Abstract
In our recent article Hörmann and coworkers have reported a role for epithelial cell-intrinsic TLR2 signaling for proliferation and renewal of the small intestinal epithelium. In this study, MyD88 and TRIF expression in the small intestine were affected by gut microbiota. Here, we report that in contrast to TLR2 and its co-receptor TLR1, TLR5 transcripts are not changed by presence of gut microbiota nor regulated through TLR2 or TLR4. Similar to TLR2 also TLR5 depends on MyD88 and TRIF adaptors. Our results indicate that TLR adaptor molecules could be determinants of TLR expression in the small intestine.
Keywords: gut microbiota, germ-free, MyD88, small intestine, TLR5, TLR2, TRIF, TOLLIP
We recently reported that colonization with a gut microbiota augments small intestinal expression and signaling of Toll-like receptor-2 (TLR2) and its co-receptor TLR1. We suggested that this is a route that supports renewal of the small intestinal epithelium through cell-intrinsic TLR2 signaling.1 In this study we analyzed germ-free (GF) mice and mice deficient for bacterial signature sensing TLRs to explore how pattern recognition of gut microbial signatures may affect renewal of the small intestinal mucosa.2-4 By decimation of gut microbial communities via administration of broad-spectrum antibiotics,5 we demonstrated that microbiota-dependent regulation of TLR2 is reversible. This finding implies that the use of antibiotics under some conditions could weaken the innate immune response to microbial signatures, which could privilege growth of resistant microbes. TLRs sense pathogen-associated molecular patterns (PAMPs) by recruiting adaptor proteins to its cytoplasmic Toll interleukin 1 receptor (TIR) domain. Adaptor molecules include TIR domain containing adaptors such as myeloid differentiation primary response gene 88 (MyD88) and/or TIR-domain-containing adaptor-inducing interferon-β (TRIF), which mediate the activation of pro-inflammatory responses.6 We reported that in addition to TLR2 and its co-receptor TLR1 also transcript levels of MyD88 were reduced by antibiotic decimation of resident gut bacteria. In contrast, transcript levels of TRIF were unchanged.1 Furthermore, we found that TLR2 and TLR1 mRNA levels in the small intestine depend on the adaptor molecules MyD88 and TRIF. In our monocolonization model with the Escherichia coli K-12 strain JP 313, we found that specifically TLR6 mRNA levels were reduced after 14 d of colonization.1 Collectively, we could show that TLR signaling in the small intestinal epithelium is orchestrated by the gut microbiota and that colonization with a single microbial strain may control expression of specific TLRs.
By the use of siRNAs that suppress TLR2 expression in MODE-K cells7 and stimulation of this mouse small intestinal epithelial cell line with the TLR2 agonist peptidoglycan (PG) we could directly address the role of epithelial cell-intrinsic TLR2 signaling for the control of intestinal epithelial proliferation. Our experiments with conventionally-raised (CONV-R) TLR-deficient mouse lines suggest that TLR2 and TLR4 impact on proliferation (Ki67), cell turn-over (cyclin D1) and apoptosis (caspase-3) in the small intestinal mucosa.1 In contrast, mice deficient in TLR5, the receptor that recognizes bacterial flagellin and is exclusively expressed on the basolateral surface of the small intestinal epithelium,6,8 did not show signs of impaired small intestinal renewal as indicated by the same markers.1
In this addendum article we address whether the gut microbiota modulates expression of TLR5 in the small intestine and if mRNA expression of this receptor is influenced through TLR receptor cross-talk. We investigate if deficiency in TLR adaptors may affect expression of other adaptor proteins and we address the impact of the adaptor molecules TRIF and MyD88 on small intestinal TLR5 transcript levels. In our recent study, we found that deficiency of both TLR adaptors suppresses mRNA levels of bacteria sensing TLR2 and TLR1, whereas transcript levels of TLR4 and TLR6 appear to depend only on TRIF.1 Based on the results of this addendum, we hypothesize that MyD88 and TRIF are important determinants of TLR5 transcript levels and that deficiency in MyD88 impacts on mRNA expression of other adaptor proteins such as TRIF and Toll-interacting protein (TOLLIP) in the small intestine.
