN, Ivanov AR, Wolf DA, Mizgerd JP. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 2009; 11:1157-63; PMID:19701194; http://dx.doi.org/ 10.1038/ncb1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 2011; 21:1450-61; PMID:21813625; http://dx.doi.org/ 10.1101/gr.118059.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knouf Ec, Wyman SK, Tewari M. The human TUT1 nucleotidyl transferase as a global regulator of microRNA abundance. PLoS One 2013; 8:e69630; PMID:23874977; http://dx.doi.org/ 10.1371/journal.pone.0069630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 2009; 23:433-8; PMID:19240131; http://dx.doi.org/ 10.1101/gad.1761509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012; 151:521-32; PMID:23063654; http://dx.doi.org/ 10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- 30. van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 2009; 139:135-48; PMID:19804759; http://dx.doi.org/ 10.1016/j.cell.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 31. Ren G, Chen X, Yu B. Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr Biol 2012; 22:695-700; PMID:22464191; http://dx.doi.org/ 10.1016/j.cub.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 2012; 22:689-94; PMID:22464194; http://dx.doi.org/ 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA 2010; 107:3906-11; PMID:20142471; http://dx.doi.org/ 10.1073/pnas.0912632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3' end processing of microRNAs in Drosophila. Curr Biol 2011; 21:1888-93; PMID:22055292; http://dx.doi.org/ 10.1016/j.cub.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3‘-to-5’ exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1. Curr Biol 2011; 21:1878-87; PMID:22055293; http://dx.doi.org/ 10.1016/j.cub.2011.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001; 15:188-200; PMID:11157775; http://dx.doi.org/ 10.1101/gad.862301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. Rna 2005; 11:459-69; PMID:15703439; http://dx.doi.org/ 10.1261/rna.7231505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 2004; 15:173-83; PMID:15260969; http://dx.doi.org/ 10.1016/j.molcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 39. Shen BZ, Goodman HM. Uridine addition after microRNA-directed cleavage. Science 2004; 306:997-; PMID:15528436; http://dx.doi.org/ 10.1126/science.1103521 [DOI] [PubMed] [Google Scholar]
- 40. Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 2006; 314:1893-; PMID:17185594; http://dx.doi.org/ 10.1126/science.1135268 [DOI] [PubMed] [Google Scholar]
- 41. Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5' RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci U S A 2014; 111:6365-70; PMID:24733911; http://dx.doi.org/ 10.1073/pnas.1405083111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rissland OS, Norbury CJ. Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 2009; 16:616-23; PMID:19430462; http://dx.doi.org/ 10.1038/nsmb.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013; 497:244-8; PMID:23594738; http://dx.doi.org/ 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. Rna 2013; 19:1632-8; PMID:24141620; http://dx.doi.org/ 10.1261/rna.040055.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. Embo J 2013; 32:1842-54; PMID:23503588; http://dx.doi.org/ 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang W, Murphy C, Sieburth LE. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc Natl Acad Sci U S A 2010; 107:15981-5; PMID:20798041; http://dx.doi.org/ 10.1073/pnas.1007060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013; 14:447-59; PMID:23732335; http://dx.doi.org/ 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- 48. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng ZP, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010; 328:1534-9; PMID:20558712; http://dx.doi.org/ 10.1126/science.1187058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ameres SL, Hung JH, Xu J, Weng ZP, Zamore PD. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. Rna 2011; 17:54-63; PMID:21106652; http://dx.doi.org/ 10.1261/rna.2498411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A-aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 2005; 19:405-19; PMID:16061186; http://dx.doi.org/ 10.1016/j.molcel.2005.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-9; PMID:21546353; http://dx.doi.org/ 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]