Small Intestinal TLR5 Expression is not Changed by Gut Microbiota and not Regulated Through TLR2 or TLR4
To further pinpoint the role of bacteria sensing TLRs expressed on the small intestinal epithelium we explored the potential role of TLR5 in the orchestration of the expression profile of other TLRs. In contrast to TLR2 and its co-receptor TLR1, which we previously found up-regulated by colonization of GF mice with a gut microbiota,1 mRNA levels of the bacterial flagellin sensing pattern recognition receptor TLR5 were not changed by presence of gut microbiota (Fig. 1A). Our results suggest that in contrast to TLR2 and TLR1, expression of TLR5 in the small intestine is not regulated by the gut microbiota. This finding is in line with unchanged small intestinal TLR5 transcript levels in small intestinal tissues of Tlr2-/- and Tlr4-/- mice (Fig. 1B, C), suggesting that small intestinal activation of TLR2 and TLR4 signaling by gut microbial communities does not impact on TLR5 mRNA expression.1 Conversely, mice deficient in TLR5 did not show decreased TLR2, TLR1, TLR6, and TLR4 mRNA levels (Fig. 2A–D) and MODE-K cells7 that we stimulated with the TLR5 agonist flagellin did not show increased TLR2, TLR1, TLR6 and TLR4 transcripts (not shown). Collectively, our results indicate that TLR5 is not involved in the regulation of TLR2 mediated proliferation responses in the small intestine.
Figure 1.
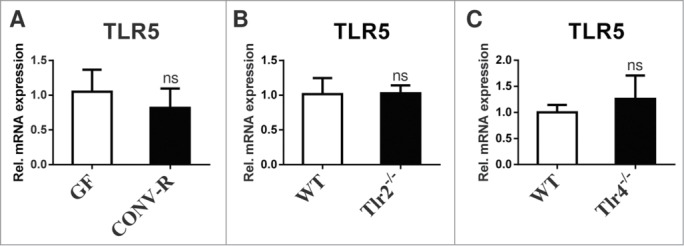
The gut microbiota does not change TLR5 transcript levels and TLR5 expression does not depend on intact TLR2 or TLR4 signaling in the small intestine. (A) Small intestinal TLR5 mRNA levels of 10–14 week old male GF and CONV-R C57BL/6 mice (N = 6–8 mice per group) were quantified by qRT-PCR relative to L32 (primers: TLR5 forward GCA GGA TCA TGG CAT GTC AAC; TLR5 reverse ATC TGG GTG AGG TTA CAG CCT). (B) and (C) Small intestinal TLR5 mRNA levels were quantified by qRT-PCR relative to L32 in female Tlr2-/- (N = 7 mice per group) and Tlr4-/- C57BL/6J mice (N = 7 mice per group). Results are shown as means +− s.e.m. ns = not significant; independent samples Student's t-test.
Figure 2.
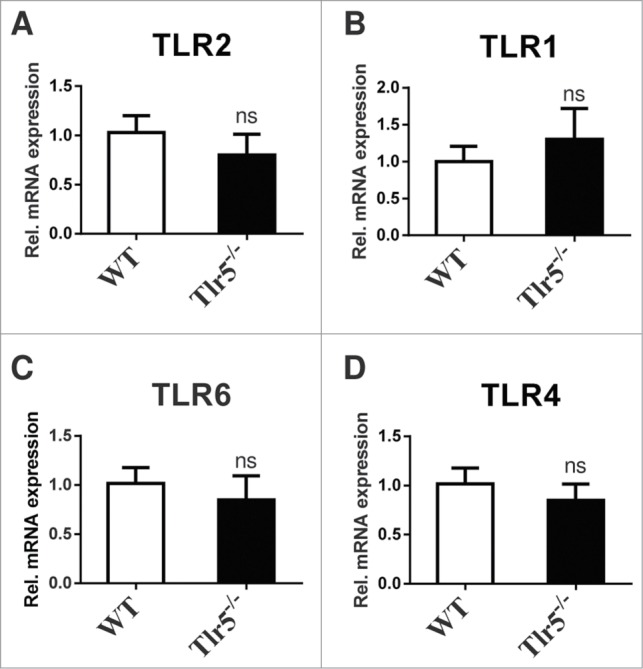
TLR5 deficiency does not impact on mRNA expression of bacteria sensing TLRs in the small intestine. Small intestinal mRNA levels of TLR2 (A), TLR1 (B), TLR6 (C), TLR4 (D) of 10–14 week old male Tlr5-/- C57BL/6 mice (N = 5–7 per group) were quantified by qRT-PCR relative to L32 (primers see ref. One). Results are shown as means +− s.e.m. ns = not significant; independent samples Student's t-test.
TLR Adaptors MyD88 and TRIF Regulate TLR5 mRNA Expression
It has been described that the number of TRIF-dependent genes far exceeds the number of MyD88-dependent genes in primary small intestinal epithelial cells and it was suggested that TRIF-mediated innate immune signaling is pivotal for homeostasis in the small intestinal mucosa.9 Therefore, it is interesting to analyze whether mRNA expression of TLRs depends on the presence of TRIF and MyD88 and whether MyD88 and TRIF expression can interfere with each other. In our previous study we reported that MyD88 transcripts in the ileum are increased by the gut microbiota, whereas TRIF transcript levels were vastly decreased.1 While MyD88 expression could be reduced by antibiotic decimation of gut bacteria expression of TRIF was not affected. We found that mRNA expression of the inhibitory adaptor molecule TOLLIP, which directly interacts with TLR2 and TLR4 and suppresses the kinase activity of IRAK,10 was increased in the small intestine of CONV-R mice compared with GF controls (Fig. 3A). Small intestinal mRNA levels of TOLLIP were dependent on the TLR adaptor MyD88 (Fig. 3B), but were independent of TRIF (Fig. 3C). Furthermore, TRIF expression levels were vastly reduced in Myd88-/- mice (Fig. 3D), but MyD88 expression levels were normal in Trif-/- mice (Fig. 3E), which is in line with a recent report indicating that epithelial TRIF signaling requires intact MyD88 signal transduction.11
Figure 3.
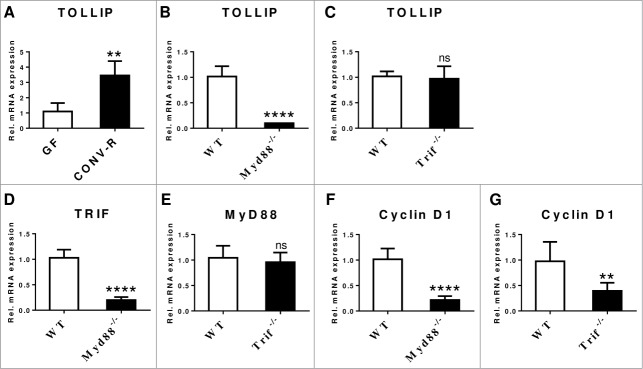
Transcripts of TLR adaptors are changed by the microbiota and MyD88 interferes with TRIF and TOLLIP. (A) Small intestinal mRNA expression levels of the TLR adaptor TOLLIP in 10–14 week old male GF and CONV-R C57BL/6 mice (N = 4–6 mice per group) was quantified by qRT-PCR relative to L32 (primers: Tollip forward CCC AGG ACT TCC TCC GTA TAA; Tollip reverse AGT CT GCC ATA ATT CTT TGC CA). (B) and (C) Small intestinal TOLLIP mRNA levels were quantified by qRT-PCR relative to L32 in female Myd88-/- (N = 7 per group) and Trif-/- C57BL/6J mice (N = 6–7 per group). (D) TRIF mRNA levels quantified by qRT-PCR relative to L32 in female Myd88-/- C57BL/6 mice (N = 7 per group). (E) MyD88 mRNA levels quantified by qRT-PCR relative to L32 in female Trif-/- C57BL/6 mice (N = 7 per group). mRNA levels of the cell cycle marker Cyclin D1 in (F) female Myd88-/- C57BL/6 mice (N = 6–7 per group) and (G) female Trif-/- C57BL/6 mice (N = 7 per group). Results are shown as means +− s.e.m. Two asterisks, P<0.01; 4 asterisks, P<0.0001; independent samples Student's t-test.
In our previous study we have shown that expression of TLR2 and its co-receptor TLR1 is reduced in the small intestine of Myd88-/- mice. Likewise, we found that mRNA levels of TLR2, its co-receptors TLR1 and TLR6, as well as TLR4 were decreased in Trif-/- mice.1 In support of a role of TLR2 in mucosal renewal and in agreement with decreased TLR2 transcripts in the small intestine of Myd88-/- and Trif-/- mice,1 cyclin D1 mRNA expression, a marker of G1/S phase transition, was reduced in the small intestine of Myd88-/- and Trif-/- mice compared with WT controls (Fig. 3F,G).
With intestinal epithelial cell models it has been established that TLR5-dependent responses depend both on MyD88 and the adaptor molecule TRIF.12 Since the TLR adaptors TRIF and MyD88 appear decisive for transcript levels of bacteria sensing TLRs,1 we wondered if TLR5 expression could be regulated through MyD88 and/or TRIF. In our previous study we found that transcriptional regulation of MyD88 and TRIF in the small intestine depends on gut microbial colonization. Similar to TLR2 we found that TLR5 mRNA levels were markedly diminished in the small intestine of Myd88-/- and Trif-/- mice (Fig. 4A,B).1 Our results indicate that TLR adaptor functions are central regulators for expression of the bacterial signature sensing TLRs in the small intestinal mucosa.
Figure 4.
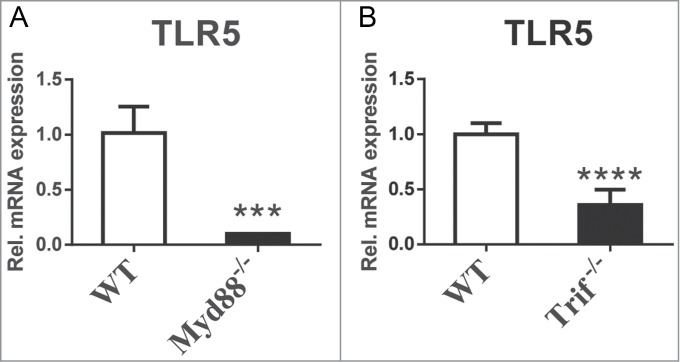
TLR5 transcript levels are dependent on TLR adaptor proteins. (A) MyD88-deficiency reduces TLR5 transcript levels in the small intestine (N = 3–7 mice per group). (B) TRIF-deficiency reduces TLR5 transcript levels in the small intestine (N = 7 mice per group). Results are shown as means +− s.e.m. Three asterisks, P<0.001; 4 asterisks, P<0.0001; independent samples Student's t-test.
The gut microbiota has emerged as an environmental factor that profoundly impacts host physiology.13 Although human intestinal epithelial cells have been considered broadly unresponsive to TLR2-dependent bacterial ligands when compared to myeloid-monocytic cells,14 we have shown that stimulation of the mouse small intestinal epithelial cell line MODE-K with PG leads to increased mRNA and protein expression of TLR2 and an increased proliferation response that depends on epithelial cell-intrinsic TLR2 function.1 Increased TLR2 mRNA and protein along with increased signs of small intestinal renewal were also detected in the small intestine of colonized mice compared with GF controls.1 Our recent study expands on reports that have implicated TLR2 and TLR5 signaling in cell migration, wound repair, proliferation and survival of human airway epithelial cells.15 The relevance of microbiota-dependent intestinal epithelial TLR2 signaling that Hörmann et al. have revealed as a determinant of small intestinal epithelial renewal under normal steady-state conditions1 is further underscored by the consecutively published article of Scheeren et al. who described a central role for the epithelial cell-intrinsic TLR2-MyD88 signaling axis in normal intestinal and mammary epithelial cells and oncogenesis.16
In this addendum, we report that small intestinal TLR5 mRNA expression under conditions of unperturbed intestinal tissue homeostasis is not affected by the gut microbiota, nor dependent on intact TLR2 or TLR4 signaling. This is in line with unaltered markers of cell turn-over and renewal in Tlr5-/- mice compared with WT controls.1 However, in the dextran sodium sulfate colitis model it has been shown that colonic flagellin administration following disruption of the epithelium results in stimulation of basolaterally localized TLR5 and severe apoptosis in the colonic epithelium indicating that epithelial barrier integrity may restrict TLR5 activation.17
We found that similar to TLR2, TLR1 and TLR4 expression1 also TLR5 transcript levels markedly depend on the adaptor molecules MyD88 and TRIF. Importantly, MyD88 impacts on TRIF expression (Fig. 3). This stresses the role of microbiota-induced pathways that may regulate expression and function of intestinal epithelial TLR adaptors and thus impact on the expression of bacteria sensing TLRs (Fig. 5). As compensatory proliferation upon injury was found impaired in Myd88-/- mice18 it is conceivable that the impaired proliferation response in Myd88-/- mice may partially result from decreased TLR expression. To further explore effects of TLR5 on the orchestration of innate immunity is clearly important since mice deficient in TLR5 are prone to develop spontaneous colitis due to a transient inability to manage proteobacteria.19,20 Since Tlr5-/- mice are prone to develop post-weaning gut inflammation19 and DSS-induced colitis is associated with a shift toward pro-inflammatory gram-negative bacteria like E. coli which are known to exacerbate colitis,21 it is essential to further explore how TLR5 signaling impacts on TLR innate immune sensing of the microbiota in the small intestine and how this may affect tissue homeostasis.
Figure 5.
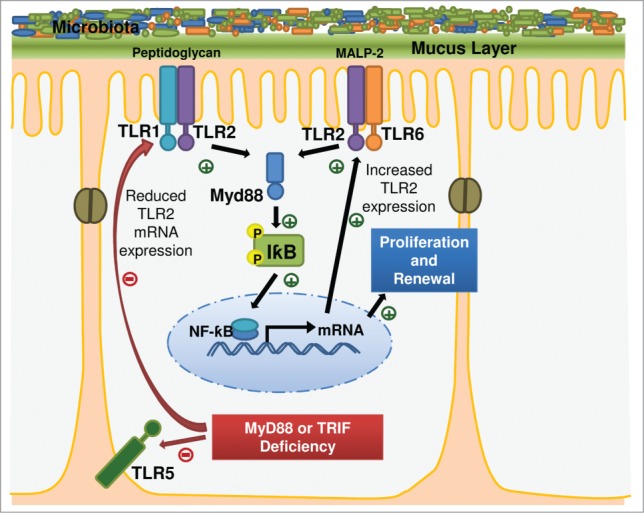
Cell-intrinsic TLR2 signaling in the small intestinal epithelium is induced by the gut microbiota. TLR adaptors impact on TLR expression.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors are grateful to Klaus-Peter Derreth for expert technical assistance, to Markus P Radsak for providing Myd88-/- and Trif-/- mice.
Funding
The project was funded by the German Federal Ministry for Education and Research (BMBF 01EO1003), a DFG Individual Grant to CR (RE 3450/5–1), intramural Stufe I project support to CR, a CTH Pre-Doctoral Fellowship to IB (BMBF 01EO1003), and a project grant from the Stiftung Pathobiochemie and Molekulare Diagnostik to CR.
Author Contributions
IB performed experiments, analyzed data and contributed to manuscript writing. CR designed experiments, analyzed data and wrote the manuscript. NH and SJ contributed with experiments.
References
- 1.Hörmann N, Brandão I, Jäckel S, Ens N, Lillich M, Walter U, Reinhardt C. Gut microbial colonization orchestrates TLR2 expression, signaling and epithelial proliferation in the small intestinal mucosa. PLoS One 2014; 9(11): e113080; PMID:25396415; http://dx.doi.org/ 10.1371/journal.pone.0113080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giraud A. Axenic mice model. Method Mol Biol 2008; 415: 321-36 [DOI] [PubMed] [Google Scholar]
- 3.Wooten RM, Ma Y, Yoder RA, Brown JP, Weis JH, Zachary JF, Kirschning CJ, Weis JJ. Toll-like receptor 2 is required for innate, but not acquired, host defense to Borrelia burgdorferi. J Immunol 2002; 168: 348-55; PMID:11751980; http://dx.doi.org/ 10.4049/jimmunol.168.1.348 [DOI] [PubMed] [Google Scholar]
- 4.Reinhardt C, Bergentall M, Grainer TU, Schaffner F, Östergren-Lundén G, Petersen LC, Ruf W, Bäckhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodeling. Nature 2012; 483: 627-31; PMID:22407318; http://dx.doi.org/ 10.1038/nature10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008; 57: 1470-81; PMID:18305141; http://dx.doi.org/ 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- 6.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol 2010; 10: 131-44; PMID:20098461; http://dx.doi.org/ 10.1038/nri2707 [DOI] [PubMed] [Google Scholar]
- 7.Vidal K, Grosjean I, Evillard JP, Gespach C, Kaiserlian D. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J Immunol Methods 1993; 166: 63-73; PMID:7693823; http://dx.doi.org/ 10.1016/0022-1759(93)90329-6 [DOI] [PubMed] [Google Scholar]
- 8.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 2001; 167: 1882-5; PMID:11489966; http://dx.doi.org/ 10.4049/jimmunol.167.4.1882 [DOI] [PubMed] [Google Scholar]
- 9.Stockinger S, Duerr CU, Fulde M, Dolowschiak T, Pott J, Yang I, Eibach D, Bäckhed F, Akira S, Sauerbaum S, et al.. TRIF signaling drives homeostatic intestinal epithelial antimicrobial peptide expression. J Immunol 2014; 193: 4223-34; PMID:25210121; http://dx.doi.org/ 10.4049/jimmunol.1302708 [DOI] [PubMed] [Google Scholar]
- 10.Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem 2002; 277: 7059-65; PMID:11751856; http://dx.doi.org/ 10.1074/jbc.M109537200 [DOI] [PubMed] [Google Scholar]
- 11.Petnicki-Ocwieja T, Chung E, Acosta DI, Ramos LT, Shin OS, Gosh S, Kobzik L, Li X, Hu LT. TRIF mediates Toll-like receptor 2-dependent inflammatory responses to Borrelia burgdorferi. Infect Immun 2013; 81: 402-10; PMID:23166161; http://dx.doi.org/ 10.1128/IAI.00890-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YJ, Im E, Chung HK, Pothoulakis C, Rhee SH. TRIF mediates Toll-like receptor 5-induced signaling in intestinal epithelial cells. J Biol Chem 2010; 285: 37570-8; PMID:20855887; http://dx.doi.org/ 10.1074/jbc.M110.158394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommer F, Bäckhed F. The gut microbiota – masters of host development and physiology. Nature Rev Microbiol 2013; 11: 227-38; PMID:23435359; http://dx.doi.org/ 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 14.Melmed G, Thomas LS, Lee N, Tesfay SY, Lukasek K, Michelsen KS, Zhou Y, Hu B, Arditi M, Abreu MT. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host-microbial interactions in the gut. J Immunol 2003; 170: 1406-15; PMID:12538701; http://dx.doi.org/ 10.4049/jimmunol.170.3.1406 [DOI] [PubMed] [Google Scholar]
- 15.Shaykhiev R, Behr J, Bals R. Microbial patterns signaling via toll-like receptors 2 and 5 contribute to epithelial repair, growth and survival. PLoS One 2008; 3: e1393; PMID:18167552; http://dx.doi.org/ 10.1371/journal.pone.0001393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheeren FA, Kuo AH, van Weele LJ, Cai S, Glykofridis I, Sikandar SS, Zabala M, Qian D, Lam JS, Johnston D, et al.. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol 2014; 16: 1238-44; PMID:25362351; http://dx.doi.org/ 10.1038/ncb3058 [DOI] [PubMed] [Google Scholar]
- 17.Rhee SH, Im E, Riegler M, Kokkotou E, O'brien M, Pothoulakis C. Pathophysiological role of toll-like receptor 5 engagement by bacterial flagellin in colonic inflammation. Proc Natl Acad Sci USA 2005; 102: 13610-5; PMID:16157881; http://dx.doi.org/ 10.1073/pnas.0502174102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004; 118: 229-41; PMID:15260992; http://dx.doi.org/ 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 19.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S, Williams IR, et al.. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest 2007; 117: 3909-21; PMID:18008007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JI, Su Y, Chassaing B, Walters WA, González A, et al.. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 2012; 12: 139-52; PMID:22863420; http://dx.doi.org/ 10.1016/j.chom.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heimesaat MM, Fischer A, Siegmund B, Kupz A, Niebergall J, Fuchs D, Jahn HK, Freudenberg M, Loddenkemper C, Batra A, et al.. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via toll-like receptors 2 and 4. PLoS One 2007; 2: e662; PMID:17653282; http://dx.doi.org/ 10.1371/journal.pone.0000662 [DOI] [PMC free article] [PubMed] [Google Scholar]


